Abstract
Escherichia coli UvrD is a non-ring-shaped model helicase, displaying a 3′–5′ polarity in DNA unwinding. Using a transverse magnetic tweezer and DNA hairpins, we measured the unwinding kinetics of UvrD at various DNA-destabilizing forces. The multiform patterns of unwinding bursts and the distributions of the off-times favour the mechanism that UvrD unwinds DNA as a dimer. The two subunits of the dimer coordinate to unwind DNA processively. They can jointly switch strands and translocate backwards on the other strand to allow slow (∼40 bp/s) rewinding, or unbind simultaneously to allow quick rehybridization. Partial dissociation of the dimer results in pauses in the middle of the unwinding or increases the translocation rate from ∼40 to ∼150 nt/s in the middle of the rewinding. Moreover, the unwinding rate was surprisingly found to decrease from ∼45 to ∼10 bp/s when the force is increased from 2 to 12 pN. The results lead to a strained-inchworm mechanism in which a conformational change that bends and tenses the ssDNA is required to activate the dimer.
Keywords: helicase, molecular motor, single molecule, unwinding mechanism, UvrD
Introduction
Helicases are vital molecular motors that use the chemical free energy derived from nucleoside triphosphate hydrolysis to directionally translocate along single-stranded (ss) nucleic acid and separate a double-stranded (ds) nucleic acid into two ss nucleic acids (Lohman and Bjornson, 1996; Ali and Lohman, 1997; Patel and Picha, 2000; von Hippel and Delagoutte, 2001). Escherichia coli UvrD is a superfamily I helicase (Maluf et al, 2003b; Fischer et al, 2004), having important functions in both methyl-directed mismatch repair and nucleotide excision repair of DNA. Despite recent progresses (Ali and Lohman, 1997; Maluf et al, 2003b; Dessinges et al, 2004; Fischer et al, 2004; Lee and Yang, 2006), the molecular mechanism of UvrD-mediated DNA unwinding is still an issue of controversy. The structure of UvrD is organized into two domains, each comprising two subdomains (named 1A, 1B and 2A, 2B, respectively) (Lee and Yang, 2006). UvrD can exist in two forms, open and closed, which differ in the 2B subdomain orientation (Lee and Yang, 2006; Lohman et al, 2008). UvrD bound to a 3′-tailed dsDNA was crystallized in the closed form (Lee and Yang, 2006). The crystal structure revealed that the 3′-tail and the substrate duplex are roughly orthogonal to each other at the ds–ssDNA junction. Binding of ATP leads to a rotation of the 1A, 1B and 2B subdomains around a hinge region connected to the 2A subdomain, possibly providing the power to separate the two strands. A series of crystal structures of UvrD complexed with partial DNA duplex and ATP hydrolysis intermediates suggested that the elemental step for helicase activity is the unwinding of a single base pair (bp). This, however, contrasts with the larger step size (∼4–5 bp) derived from bulk kinetic measurements and single molecule study (Ali and Lohman, 1997; Dessinges et al, 2004). Moreover, solution studies on UvrD have indicated that at least a dimer is required for its helicase activity in vitro (Maluf et al, 2003b). Apparently, new experimental evidences are needed to test the different models for UvrD and, ultimately, to build a new model that reconciles the crystal structures and the kinetic measurements.
Single-molecule study can provide new insight into the mechanism of helicases. For example, single-molecule fluorescence measurements (Ha et al, 2002; Myong et al, 2005) showed that a monomeric Rep, which is structurally homologous to UvrD, can bind to the ds–ssDNA junction but does not unwind DNA. The unwinding is initiated only when a functional helicase is formed through additional protein binding. This result is consistent with bulk kinetic assays. One of us, in collaboration with Dessinges et al (2004), performed a single molecule study on UvrD-mediated unwinding of nicked DNA. Although the study did not distinguish whether a monomer or a dimer is required to unwind DNA, it revealed that UvrD might switch strands and translocate backwards on the other strand after unwinding a certain amount of base pairs. The unwinding rate was measured to be 248(±74) bp/s, much larger than that measured in bulk kinetic assays (68(±9) bp/s). But it was questioned (Fischer et al, 2004) that the large force (35 pN) applied to the two ends of the nicked DNA may destabilize the DNA so that the measured rate was actually the translocation rate along ssDNA.
Herein we report a single-molecule study on the unwinding kinetics of UvrD using DNA hairpins rather than linear DNA molecules. In our study, the two tails of a ds–ssDNA fork is grasped and the distance between the ends of the two tails is monitored, as has been done in previous studies on other helicases (Dumont et al, 2006; Johnson et al, 2007; Lionnet et al, 2007). We observed individual unwinding bursts similar to the ones reported by Dessinges et al (2004). Working with the DNA hairpins allowed us to gain a deeper insight into the unwinding behaviour of UvrD. We found evidences that favour the model that UvrD unwinds DNA as a dimer. We also found that the force exerted on the two tails of the DNA hairpin slows down the unwinding rate of UvrD. Our results, when combined with the available biochemical and structural data, lead us to propose a new model—a ‘strained-inchworm' model—to interpret the UvrD-mediated DNA unwinding. In our model, two UvrD molecules dimerize at the ds–ssDNA junction and coordinate to unwind DNA. The dimerization bends and tenses the ssDNA inside the dimer. The force inhibits the bending of the ssDNA and, consequently, compromises the UvrD–UvrD interaction, thereby reducing the unwinding rate.
Results
Processive unwinding by UvrD results in bursts of DNA extension
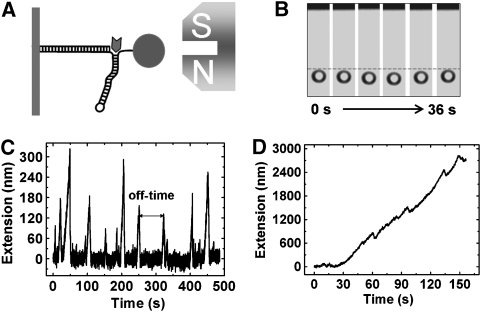
We used a transverse magnetic tweezer to analyse the UvrD-mediated unwinding of DNA. A magnetic bead was tethered through a DNA hairpin onto the sidewall of a fluidic chamber (Figure 1A, see also Supplementary Figure S1). A constant force was exerted on the bead connected to the 3′-ssDNA loading tail. The DNA is very stable when the force is smaller than 13 pN, beyond which it can be unzipped, displaying very large stepwise increase in the bead-surface distance (Essevaz-Roulet et al, 1997). When UvrD (⩾1 nM) and saturated ATP (⩾1 mM) were injected into the flow chamber, the DNA was unwound by UvrD. The molecular geometry of the DNA hairpin results in the release of 2 nt for each base pair unwound. The effective length per base pair released at a certain force was determined by the extension versus force curve of ssDNA, which can be well described by the freely jointed chain (FJC) model at low forces (Smith et al, 1996; Dessinges et al, 2004; Dumont et al, 2006; Johnson et al, 2007; Lionnet et al, 2007). When the force is over 4 pN and the concentration of UvrD is lower than 10 nM, the bead-surface distance, that is, the DNA extension, displays isolated bursts (Figure 1C), which begin with a constant-rate increase in extension as the DNA is unwound, followed by a fast decrease. The processivity of the helicase (Supplementary Figure S2) is in agreement with that measured by bulk kinetic assays (Maluf et al, 2003b). As the time span of two isolated bursts is much larger than the time of single unwinding burst, one isolated burst can be considered as single enzyme working on the DNA substrate. The shape of the bursts is similar to, but the slope is much smaller than, that in the previous study (Dessinges et al, 2004). It is noteworthy that the observed increase in extension stems from the separation of the DNA strands at the ds–ssDNA fork junction rather than from the unwinding of the DNA near the nicks on the linker arm. To demonstrate this, we performed similar measurements with a linear λ-DNA to which no DNA hairpin was attached. In agreement with the observation of Dessinges et al (2004), we found that the unwinding bursts cannot be observed if the force is lower than 30 pN. Moreover, to study the effect of the 3′-ssDNA loading tail on the activity of UvrD, we constructed two types of DNA hairpins with a loading length of 8 and 15 nt, respectively. No unwinding can be observed with the 8-nt-long loading tail. The result is consistent with the bulk assay of Maluf et al (2003b) that a 3′-ssDNA tail of at least 15 nt is required to observe optimal unwinding by UvrD dimers even though a DNA substrate possessing a 3′-ssDNA tail of 4–10 nt is sufficient to bind a single UvrD monomer with high affinity.
Figure 1.
Magnetic tweezer assay of UvrD. (A) The DNA construct is composed of two covalently connected lambda-DNAs (48.5 kbp), one behaving as a long linker arm and the second mimicking a ds–ssDNA fork. The linker arm is anchored to the sidewall of a fluid chamber and the 3′-loading tail is connected to the magnetic bead. Note that the figure is not to scale. (B) A series of images during a typical unwinding burst. (C) A trace of extension against time measured at F=9 pN and [UvrD]=5 nM. (D) Bunched unwinding events at F=4 pN and [UvrD]=20 nM.
Distributions of the off-times are not exponential
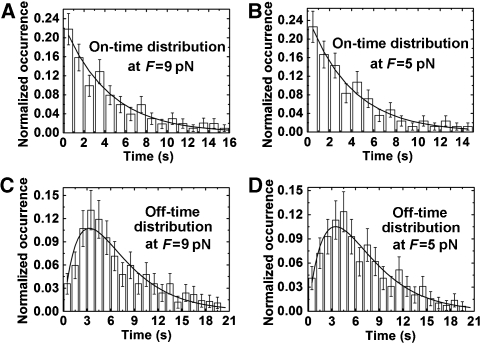
The enzyme unbinds the ds–ssDNA fork junction after unwinding a certain number of base pairs, resulting in a decrease in DNA extension. We define the on-time as the time span of an unwinding burst and the off-time as the time between two adjacent bursts. The distributions of the on-times are exponential (Figure 2A and B). But the distributions of the off-times are not (Figure 2C and D). The result is not consistent with the mechanism that UvrD unwinds DNA as a monomer because such a mechanism would lead to an exponential off-time distribution. This can be understood through the following analysis: if UvrD works as a monomer, the binding kinetics of UvrD to DNA can be described by the minimal chemical reaction pathway shown in scheme 1. 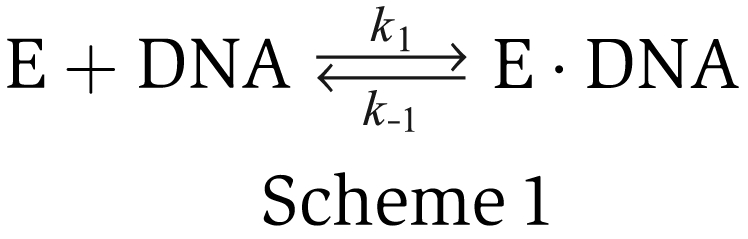
Figure 2.
Distributions of the on-times and the off-times. The histograms are extracted from the measurements at F=9 pN (A, C) and F=5 pN (B, D), respectively, at [ATP]=1 mM and [UvrD]=5 nM. The distributions of the on-times (A, B) are exponential and can be fit with a single exponential function f(t)=ku exp(−kut) with ku=0.23±0.03/s for panel A and ku=0.25±0.04/s for panel B. The distributions of the off-times (C, D) are not exponential and can be fit with the function in Equation (1). The solid lines are the fits to the data.
An off-time begins when the helicase dissociates from the substrate, allowing the separated ssDNA to rehybridize such that the experimental condition returns to the one with free UvrD molecules and a single tethered DNA hairpin. This is followed by a diffusion-limited wait for the same, or a different, helicase to bind to the DNA with rate constant k1. As it is a Poisson process, the distribution of the waiting time, or equivalently the off-time, follows a single exponential function f1(t)=k1 exp(−k1t), which is obviously not the one we observed.
Multiform unwinding events are observed
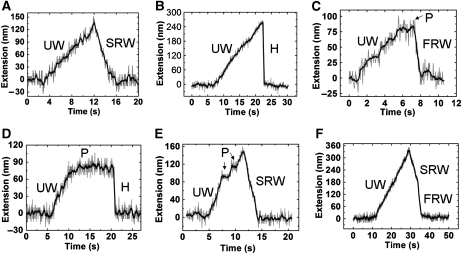
The majority of the unwinding events are similar to those reported previously (Dessinges et al, 2004), that is, a constant velocity increase of the DNA extension followed by either a slow (through strand switching; Figure 3A) or an abrupt (through complete unbinding; Figure 3B) recovery back to the initial extension. But we also observed new events such as those shown in Figure 3C–F, wherein an unwinding process is interrupted by a long time pause that is either followed by rezipping of the DNA (Figure 3C and D) or re-initiation of the unwinding (Figure 3E). The rezipping profiles are also multifarious. Besides the constant rate rewinding (see Figure 3A and E for slow rewinding and Figure 3C for fast rewinding) and the abrupt rehybridization (Figure 3B and D), we also observed rewinding processes that exhibit mixed speed: first slow, then fast (Figure 3F). Statistically, of the total unwinding processes observed at [UvrD]=5 nM, ∼30% are followed by abrupt rehybridization, ∼59% by slow rewinding and ∼11% by fast rewinding. Moreover, the percentage to see an unwinding with a pause is ∼25%, whereas the percentage to see a rewinding with mixed speed is ∼2%.
Figure 3.
Event phenomenology. Three distinct unwinding profiles are recorded: constant rate unwinding (A, B, F), unwinding ended with a pause (C, D), unwinding interrupted by pauses followed by unwinding re-initiation (E). Four rezipping profiles are recorded: slow rewinding (A, E), fast rewinding (C), abrupt rehybridization (B, D) and slow rewinding followed by fast rewinding (F). The curves are smoothed by 10-point adjacent averaging to guide the eyes (black lines). The specific processes are labelled as follows: ‘UW' for unwinding, ‘H' for abrupt rehybridization, ‘FRW' for fast rewinding, ‘SRW' for slow rewinding and ‘P' for pause.
Unwinding rate decreases with the increase of force
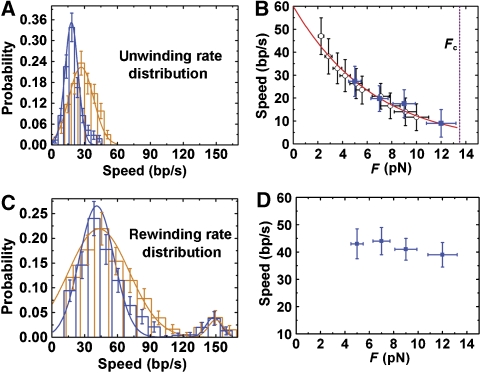
We can directly deduce the unwinding rate from the slope of the unwinding profile. Figure 4A depicts the distributions of the unwinding rates measured at F=5 and 9 pN, respectively. We noticed that the unwinding rates are much lower than that from bulk assays (Fischer et al, 2004). As the force may significantly affect the behaviour of the helicase (Dessinges et al, 2004; Dumont et al, 2006; Lionnet et al, 2007), we measured the unwinding rate at various forces (Figure 4B). Owing to the finite spatial resolution of our apparatus, isolated unwinding bursts can be observed less frequently if the force is lower than 4 pN. We therefore increased the concentration of UvrD so that we are able to measure the unwinding even at a force as low as 2 pN. When [UvrD] is higher than 10 nM, successive actions of several helicases lead to burst bunching (Figure 1D). But the unwinding rate is still well defined and is independent of [UvrD] (Supplementary Figure S3), implying that the unwinding events are due to individual UvrD complexes located at the DNA fork junction even if the [UvrD] is high. This argument is also supported by the fact that the rate obtained in this way is equal to the one measured from the bursts (see the squares and the circles in Figure 4B).
Figure 4.
Unwinding kinetics. (A) Distributions of unwinding rate at F=9 pN (black, 〈v〉=18±3 bp/s with a standard deviation σ=6 bp/s) and F=5 pN (grey, 〈v〉=27±4 bp/s with σ=11 bp/s). (B) Unwinding rate at different forces. The black squares are derived from individual unwinding bursts and the circles from curves exhibiting event bunching. The vertical dashed line marks the mechanical unwinding force Fc of dsDNA. (C) Distributions of rewinding rate at F=9 pN (black, 〈v〉=41±3 bp/s with σ=15 bp/s) and F=5 pN (grey, 〈v〉=44±3 bp/s with σ=21 bp/s). But there are also unwinding rates that distribute around 150 bp/s. The total number of such high rate events is too small to give a good statistics. (D) Rewinding rate at different forces.
The rate–force curve in Figure 4B reveals that the unwinding rate of UvrD decreases with the increasing force, indicating that the force kinetically impedes the UvrD-mediated DNA unwinding. We had expected that the force exerted on the two tails of the fork would assist the helicase to unwind DNA, that is, to increase the unwinding rate, because the force may destabilize DNA (Essevaz-Roulet et al, 1997) by reducing the activation energy for DNA base-pair melting. The counterintuitive rate–force relationship observed for UvrD should therefore provide a new insight into the molecular mechanism of it.
Two rewinding rates are observed
Two main types of rezipping profiles similar to those reported in previous studies were observed (Dessinges et al, 2004; Dumont et al, 2006; Lionnet et al, 2007). The first type (Figure 3B) is an abrupt jump: the DNA rehybridizes rapidly when an active complex unbinds the fork junction. The second type (Figure 3A) is a slower, constant-rate rewinding, which is caused by a single complex that performs strand switching and moves away from the fork junction (Dessinges et al, 2004; Dumont et al, 2006). The rewinding rate can be deduced from the slope of the rewinding profile. The most probable rewinding rate (Figure 4C) is much lower than the ssDNA translocation rate of monomeric UvrD (∼190 nt/s) deduced from bulk assay (Fischer et al, 2004). This is different from those obtained for ring-shaped helicases T4 and T7 and non-ring-shaped helicase HCV NS3 of which the rewinding rates are almost equal to the ssDNA translocation rates (Dumont et al, 2006; Johnson et al, 2007; Lionnet et al, 2007). We also noticed that the rewinding rates measured in the force range from 4 to 12 pN are lower than the unwinding rate at zero force. It is most likely that the force may also reduce the rewinding rate. Unfortunately, we are not able to obtain the rewinding rate when the force is lower than 4 pN because individual unwinding bursts are hardly observable when the force is small.
A small amount (∼15%) of the rewinding profiles exhibit a high rate of ∼150 bp/s (Figure 4C, small peak). In agreement with this, we observed more complicated rewinding behaviours than those shown in Figure 3A and B. For example, although the majority of the rewinding processes are slow with a rate of ∼40 bp/s, we also observed events with higher rewinding rate that is close to the translocation speed of monomeric UvrD on ssDNA (Fischer et al, 2004). More surprisingly, we observed rewinding events in which the rewinding rate may increase suddenly to ∼150 bp/s after a slow rewinding process (∼40 bp/s) (Figure 3F). Although such mixed rewinding events occur less frequently (∼4% of the slow rewinding processes), they do imply that there are two types of translocases.
Discussion
Off-time distributions reveal a subsequent binding of two UvrD molecules
We have shown that the non-exponential distributions of the off-times in Figure 2 cannot be interpreted by the single binding kinetics in scheme 1. As has been pointed out by Xie (2001) and Ha et al (2002), such a distribution implies more than one sub-event, in general. We also found that a 3′-ssDNA tail of 8 nt does not provide sufficient space for an active UvrD complex, even though a tail of 4–10 nt is long enough for a UvrD monomer (Maluf et al, 2003b), indicating that an active UvrD complex contains at least two UvrD molecules. To be consistent with the previous bulk assays (Maluf et al, 2003a, 2003b), we assume that the complex is composed of two UvrD molecules only. We will show in the following that such a model can interpret the observed off-time distribution very well. As free UvrD molecules exist as monomers under the solution conditions of this study (Maluf et al, 2003a), it is reasonable to assume that two UvrD molecules bind to the DNA subsequently and meet at the ds–ssDNA junction to form a dimer. We remind that an off-time begins when an unwinding complex unbinds the DNA completely. The ending of the off-time is therefore determined by the binding kinetics of the enzyme, which is represented by scheme 2. 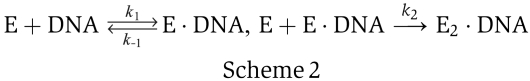
The first-half reaction in scheme 2 represents the binding of the first enzyme (E) to the DNA substrate and the second half represents the binding of the second enzyme. Each of the two halves is characterized by a single exponential function fi(t)=ki exp(−kit), i=1, 2. The time distribution for scheme 2 is hence the convolution of two exponential functions, f1(t′)=k1 exp(−k1t′) and f2(t–t′)=k2 exp(−k2(t–t′)), leading to the following equation (Xie, 2001): 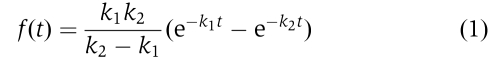
It has an exponential rise followed by an exponential decay, quite similar to the ones shown in Figure 2C and D. We used Equation (1) to fit the measured non-exponential distributions and obtained the binding rate constants k1=0.23±0.05/s and k2=0.38±0.08/s for F=9 pN and k1=0.27±0.04/s and k2=0.33±0.09/s for F=5 pN. They are almost independent of the force. We therefore will use the averaged values in the following discussion, that is, we will take k1=0.25±0.05/s and k2=0.36±0.09/s. Note that the rate constants were obtained for [UvrD]=5 nM. They are comparable with those obtained by quenched-flow experiments that gave k1=0.6/s and k2⩾0.75/s for the same UvrD concentration (Maluf et al, 2003a), further supporting the kinetics in scheme 2. It is worth pointing out that the distributions of the on-times (the time during which a dimer works) are exponential (Figure 2A and B) because a single event, for example, the complete unbinding of the dimer off the DNA or the disassembly of the dimer, can simply lead to an end of the unwinding action.
It is well known that the binding rate constants are proportional to the concentration of UvrD. The two binding rate constants therefore become k1′=0.050/s and k2′=0.074/s at [UvrD]=1 nM. The mean initiation time for a UvrD molecule to bind to the DNA substrate can be estimated to be 1/k1=4 s at [UvrD]=5 nM and 1/k1′=20 s at [UvrD]=1 nM. We found that the unwinding rarely occurs if [UvrD]<1 nM. To interpret this phenomenon, we adopt the unbinding rate constant for UvrD monomers off the DNA substrate from a quenched-flow kinetics measurement (Maluf et al, 2003a), namely we take k−1=0.12/s in scheme 2. The average lifetime of a monomer–DNA complex is accordingly 1/k−1=8.3 s. It is obviously not long enough to ensure the formation of a dimer on the DNA substrate because the unbinding of the first UvrD monomer occurs before the second UvrD monomer binds to the substrate, which takes about 1/k2′=13.5 s on average at [UvrD]=1 nM.
The multiform unwinding bursts can be explained by the dimeric model
Before we discuss the rewinding process, we would like to point out that the pauses in the unwinding profiles (Figure 3) are all consistent with the dimeric model. For example, dissociation of a monomer from the functional complex would inactivate the helicase, resulting in a pause in the middle of an unwinding process. The pause can be ended by the re-formation of a functional complex, which re-initiates the unwinding (Figure 3E). It can also be directly followed by an abrupt rehybridization, if, after the pause, the remanent UvrD unbinds the substrate, too (Figure 3D). If the two subunits of the dimer switch strands together or unbind the substrate simultaneously, no pause would be observed in the unwinding process (Figure 3A and B). One may think of other reasons for the pauses such as the one that depends on the DNA sequence or length. Indeed, other helicases (Dumont et al, 2006) have been reported to pause after each kinetic step. But our measurements favour the dissociation mechanism for the following reason. As there is one and only one active complex at the junction when the DNA is being unwound, the frequency to see a pause should be independent of [UvrD] if the pause is sequence or length dependent only. But we found that the frequency was reduced more than two-fold when [UvrD] was increased from 5 to 10 nM. The reduction of the frequency can be well accounted for by the dissociation mechanism. The binding rate constant k2 (see scheme 2), which determines how fast a new monomer binds to the substrate to form a new dimer after the old one has dissociated, increases with the increase in [UvrD]. Therefore, the durations of a large number of pauses become very short at the increased [UvrD], so they are not recognizable by our instrument.
As has been proposed by Dessinges et al (2004), the rewinding process with constant rate can be explained by a mechanism that the unwinding is followed by an enzyme-translocation-limited rezipping. We observed two rewinding rates, one is ∼40 bp/s and another is ∼150 bp/s (Figure 4C). The two rates can be easily explained by assuming that there are two kinds of translocases. We can exclude the possibility that the two rates are due to two kinds of UvrD monomers, with somewhat different activities, for the following two reasons. (1) Two kinds of UvrD monomers will also result in two unwinding rates. But only one unwinding rate (Figure 4A) is observed whose distribution is even narrower than that of the rewinding rate; (2) The proteins produced with the same protocol were studied previously with a linear λ-DNA without the attached hairpin. But only one rewinding rate was observed there (Dessinges et al, 2004). We propose that the two kinds of translocases are the UvrD dimers and monomers, respectively. It means that both the UvrD dimer and monomer can switch from the 3′-ssDNA strand to the 5′-ssDNA strand and moves away from the fork junction to allow the DNA to rezip.
The high rewinding rate (∼150 bp/s) is close to the monomeric UvrD translocation rate along ssDNA determined from bulk assay (∼190 nt/s) (Fischer et al, 2004). It can hence be attributed to a monomer-translocation-limited process. The low rewinding rate (∼40 bp/s) has not been reported before. But the assumption that it corresponds to the translocation of a dimer would help us to explain the rewinding profiles. We give three examples here. (1) The rewinding process in Figure 3F is composed of two events: a low-rate translocation followed by a high-rate one. Such a profile can be rationalized if we assume that, after strand switching, the dimer first translocates on the ssDNA as a whole and then dissociates, leaving a monomer to translocate on the ssDNA with a faster rate. Note that the dissociation of the dimer does not block the translocation but blocks the unwinding because the monomer left on the substrate is able to translocate on ssDNA but is not able to unwind dsDNA. This explains why a dissociation in the middle of an unwinding process results in a pause, whereas a dissociation in the middle of a rewinding process results in an increase in speed. (2) The pauses are followed by fast rewinding or abrupt rehybridization processes. The pauses have been attributed to the inactive monomer at the fork junction after its partner unbinds the substrate. The fast rewinding is therefore due to the strand switching of the monomer, whereas the abrupt rehybridization is due to the unbinding of it. (3) We have never observed a rewinding process that is the reverse of that shown in Figure 3F, namely a fast translocation followed by a slow one. This cannot be explained by the monomeric model because it is hard to imagine that a monomeric translocase would suddenly increase its average translocation speed almost three-fold but never change backwards when its environment remains unchanged. On the other hand, a monomer being translocating on the 5′-ssDNA tail would not be caught up by another monomer that moves in the same direction with a similar speed, unless the first one is blocked. This excludes the possibility that they will dimerize again during the rewinding process, hence explaining why we did not observe a fast rewinding process being followed by a slow one.
Phenomenological models for the force dependence of unwinding rate
It seems counterintuitive that the force can slow down the unwinding rate because the force exerted on the two tails of the DNA fork may perform positive work by W=2lntF, where lnt=0.57 nm is the contour length of 1 nt, when a base pair is separated and released. Nevertheless, one can construct a phenomenological model to account for the impediment of UvrD by force if one keeps in mind that the enzymatic cycle of a helicase-mediated DNA unwinding is usually composed of two basic reactions, namely melting of the base pairs and releasing of the displaced nucleotides, and that there may exist a delay between the two reactions. The force lowers the effective energy barrier to the base-pair melting only when the melting of the base pair and the releasing of the displaced nucleotides occur simultaneously, in which case the time consumed in the melting reaction is reduced to τ0′=τ0exp[−Fδ/kBT], where τ0 is the reaction time at zero force and δ is the distance over which the force does positive work. This is true for ring-shaped helicases that unwind the DNA by rectifying thermal fluctuations, which spontaneously open base pairs. Indeed, two recent single molecule assays showed that, for T4 (Lionnet et al, 2007) and T7 (Johnson et al, 2007) helicases, the unwinding rate increases about 10-fold when the force is increased from 4 to 12 pN. If the delay is not zero, that is, if the base-pair melting and the nucleotide releasing are separated in time, the force may not perform work in the base-pair melting reaction so that the unwinding rate is not affected. This might be the case for the non-ring-shaped helicase HCV NS3 whose unwinding rate was shown by optical tweezers to be almost independent of force (Dumont et al, 2006). However, as will be discussed in the next subsection, the force can also perform negative work in the base-pair melting reaction so that the unwinding rate can be lowered by the force. The time consumed in this reaction is increased to τ1 exp[Fd/kBT], where τ1 is the time of the rate-limiting reaction at zero force and d is the retraction of the DNA induced by the mechanical transition.
In general, the overall time to complete an enzymatic cycle is the sum of three times, 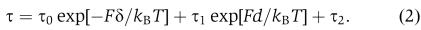
When τ0 is much longer than the other two times, that is, when τ0 is rate limiting, the unwinding rate increases with force. Similarly, when τ1 is much longer than the other two times, the unwinding rate decreases with force. The latter is true for our experiments. We introduce a small parameter A to represent the two short times (i.e., τ0 and τ2) and calculate the unwinding rate, which is the reciprocal of Equation (2), 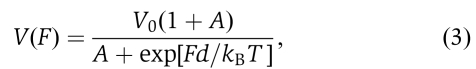 where V0 is the rate at zero force. Fitting to the rate–force relationship in Figure 4B with Equation (3) gives V0=60±7 bp/s and d=0.7±0.2 nm. The fitting is not sensitive to A so long as it is much smaller than 1. It is interesting to note that V0 is within experimental error in agreement with that (68±9 bp/s) given by quenched-flow measurements in which no external force was applied. The characteristic length d is a value we must reproduce when constructing a mechanism for UvrD. It represents the reduction of the ssDNA extension induced by the helicase during the force-dependent reaction (Figure 5).
where V0 is the rate at zero force. Fitting to the rate–force relationship in Figure 4B with Equation (3) gives V0=60±7 bp/s and d=0.7±0.2 nm. The fitting is not sensitive to A so long as it is much smaller than 1. It is interesting to note that V0 is within experimental error in agreement with that (68±9 bp/s) given by quenched-flow measurements in which no external force was applied. The characteristic length d is a value we must reproduce when constructing a mechanism for UvrD. It represents the reduction of the ssDNA extension induced by the helicase during the force-dependent reaction (Figure 5).
Figure 5.
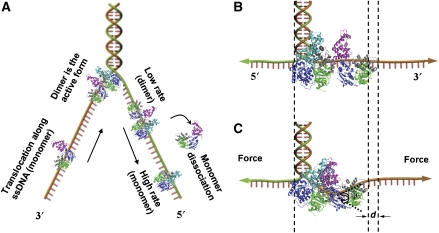
Proposed mechanism of UvrD unwinding. (A) A dimer at the ds-ssDNA junction is required to initiate DNA unwinding. The dimer can switch strands and translocate on ssDNA (see the main text for details). (B) A dimer in inactive state. The interaction between the two 2B subdomains is relaxed. (C) A dimer in active state. Close contact between the two 2B subdomains results in a conformational change that bends and tenses the ssDNA, leading to a retraction of the ssDNA by d. The work done by the external force is W=Fd. The retraction d can be calculated by d=LssDNA × (1−cos(ψ/2)), where LssDNA is the length of the ssDNA in the dimer and ψ (ranging from ∼45 to 65°, see Supplementary Figure S5) is the bending angle of the ssDNA induced by the dimer. Domain 1A, 1B and 2A are coloured green, grey and blue, respectively. Domain 2B is coloured cyan in the closed configuration (PDB entry code 2IS2) and magenta in the open configuration (PDB entry code 1UAA). Note that we used the open-form Rep to replace the open-form UvrD because the latter is not yet available (see the main text).
A mechanistic model for UvrD unwinding
The crystal structure shows that, in the complex of UvrD with a 3′-ssDNA-flanked DNA duplex, only one molecule can be accommodated at the ds–ssDNA junction (Lee and Yang, 2006). The 3′-ssDNA tail is bound across the 1A and 2A subdomains, with one ATP binding site in a cleft between the two subdomains. Binding of ATP leads to the closure of the cleft and a rotation of the 1A, 1B and 2B subdomains around a hinge region connected to the 2A subdomain. We failed to reproduce the required reduction of ssDNA extension using the monomeric model. For example, one may imagine that the ATP binding-induced change in conformation will force the ssDNA to rotate round the hinge so that the effective length of the ssDNA is reduced. The force exerted on the 3′-tail may hamper the rotation of the 3′-ssDNA tail so that the unwinding is slowed down. Such a model is attractive, but it does not quantitatively interpret the observed force–rate relationship because the maximum rotation angle is only 20° according to the crystal structures. Moreover, only 6 nt on the 3′-ssDNA tail are embedded in a monomer (Lee and Yang, 2006). The maximum reduction is therefore only 6 × lnt × (1−cos 20°)=0.2 nm, much less than that needed to interpret the data.
A dimeric inchworm mechanism has been proposed for UvrD in which the leading subunit of the dimer functions as the helicase, interacting directly with the dsDNA, whereas the trailing subunit functions as an ssDNA translocase (Maluf et al, 2003a, 2003b). On the basis of observations that the helicase activity of Rep monomer is activated when the subdomain 2B is removed, and that in the crystal structure of the RecBCD helicase (Singleton et al, 2004), the subdomain 2B of RecB interacts with the subdomain 2B of RecC, rather than with the dsDNA, Lohman et al (2008) proposed that the leading subunit can be autoinhibited by one of its subdomains (possibly the subdomain 2B). The autoinhibition can be relieved by the interaction with the trailing subunit of the dimer. Following the same idea, we propose that the two 2B subdomains in the UvrD dimer must contact with each other to relieve the autoinhibition. To do so, the 3′-ssDNA tail must be bent and tensed because the two enzymes are directed to the same direction on the ssDNA in a head-to-tail configuration (Figure 5). We tried to model the joint of the two molecules on ssDNA using the crystal structure of the UvrD–DNA complex and assuming that the 2B subdomain of the leading subunit is in the closed configuration, whereas the 2B subdomain of the trailing subunit is open. As the structure of UvrD in the open form is not yet available (Tomko et al, 2007; Lohman et al, 2008), we used the structure of E. coli Rep (Korolev et al, 1997) to replace the open form UvrD in the modelling process. E. coli Rep is closely related to E. coli UvrD with ∼40% sequence homology and can form heterodimers with UvrD (Wong et al, 1993). The modelling was done by simply matching the two molecules on the ssDNA according to their outer contours. The interaction of the two subdomains could be formed by a relative rotation of the two molecules, satisfying the constraints that (1) the ssDNA runs across the 1A and 2A subdomains of both molecules and moves together with them, (2) the atoms of one molecule does not penetrate into the other and (3) the number of the interfacing atoms is the largest. We tried to generate several possible binding modes by manually rotating the molecules using the software PyMol (PyMol 2005, Delano Scientific LLC). For each mode, the size of the interface was calculated by counting the number of the interfacing residues. The process resulted in a bending by Ψ≈45−65° of the ssDNA within the dimer (Figure 5C). On the other hand, Maluf et al (2003b) have pointed out that 12 nt is the minimum length to accommodate an active UvrD dimer. The retraction of the ssDNA with respect to the configuration in which the two molecules are in loose contact (Figure 5B) can be calculated by d=12 × lnt × (1−cos(ψ/2)), resulting in a retraction length ranging from 0.52 to 1.07 nm. The parameter d deduced from Equation (3) lies within this range, indicating that such a retraction is possible from the structural point of view.
The above calculations encouraged us to construct a molecular mechanism of UvrD-mediated DNA unwinding. First of all, UvrD unwinds DNA as a dimer. Second, in an enzymatic cycle, a few base pairs (∼4 bp, Supplementary Figure S4) are broken by the leading UvrD. The displaced nucleotides are stacked in the dimer before they are released by the trailing UvrD. Such a detail is borrowed from the non-uniform-stepping mechanism for ssDNA translocation of UvrD proposed by Tomko et al (2007), in which ∼4 nt are reeled into the leading subdomain of a UvrD molecule in each enzymatic cycle, with each nucleotide translocation coupled to the hydrolysis of one ATP, followed by a release of the stretch through the trailing subdomain. Third, the two subunits of the dimer can either switch strands jointly or unbind the DNA substrate simultaneously after unwinding a certain number of base pairs. The dimer can also occasionally disassemble, resulting in either a pause during the unwinding process or an increase in translocation rate during the rewinding process. Finally, the dimer must be activated before it is able to unwind DNA. This may involve the interaction of the two 2B subdomains. To do so, the 3′-ssDNA tail must be bent so that its effective length is reduced (Figure 5).
We call the above-mentioned mechanism a strained-inchworm mechanism because the two subunits of the inchworm are under strain by the ssDNA. The dimerization reaction is hindered by the force because it increases the tension on the ssDNA. Two reaction pathways can be imagined to interpret the rate–force relationship: (1) the two subunits disassemble after each enzymatic cycle, and the trailing subunit must rebind to the leading subunit to reactivate the helicase; and (2) the two subunits are always in contact to remain active during the unwinding action until they disassemble spontaneously or are separated by the external force. The first scheme is the on-pathway scheme in which the force impedes the conformational change of the dimer. The second is the off-pathway scheme in which the force disrupts the dimer to pull the complex off the reaction pathway. In either case, the assembling efficiency is reduced by the force so that the unwinding rate is reduced.
Comparison with previous results
In the previous single molecule study on UvrD-mediated unwinding (Dessinges et al, 2004), the force was exerted on the two extremes of the same ssDNA strand of a nicked dsDNA. The apparent rate of DNA unwinding at F=35 pN was measured to be 248(±74) bp/s. This value is much larger than the one measured by quenched-flow methods (Maluf et al, 2003b); however, it is close to the rate for UvrD translocation along ssDNA (189.0(±0.7) nt/s) (Fischer et al, 2004). The rewinding rate (298(±88) bp/s) was also measured. It is, again, close to the one for UvrD translocation along ssDNA, but much larger than the rewinding rate reported here. Fischer et al (2004) have given a plausible explanation for the apparent discrepancy. They suspected that the single-molecule experiments reported by Dessinges et al (2004) were most likely monitoring UvrD translocation along the ssDNA near a nick. The stretching of the duplex DNA at F=35 pN may have lowered the activation energy for DNA base-pair melting such that simple translocation by UvrD is able to unwind the duplex DNA through the thermal fluctuation-rectifying mechanism. However, as has been made clear by Johnson et al (2007), the sensitivity of the unwinding rate on force in such a mechanism depends actually on the step size of the helicase. Using the formula given by them, we calculated the translocation-limited unwinding of UvrD and found that the unwinding rate exhibits little dependence on the force because of its large step size (∼6 bp as given by Dessinges et al, 2004). The calculation is consistent with Dessinges et al's measurement. It is noteworthy that the external tension is directly exerted on the 3′-ssDNA tail in our study, whereas the displaced 3′-ssDNA tail near the nick is free of tension in Dessinges et al (2004) so that the bending of the 3′-ssDNA is not affected by the force.
Four observations support the dimeric model. (1) A 15 nt 3′-ssDNA loading tail is necessary for an active dimer; UvrD does not unwind the DNA with a 8-nt-long tail. This point has also been used to judge if a monomer is able to unwind DNA in bulk assays (Maluf et al, 2003b; Zhang et al, 2006). (2) If [UvrD]<1 nM, the unwinding rarely occurs. Similarly, Ha et al (2002) found that Rep-mediated DNA unwinding cannot be detected by single-molecule technique when [Rep]<2 nM. The observation was used by them to support the dimeric model of Rep. (3) The non-exponential distribution of the off-time is indicative of more than one binding event. Similar distributions have been used by Myong et al (2007) and Xie (2001) to demonstrate the existence of multi-event processes. (4) The multiform patterns of the unwinding bursts, especially the one with two rewinding speeds, can be readily accounted for by the dimeric model. Such a feature has not been reported before for helicases.
The kinetic features of UvrD reported here are similar to those measured previously (see Table I). We hence tend to believe that other features deduced from our present assay are also credible. Two new features are observed in this study. (1) The rewinding rate is in general much lower than the ssDNA translocation rate of monomeric UvrD. This, together with the observed multiform unwinding patterns, has led us to propose that the two subunits of a dimer can jointly switch strands and translocate backwards on the other strand to allow slow rewinding, or unbind simultaneously to allow quick rehybridization. (2) The unwinding rate is found to decrease with the increasing force. This seemingly counterintuitive result has led to a strained-inchworm mechanism in which a conformational change that bends and tenses the ssDNA is required to activate the dimer.
Table 1.
Comparison of the kinetics parameters measured in this study and in the previous ones
| Single-molecule assay (this study) | Bulk assay by Maluf et al | Single-molecule assay by Dessinges et al | |
|---|---|---|---|
| Solution condition | [ATP]=1 mM, [UvrD]⩾5 nM | Various [ATP] and [UvrD] | [ATP]=0.5 mM, [UvrD]⩽1 nM |
| Unwinding rate | 60±7 bp/s @ F=0 pN | 68±9 bp/s @ F=0 pN | 248 bp/s @ F=35 pN |
| Rewinding rate | ∼40 bp/s @ F=9 pN | N/A | 298 bp/s @ F=35 pN |
| Translocation rate | ∼150 nt/s @ F=9 pN | 189.0±0.7 nt/s @ F=0 pN | N/A |
| Step size | 4.2±1.7 bp | 4–5 bp | 6±1.5 bp |
| Processivity | 92±9 bp @ F=9 pN | 40–50 bp @ F=0 pN | 234 bp @ F=35 pN |
Materials and methods
Protein and substrate DNA
His-6-tagged E. coli UvrD helicase is expressed from pET-15b expression plasmid in E. coli strain BL21 (DE3). The overexpressed protein is purified under native conditions by using chromatography on Ni2+-nitrilotriacetic acid columns (Qiagen, Valencia, CA, USA), followed by FPLC size exclusion chromatography (Superdex 200, Pharmacia) and an ion-exchange chromatography (DEAE Sephadex A-50).
A DNA construction similar to the one reported by Danilowicz et al (2003) and Koch et al (2002) was prepared, as shown in Supplementary Figure S1. It was composed of two covalently connected lambda-DNAs (48.5 kbp, New England Biolabs), one behaving as a long linker arm and the second as the DNA hairpin to be opened. The linker DNA was hybridized and ligated with a digoxigenin-labelled oligonucleotide, whereas the DNA to be opened was hybridized and ligated with a biotinylated oligonucleotide. The DNA construction connected a superparamagnetic bead to the polished edge of a coverslip using specific interactions.
Single-molecule assay
We used a homemade transverse magnetic tweezer to monitor the UvrD-mediated unwinding of DNA. The DNA construct was immobilized in a flow chamber fabricated by sandwiching a 0.1-mm-thick coverslip between two glass slides (Figure 1A). The open side of the chamber was sealed using polydimethylsiloxane. The experiments were performed at 25°C in a buffer of 25 mM Tris–HCl (pH 7.4), 50 mM NaCl, 3 mM MgCl2, 0.2 μg/μl BSA, 1 mM DTT and 1 mM ATP. The flow chamber is located above the 60 × objective of an inverted microscope (IX71, Olympus). A magnetic bead was tethered through a DNA hairpin on the sidewall of the fluidic chamber. The magnetic field gradient in the focal plane was produced by a permanent magnet, which was held in a lateral position with respect to the flow chamber to exert a constant force on the magnetic bead. The distance between the bead and the surface of the sidewall (the extension of DNA) was directly monitored by a video camera (Figure 1B). After checking the state of the DNA connection, UvrD was added to a final concentration of 5–100 nM. The extension of the DNA as a function of time was then recorded at different forces.
Data analysis
The helicase unwinding and rewinding events are monitored by recording the bead-surface distance as a function of time. The force versus extension curve of ssDNA is well modelled as a FJC at low forces (Smith et al, 1996). It was used to convert the measured extension into the number of base pairs unzipped, that is, the effective length per base pair at a certain force. The molecular geometry results in the release of 2 nt for each base pair unwound, thereby amplifying the unwinding signal.
Supplementary Material
Supplementary Information
Acknowledgments
This work was supported by the National Key Basic Research Program of China (Grant No. 2006CB910302), the National Natural Science Foundation of China (Grant No. 30870590), the Centre National de la Recherche Scientifique and Institut Curie. We are grateful to Dr G Baldacci for his continuous support.
References
- Ali JA, Lohman TM (1997) Kinetic measurement of the step size of DNA unwinding by Escherichia coli UvrD helicase. Science 275: 377–380 [DOI] [PubMed] [Google Scholar]
- Danilowicz C, Coljee VW, Bouzigues C, Lubensky DK, Nelson DR, Prentiss M (2003) DNA unzipped under a constant force exhibits multiple metastable intermediates. Proc Natl Acad Sci USA 100: 1694–1699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dessinges MN, Lionnet T, Xi XG, Bensimon D, Croquette V (2004) Single-molecule assay reveals strand switching and enhanced processivity of UvrD. Proc Natl Acad Sci USA 101: 6439–6444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont S, Cheng W, Serebrov V, Beran RK, Tinoco I Jr, Pyle AM, Bustamante C (2006) RNA translocation and unwinding mechanism of HCV NS3 helicase and its coordination by ATP. Nature 439: 105–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essevaz-Roulet B, Bockelmann U, Heslot F (1997) Mechanical separation of the complementary strands of DNA. Proc Natl Acad Sci USA 94: 11935–11940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer CJ, Maluf NK, Lohman TM (2004) Mechanism of ATP-dependent translocation of E. coli UvrD monomers along single-stranded DNA. J Mol Biol 344: 1287–1309 [DOI] [PubMed] [Google Scholar]
- Ha T, Rasnik I, Cheng W, Babcock HP, Gauss GH, Lohman TM, Chu S (2002) Initiation and re-initiation of DNA unwinding by Escherichia coli Rep helicase. Nature 419: 638–641 [DOI] [PubMed] [Google Scholar]
- Johnson DS, Bai L, Smith BY, Patel SS, Wang MD (2007) Single-molecule studies reveal dynamics of DNA unwinding by the ring-shaped T7 helicase. Cell 129: 1299–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch SJ, Shundrovsky A, Jantzen BC, Wang MD (2002) Probing protein–DNA interactions by unzipping a single DNA double helix. Biophys J 83: 1098–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korolev S, Hsieh J, Gauss GH, Lohman TM, Waksman G (1997) Major domain swiveling revealed by the crystal structures of complexes of E. coli Rep helicase bound to single-stranded DNA and ADP. Cell 90: 635–647 [DOI] [PubMed] [Google Scholar]
- Lee JY, Yang W (2006) UvrD helicase unwinds DNA one base pair at a time by a two-part power stroke. Cell 127: 1349–1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lionnet T, Spiering MM, Benkovic SJ, Bensimon D, Croquette V (2007) Real-time observation of bacteriophage T4 gp41 helicase reveals an unwinding mechanism. Proc Natl Acad Sci USA 104: 19790–19795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohman TM, Bjornson KP (1996) Mechanisms of helicase-catalyzed DNA unwinding. Annu Rev Biochem 65: 169–214 [DOI] [PubMed] [Google Scholar]
- Lohman TM, Tomko EJ, Wu CG (2008) Non-hexameric DNA helicases and translocases: mechanisms and regulation. Nat Rev Mol Cell Biol 9: 391–401 [DOI] [PubMed] [Google Scholar]
- Maluf NK, Ali JA, Lohman TM (2003a) Kinetic mechanism for formation of the active, dimeric UvrD helicase-DNA complex. J Biol Chem 278: 31930–31940 [DOI] [PubMed] [Google Scholar]
- Maluf NK, Fischer CJ, Lohman TM (2003b) A dimer of Escherichia coli UvrD is the active form of the helicase in vitro. J Mol Biol 325: 913–935 [DOI] [PubMed] [Google Scholar]
- Myong S, Bruno MM, Pyle AM, Ha T (2007) Spring-loaded mechanism of DNA unwinding by hepatitis C virus NS3 helicase. Science 317: 513–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myong S, Rasnik I, Joo C, Lohman TM, Ha T (2005) Repetitive shuttling of a motor protein on DNA. Nature 437: 1321–1325 [DOI] [PubMed] [Google Scholar]
- Patel SS, Picha KM (2000) Structure and function of hexameric helicases. Annu Rev Biochem 69: 651–697 [DOI] [PubMed] [Google Scholar]
- Singleton MR, Dillingham MS, Gaudier M, Kowalczykowski SC, Wigley DB (2004) Crystal structure of RecBCD enzyme reveals a machine for processing DNA breaks. Nature 432: 187–193 [DOI] [PubMed] [Google Scholar]
- Smith SB, Cui Y, Bustamante C (1996) Overstretching B-DNA: the elastic response of individual double-stranded and single-stranded DNA molecules. Science 271: 795–799 [DOI] [PubMed] [Google Scholar]
- Tomko EJ, Fischer CJ, Niedziela-Majka A, Lohman TM (2007) A nonuniform stepping mechanism for E. coli UvrD monomer translocation along single-stranded DNA. Mol Cell 26: 335–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Hippel PH, Delagoutte E (2001) A general model for nucleic acid helicases and their ‘coupling' within macromolecular machines. Cell 104: 177–190 [DOI] [PubMed] [Google Scholar]
- Wong I, Amaratunga M, Lohman TM (1993) Heterodimer formation between Escherichia coli Rep and UvrD proteins. J Biol Chem 268: 20386–20391 [PubMed] [Google Scholar]
- Xie S (2001) Single-moleucle approach to enzymology. Single Mol 2: 229–236 [Google Scholar]
- Zhang XD, Dou SX, Xie P, Hu JS, Wang PY, Xi XG (2006) Escherichia coli RecQ is a rapid, efficient, and monomeric helicase. J Biol Chem 281: 12655–12663 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information