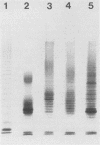
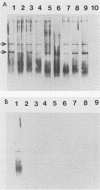
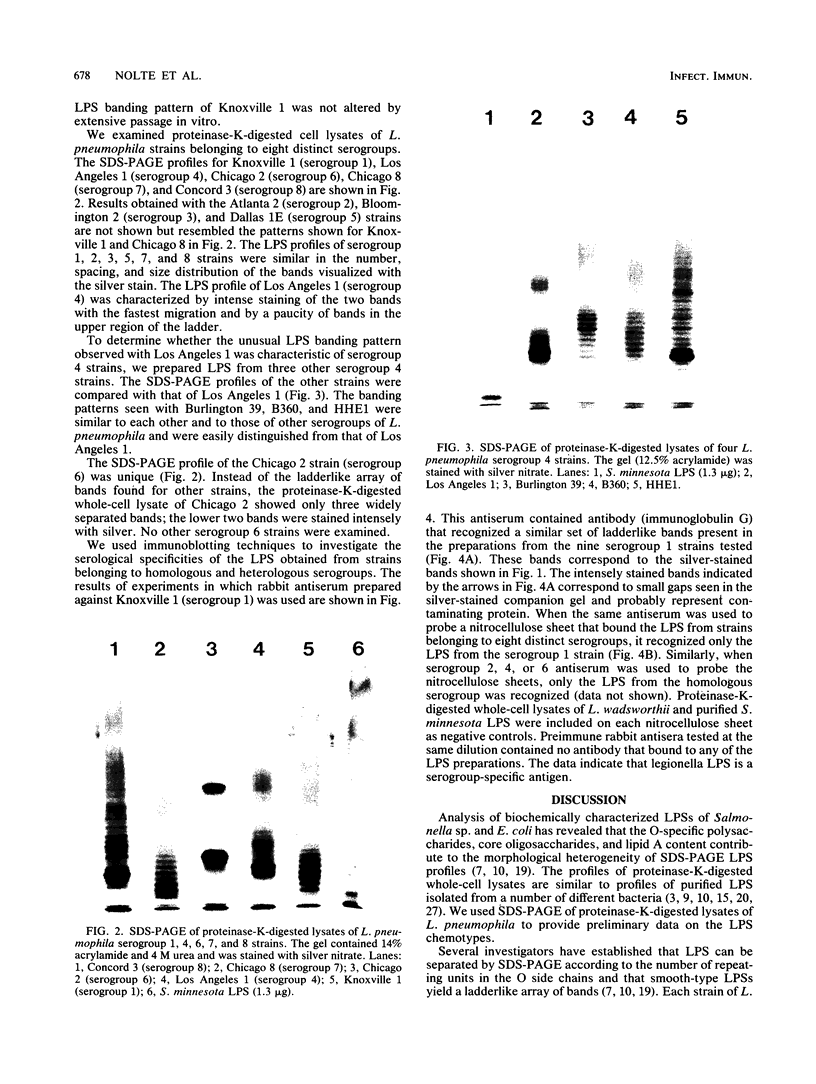
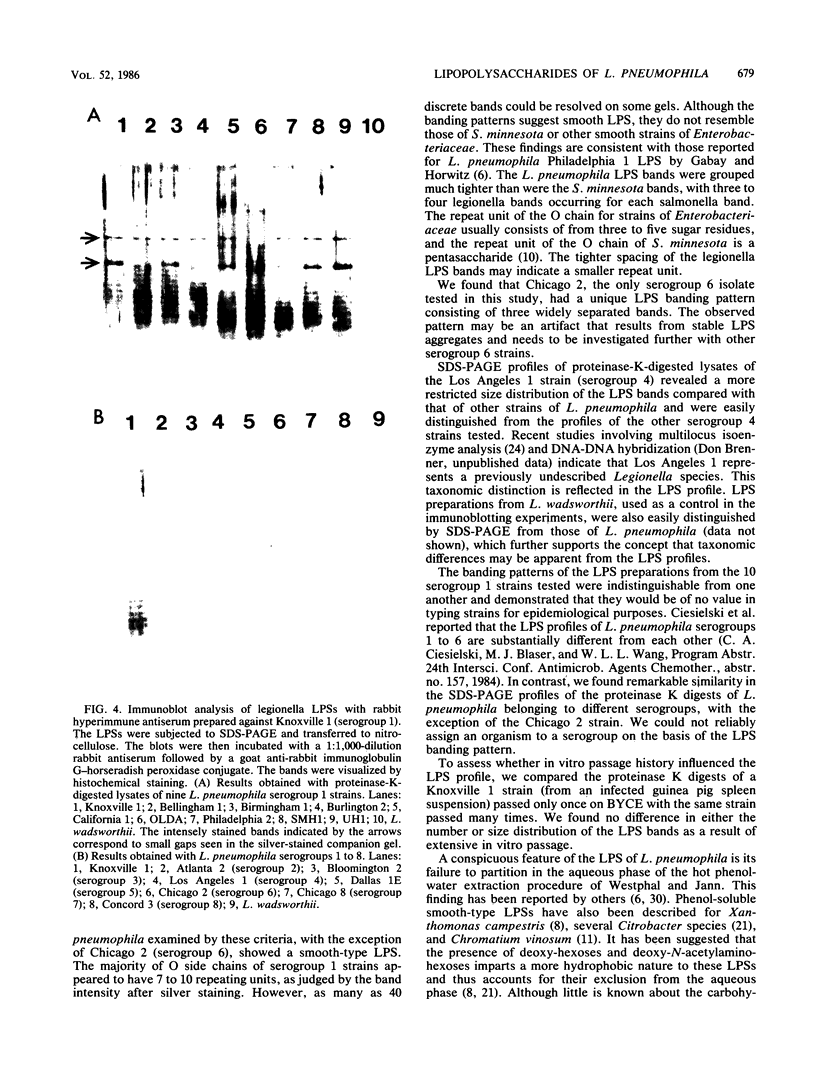
Abstract
Lipopolysaccharide (LPS) is a major constituent of the outer membrane of gram-negative bacteria. We used sodium dodecyl sulfate-polyacrylamide gel electrophoresis of proteinase-K-digested cell lysates to provide preliminary data on the LPS chemotypes for 20 strains of Legionella pneumophila (serogroups 1 to 8). The profiles of all strains except Chicago 2 (serogroup 6) were similar in the number, spacing, and size distribution of the bands visualized on silver-stained gels and were indicative of smooth LPS. However, compared with the bands from Salmonella minnesota smooth LPS, their banding pattern was much tighter, with three to four legionella bands for every salmonella band. The proteinase K digest of Chicago 2 was unique in that only two widely separated silver-stained bands were seen. LPS profiles of 10 serogroup 1 strains were identical, and the profile of Knoxville 1 was not altered by extensive in vitro passage. We used immunoblotting to investigate the serological specificities of the LPSs. When a rabbit antiserum prepared against a serogroup 1 strain was used to probe nitrocellulose sheets that bound LPS from strains belonging to eight different serogroups, it recognized only the LPS from the homologous serogroup. Similar results were observed with serogroup 2, 4, and 6 antisera. Our data indicate that L. pneumophila has a smooth-type LPS with an unusual banding pattern and that it is a serogroup-specific antigen.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bradbury W. C., Mills S. D., Preston M. A., Barton L. J., Penner J. L. Detection of lipopolysaccharides in polyacrylamide gels by transfer to nitrocellulose followed by immunoautoradiography with antibody and 125I-protein A: "LPS blotting". Anal Biochem. 1984 Feb;137(1):129–133. doi: 10.1016/0003-2697(84)90358-0. [DOI] [PubMed] [Google Scholar]
- Butler C. A., Street E. D., Hatch T. P., Hoffman P. S. Disulfide-bonded outer membrane proteins in the genus Legionella. Infect Immun. 1985 Apr;48(1):14–18. doi: 10.1128/iai.48.1.14-18.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldwell H. D., Hitchcock P. J. Monoclonal antibody against a genus-specific antigen of Chlamydia species: location of the epitope on chlamydial lipopolysaccharide. Infect Immun. 1984 May;44(2):306–314. doi: 10.1128/iai.44.2.306-314.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciesielski C. A., Blaser M. J., Wang W. L. Serogroup specificity of Legionella pneumophila is related to lipopolysaccharide characteristics. Infect Immun. 1986 Feb;51(2):397–404. doi: 10.1128/iai.51.2.397-404.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelstein P. H. Improved semiselective medium for isolation of Legionella pneumophila from contaminated clinical and environmental specimens. J Clin Microbiol. 1981 Sep;14(3):298–303. doi: 10.1128/jcm.14.3.298-303.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flesher A. R., Ito S., Mansheim B. J., Kasper D. L. The cell envelope of the Legionnaires' disease bacterium. Morphologic and biochemical characteristics. Ann Intern Med. 1979 Apr;90(4):628–630. doi: 10.7326/0003-4819-90-4-628. [DOI] [PubMed] [Google Scholar]
- Gabay J. E., Horwitz M. A. Isolation and characterization of the cytoplasmic and outer membranes of the Legionnaires' disease bacterium (Legionella pneumophila). J Exp Med. 1985 Feb 1;161(2):409–422. doi: 10.1084/jem.161.2.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman R. C., Leive L. Heterogeneity of antigenic-side-chain length in lipopolysaccharide from Escherichia coli 0111 and Salmonella typhimurium LT2. Eur J Biochem. 1980;107(1):145–153. doi: 10.1111/j.1432-1033.1980.tb04635.x. [DOI] [PubMed] [Google Scholar]
- Hickman J., Ashwell G. Isolation of a bacterial lipopolysaccharide from Xanthomonas campestris containing 3-acetamido-3,6-dideoxy-D-galactose and D-rhamnose. J Biol Chem. 1966 Mar 25;241(6):1424–1428. [PubMed] [Google Scholar]
- Hitchcock P. J. Analyses of gonococcal lipopolysaccharide in whole-cell lysates by sodium dodecyl sulfate-polyacrylamide gel electrophoresis: stable association of lipopolysaccharide with the major outer membrane protein (protein I) of Neisseria gonorrhoeae. Infect Immun. 1984 Oct;46(1):202–212. doi: 10.1128/iai.46.1.202-212.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitchcock P. J., Brown T. M. Morphological heterogeneity among Salmonella lipopolysaccharide chemotypes in silver-stained polyacrylamide gels. J Bacteriol. 1983 Apr;154(1):269–277. doi: 10.1128/jb.154.1.269-277.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurlbert R. E., Weckesser J., Mayer H., Fromme I. Isolation and characterization of the lipopolysaccharide of Chromatium vinosum. Eur J Biochem. 1976 Sep 15;68(2):365–371. doi: 10.1111/j.1432-1033.1976.tb10823.x. [DOI] [PubMed] [Google Scholar]
- Jann B., Reske K., Jann K. Heterogeneity of lipopolysaccharides. Analysis of polysaccharide chain lengths by sodium dodecylsulfate-polyacrylamide gel electrophoresis. Eur J Biochem. 1975 Dec 1;60(1):239–246. doi: 10.1111/j.1432-1033.1975.tb20996.x. [DOI] [PubMed] [Google Scholar]
- Karkhanis Y. D., Zeltner J. Y., Jackson J. J., Carlo D. J. A new and improved microassay to determine 2-keto-3-deoxyoctonate in lipopolysaccharide of Gram-negative bacteria. Anal Biochem. 1978 Apr;85(2):595–601. doi: 10.1016/0003-2697(78)90260-9. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Logan S. M., Trust T. J. Structural and antigenic heterogeneity of lipopolysaccharides of Campylobacter jejuni and Campylobacter coli. Infect Immun. 1984 Jul;45(1):210–216. doi: 10.1128/iai.45.1.210-216.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayberry W. R. Dihydroxy and monohydroxy fatty acids in Legionella pneumophila. J Bacteriol. 1981 Aug;147(2):373–381. doi: 10.1128/jb.147.2.373-381.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney R. M., Thomason B. M., Harris P. P., Thacker L., Lewallen K. R., Wilkinson H. W., Hebert G. A., Moss C. W. Recognition of a new serogroup of Legionnaires disease bacterium. J Clin Microbiol. 1979 Jan;9(1):103–107. doi: 10.1128/jcm.9.1.103-107.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meenhorst P. L., Reingold A. L., Groothuis D. G., Gorman G. W., Wilkinson H. W., McKinney R. M., Feeley J. C., Brenner D. J., van Furth R. Water-related nosocomial pneumonia caused by Legionella pneumophila serogroups 1 and 10. J Infect Dis. 1985 Aug;152(2):356–364. doi: 10.1093/infdis/152.2.356. [DOI] [PubMed] [Google Scholar]
- Palva E. T., Mäkelä P. H. Lipopolysaccharide heterogeneity in Salmonella typhimurium analyzed by sodium dodecyl sulfate polyacrylamide gel electrophoresis. Eur J Biochem. 1980;107(1):137–143. doi: 10.1111/j.1432-1033.1980.tb04634.x. [DOI] [PubMed] [Google Scholar]
- Peppler M. S., Schrumpf M. E. Isolation and characterization of Bordetella pertussis phenotype variants capable of growing on nutrient agar: comparison with phases III and IV. Infect Immun. 1984 Jan;43(1):217–223. doi: 10.1128/iai.43.1.217-223.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raff R. A., Wheat R. W. Carbohydrate composition of the phenol-soluble lipopolysaccharides of Citrobacter freundii. J Bacteriol. 1968 Jun;95(6):2035–2043. doi: 10.1128/jb.95.6.2035-2043.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reisner A. H., Nemes P., Bucholtz C. The use of Coomassie Brilliant Blue G250 perchloric acid solution for staining in electrophoresis and isoelectric focusing on polyacrylamide gels. Anal Biochem. 1975 Apr;64(2):509–516. doi: 10.1016/0003-2697(75)90461-3. [DOI] [PubMed] [Google Scholar]
- Ristroph J. D., Hedlund K. W., Allen R. G. Liquid medium for growth of Legionella pneumophila. J Clin Microbiol. 1980 Jan;11(1):19–21. doi: 10.1128/jcm.11.1.19-21.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selander R. K., McKinney R. M., Whittam T. S., Bibb W. F., Brenner D. J., Nolte F. S., Pattison P. E. Genetic structure of populations of Legionella pneumophila. J Bacteriol. 1985 Sep;163(3):1021–1037. doi: 10.1128/jb.163.3.1021-1037.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin H., Staehelin T., Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai C. M., Frasch C. E. A sensitive silver stain for detecting lipopolysaccharides in polyacrylamide gels. Anal Biochem. 1982 Jan 1;119(1):115–119. doi: 10.1016/0003-2697(82)90673-x. [DOI] [PubMed] [Google Scholar]
- Watanabe H., Nakamura A., Timmis K. N. Small virulence plasmid of Shigella dysenteriae 1 strain W30864 encodes a 41,000-dalton protein involved in formation of specific lipopolysaccharide side chains of serotype 1 isolates. Infect Immun. 1984 Oct;46(1):55–63. doi: 10.1128/iai.46.1.55-63.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong K. H., Feeley J. C. Lipopolysaccharide of Legionella as adjuvant for intrinsic and extrinsic antigens. Proc Soc Exp Biol Med. 1984 Dec;177(3):475–481. doi: 10.3181/00379727-177-41975. [DOI] [PubMed] [Google Scholar]
- Wong K. H., Moss C. W., Hochstein D. H., Arko R. J., Schalla W. O. "Endotoxicity" of the Legionnaires' disease bacterium. Ann Intern Med. 1979 Apr;90(4):624–627. doi: 10.7326/0003-4819-90-4-624. [DOI] [PubMed] [Google Scholar]