Abstract
Type I interferon (IFN) inhibits, by an unknown mechanism, the replication of human papillomaviruses (HPV), which are major human pathogens, Here, we present evidence that P56 (a protein), the expression of which is strongly induced by IFN, double-stranded RNA and viruses, mediates the anti-HPV effect of IFN. Ectopic expression of P56 inhibited HPV DNA replication and its ablation in IFN-treated cells alleviated the inhibitory effect of IFN on HPV DNA replication. Protein–protein interaction and mutational analyses established that the antiviral effect of P56 was mediated by its direct interaction with the DNA replication origin-binding protein E1 of several strains of HPV, through the tetratricopeptide repeat 2 in the N-terminal region of P56 and the C-terminal region of E1. In vivo, the interaction with P56, a cytoplasmic protein, caused translocation of E1 from the nucleus to the cytoplasm. In vitro, recombinant P56, or a small fragment derived from it, inhibited the DNA helicase activity of E1 and E1-mediated HPV DNA replication. These observations delineate the molecular mechanism of IFN's antiviral action against HPV.
Keywords: antiviral action of interferons, HPV E1, human papillomavirus, ISG56, viral DNA replication
Introduction
The interferon (IFN) system is the first line of defence against virus infection in the vertebrates (Stark et al, 1998; Sen, 2001). The system is designed to contain the spreading of virus infection not only by impairing virus replication but also by elimination of the infected cells through direct apoptosis or by the action of cells of the immune system (Samuel, 2001). The infection itself triggers IFN synthesis and secretion. The secreted IFNs exert an effect on as yet uninfected cells and forearm them to combat with subsequent virus infection. The actions of IFNs extend to certain parasitic infection and malignant tumour. Consequently, IFNs are used clinically for managing specific types of malignancies. They are also used to treat chronic infections with hepatitis B and hepatitis C viruses (Lin and Keeffe, 2001) and to treat infections with herpes viruses and papillomaviruses (Koromilas et al, 2001; Lin and Keeffe, 2001; Whitley, 2001).
The actual antiviral actions are carried out by the products of IFN-stimulated genes (ISGs), which number in the hundreds (Der et al, 1998). A subset of the ISGs, the viral stress-inducible genes (VSIGs), is also induced by double-stranded (ds) RNA, a common by-product of virus infection, and by other viral gene products (Geiss et al, 2001; Grandvaux et al, 2002). Thus, several consequences of viral infection lead to the induction of a common set of genes (Sen and Peters, 2007). Although the same cis-acting sequence, the IFN-stimulated response element (ISRE), present in the promoters of these genes, receives signals from all inducers, the transcription factors responsible and their modes of activation are quite distinct. IFNs stimulate transcription of VSIG by activating the Jak–STAT pathway of signal transduction. The critical transcription factor used by IFNs to stimulate VSIG transcription is ISGF3, composed of STAT1, STAT2 and IRF9 (Stark et al, 1998). In contrast, induction of the same genes by dsRNA or virus infection does not use the Jak–STAT pathway (Elco et al, 2005). The critical transcription factor in this case is IRF-3, which is activated by signalling pathways triggered by different Toll-like receptors or cytoplasmic RNA helicases, RIG-I and Mda-5.
Although IFN's antiviral activities are well known, it has been difficult to delineate the underlying molecular basis of the observed block in replication of most viruses. Moreover, for no virus, a single IFN-induced protein can account for the entire antiviral effect; usually several such proteins inhibit distinct steps of virus replication. In this context, we have been studying the properties of P56, the product of the ISG56 (IFIT1) gene, which is highly induced in response to IFN, dsRNA and many viruses (Der et al, 1998; Geiss et al, 2001; Grandvaux et al, 2002). P56 belongs to a family of structurally related proteins that are induced by viral stresses. In humans, there are three other members, P60, P58 and P54 (Sarkar and Sen, 2004). Most untreated cells do not express P56 at a detectable level, but viral and other stresses induce transcription of the ISG56 gene rapidly and strongly. All of these proteins contain multiple tetratricopeptide (TPR) motifs that are known to mediate protein–protein interactions through scaffolds formed among tandem TPR repeats (Lamb et al, 1995). The exact sequences of TPRs are not highly conserved, but there are invariant residues present in same locations in all TPRs and their substitutions destroy the TPR structure. We have been investigating the functional properties of P56 and other proteins of this family (Guo et al, 2000a). Using yeast two-hybrid screens, we have identified several cellular proteins that interact with P56, the most well characterized of which is the Int-6 protein that is encoded by the Int-6 gene, whose disruption by the integration of the mouse mammary tumour virus genome, causes breast cancer in mice (Marchetti et al, 1995); Int-6 is identical to the eIF-3e subunit of the eukaryotic translation initiation factor 3 (Asano et al, 1997). The C-terminal region of P56 mediates its interaction with eIF-3e (Guo and Sen, 2000) and causes an impairment of eIF-3 function and resultant inhibition of protein synthesis (Guo et al, 2000a). Other members of the P56 family can also interact with the ‘e' or the ‘c' subunit of eIF-3 and cause inhibition of translation initiation (Guo et al, 2000a; Hui et al, 2003, 2005). The P56 families of proteins have been implicated in IFN's antiviral actions against HCV, West Nile virus and LCMV (Wang et al, 2003; Wacher et al, 2007).
Human papillomaviruses (HPVs) infect squamous epithelial cells and induce hyperproliferative lesions (Howley and Lowy, 2001). The high-risk groups, such as HPV16, 18 or 31, are associated with anogenital cancer and the primary aetiological agent for cervical carcinoma (zur Hausen, 1996). The low-risk groups, such as HPV6 or 11, cause genital warts (Lowy et al, 1994; zur Hausen and de Villiers, 1994). HPV replication is tightly dependent on the state of terminal differentiation of the infected cells (Dollard et al, 1992). For this reason, organotypic or raft cultures, instead of monolayer tissue cultures of dividing cells, need to be used for studying HPV replication in vitro (Chow and Broker, 1997). Two viral proteins, E1 and E2, are required, along with many host factors, for HPV DNA replication that originates at the viral Ori sequence; Ori-containing viral DNA or plasmids can replicate episomally and this process can be studied either in cells expressing ectopic E1 and E2 or in cell-free systems containing these proteins (Sverdrup and Khan, 1994). E1, a phosphoprotein that shuttles between the nucleus and the cytoplasm, has ATPase and DNA helicase activities (Hughes and Romanos, 1993); E1 and E2, the two nuclear phosphoproteins, form heteromeric complexes with the Ori DNA (Chen and Stenlund, 2002). IFNs have been found to be effective in treating genital warts, an HPV-associated disease (Lowy and Howley, 2001). However, the underlying mechanism has been difficult to discern because of the paucity of good in vitro models. In an early study, Turek et al (1982) observed complete elimination of BPV genome on long-term IFN treatments of BPV-transformed mouse cells, which caused their reversion to a non-transformed phenotype. Khan et al (1993) observed IFN-mediated inhibition of keratinocyte transformation by HPV16 and Chang and Laimins (2001) observed that the basal level expression of ISGs is repressed by HPV31 gene products in proliferating cultures, causing delayed response to IFN. The same group found that long-term treatment with IFN causes growth arrest and apoptosis of HPV-positive squamous carcinoma cells and episomal HPV DNA was eliminated from the surviving cells (Chang et al, 2002).
The current study delineates the molecular basis of IFN's actions against HPV replication. We observed, by yeast two-hybrid assays, that P56 interacted with HPV18 E1. Further experiments with human cells confirmed this interaction and allowed us to map the interacting domains. In a cell-based replication system, IFN pretreatment inhibited HPV DNA replication, an effect that was reversed by the ablation of P56 expression. In vitro, P56 bound to E1 directly and inhibited its DNA helicase activity, as well as its ability to support HPV DNA synthesis in a cell-free system. These results indicate that P56 is the major, if not the only, IFN-induced protein that is responsible for inhibiting HPV replication.
Results
Interaction of P56 with HPV E1 protein
To identify P56-interacting human proteins, we conducted a yeast two-hybrid screen of a cDNA library from HeLa cells, which happen to express HPV18 genes as well. Surprisingly, P56 interacted with a clone containing HPV DNA (Figure 1A, slot 5). Appropriate controls (slots 1–4 and 6) confirmed that the observed interaction was genuine. The P56-interacting clone encoded a protein consisting of the GAL-4 activation domain fused to the C-terminal residues 360–657 of HPV18 E1 protein. The suggested interaction between P56 and HPV18 E1 was confirmed by co-transfecting plasmids expressing the two full-length proteins in mammalian cells; when P56 was immunoprecipitated, HPV18 E1 was bound to it (Figure 1B, left top panel). Similar results were obtained when endogenous P56 was induced by treating cells with IFN-β (Figure 1B, right top panel). Interaction between HPVE1 and P56 was specific; other proteins of the human ISG56 family, P54, P58 and P60, did not interact with HPVE1 (Figure 1C). Moreover, the interaction between P56 and HPV18 E1 was direct as shown by co-immunoprecipitation of the two purified proteins (Figure 1D). Similar interactions were also observed between P56 and HPV11 E1 (Supplementary Figure S1) and HPV31 E1 proteins (data not shown), indicating that P56 may interact with E1 proteins of many strains of HPV. The observed interaction between P56 and E1 was confirmed using immunofluorescence assays. Because E1 is a nuclear protein, a GFP–E1 fusion protein was located in the nucleus, (Figure 1E, panel 1, middle) and as expected, transfected P56 was in the cytoplasm. When co-expressed, through its interaction with E1, P56 translocated E1 to the cytoplasm (Figure 1E, panel 2, middle). Similar results were obtained with P56 induced by IFN treatment of cells (Figure 1E, panel 3). However, P54, which does not interact with E1 as judged by the lack of their co-immunoprecipitation (Figure 1C), did not translocate E1 to the cytoplasm (Figure 1E, panel 4).
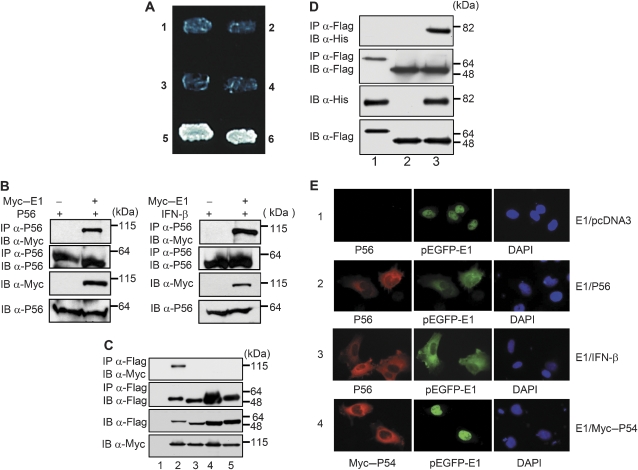
Figure 1.
Interaction between E1 and P56 proteins. (A) Yeast two-hybrid interaction between P56 and the C-terminal (360–657 aa) domain of HPV18 E1. Yeast strain Y190 was transformed with the following pairs of expression vectors, and plated onto the selection medium without histidine: (1) BD vector+AD-360–657 HPV18 E1; (2) BD-P56+AD-SV40 large T-antigen; (3) BD-P56+AD vector; (4) BD-P53+AD-360–657 HPV18 E1; (5) BD-P56+AD-360–657 HPV18 E1; (6) BD-P53+AD-SV40 large T-antigen. (B) Co-immunoprecipitation of E1 and P56. HT1080 cells were transfected with empty vector or a plasmid expressing Myc-fused E1; in addition, a P56-expressing plasmid was transfected (left panel) or cells were treated with IFN to induce the expression of P56 (right panel). The cell extracts were immunoblotted (IB) directly or after immunoprecipitation (IP) using indicated antibodies. Molecular weight markers are shown on the right. (C) Specific interaction between E1 and P56. HT1080 cells were co-transfected with plasmids expressing Flag-fused P56 family member proteins (lane 2: P56; lane 3: P54; lane 4: P58 and lane 5, P60) and Myc-fused E1. The cell extracts were immunoblotted (IB) directly or after immunoprecipitation (IP) using indicated antibodies. Lane 1 represents empty Myc vector-transfected cell extracts. (D) Direct interaction between E1 and P56. Equimolar amounts of purified polyhistidine-tagged E1, purified Flag–P56 or purified Flag–PKR were mixed. After 2 h, the samples were processed as indicated. Lane 1: PKR and E1; lane 2: only P56; lane 3: P56 and E1. (E) Cytoplasmic translocation of HPV11 E1 by P56 interaction. P2.1 cells were transfected with an expression vector of E1 (pEGFP-E1); in addition, the indicated vectors were co-transfected in panels 1, 2 and 4 or cells were treated with IFN in panel 3. After 18 h, cells were stained with DAPI and antibodies against P56 or Myc and then analysed by immunofluorescence. Images shown here are representative of three independent experiments.
Mapping of the interaction domains
Deletion and point mutants of P56 and E1 were generated to identify their interacting domains. Full-length E1 or E 360–657 C-terminal fragment interacted with P56 (data not shown). But further deletions of E1 from either the N- or C-terminal end eliminated the interaction (data not shown). Similar deletion analyses of P56 revealed that the N-terminal residues 1–179 of P56 were sufficient for interacting with E1 (Figure 2A). Further deletions showed that proteins containing P56 residues 1–152 or 29–179 fused with MBP, could interact with E1 (Figure 2B). These results suggested that P56 residues 29–152 were sufficient for interacting with E1. This conclusion was confirmed by expressing P56 1–152 (Figure 2C, lane 2), P56 29–179 (Figure 2C, lane 3) and P56 29–152 (Figure 2C, lane 4). As expected, E1 interacted with all of these proteins (Figure 2C, upper panel). The N-terminal region of P56, which interacted with E1, contains three TPR motifs that mediate protein–protein interactions (Figure 3A). To determine which of these TPR motifs mediated P56–E1 interaction, we mutated each of them individually; three residues, the mutations of which are known to perturb the TPR structure, were substituted for this purpose. Mutation of TPR2 (lane 1), but not TPR1 (lane 2), TPR3 (lane 3) or both TPR1 and TPR3 (lane 4), eliminated the ability of P56 1–179 to interact with E1 (Figure 3B), indicating that the interaction was through TPR2. As expected, E1 did not co-immunoprecipitated with full-length P56 that had TPR2 mutated (M2) (Supplementary Figure S2). Immunofluorescence data confirmed the lack of interaction between M2P56 and E1; unlike WtP56, M2P56 could not translocate pEGFP-E1 from the nucleus to the cytoplasm (Figure 3C, upper panel). The results presented above demonstrated that the N-terminal region of P56 interacted with the C-terminal region of E1 and the interaction required TPR2 of P56, but not other TPRs (Figure 3D). Although the M2 mutant of P56 could not interact with E1, it could still bind to eIF-3e (data not shown); consequently, this mutant was as potent an inhibitor of translation as WtP56 (Figure 3E).
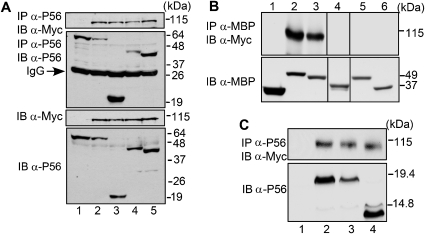
Figure 2.
Mapping of the E1-binding domain of P56. (A) Several deletion mutants of P56 were co-expressed with Myc–E1. The cell extracts were immunoblotted (IB) directly or after immunoprecipitation (IP) using indicated antibodies. Lane 1: only P56; lane 2: full-length P56 and E1; lane 3: P56 1–179 and E1; lane 4: P56 lacking 179–335 and E1; lane 5: P56 1–345 and E1. (B) Nested deletions of P56 1–179 were expressed as MBP-tagged proteins along with Myc–E1. Cell lysates were immunoprecipitated with MBP antibody and western blotted with Myc antibody (upper panel) or directly western blotted with MBP antibody (lower panel). Lane 1: MBP; lane 2: MBPP56 1–179; lane 3: MBPP56 1–152; lane 4: MBPP56 1–46; lane 5: MBPP56 86–179; lane 6: MBPP56 132–179. Because two additional deletion mutants showed very little expression, the corresponding lanes have been deleted from the figure. (C) Three deletion mutants of P56 1–179 were expressed, without the MBP tag, along with Myc–E1. Cell lysates were analysed as indicated. Lane 1: only E1; lane 2: P56 1–152 and E1; lane 3: P56 29–179 and E1; lane 4: P56 29–152 and E1.
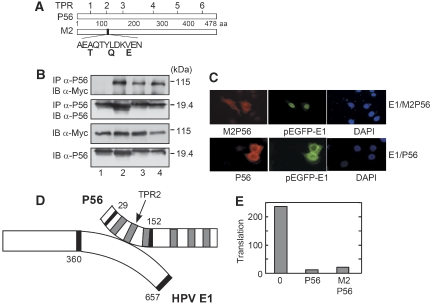
Figure 3.
Requirement of TPR2 for P56 interaction with E1. (A) Schematic diagram of P56 structure: the locations of the TPR domains are shown on the top and the specific mutations in TPR2 of the M2 mutant are shown below. (B) Point mutants of P56 1–179 at TPR1 (lane 2), TPR2 (lane 1), TPR3 (lane 3) and a double mutant at TPR1 and 3 (lane 4) were expressed together with Myc–E1. The cell extracts were immunoblotted (IB) directly or after immunoprecipitation (IP) using indicated antibodies. (C) Cells were co-transfected with expression vector of E1 (pEGFP-E1) and wild-type P56 (P56, lower panel) or the TPR2 mutant of P56, M2 (M2P56, upper panel). Cells were stained with DAPI and antibody against P56 and analysed by immunofluorescence. (D) Schematic diagram of the E1–P56 interacting domains: the P56-binding site is from residue 29 to 152 with TPR2 being involved in the interaction and the cognate domain of E1 is from residue 360 to 657. The sketch is not to scale. (E) WtP56 or M2P56 was tested for inhibiting in vitro translation of luciferase mRNA. Newly synthesized radiolabelled luciferase was separated by gel electrophoresis and quantified by Phosphorimager analysis. The data are presented in arbitrary units.
Inhibition of HPV DNA replication by IFN and dsRNA
Transcription of ISG56, which encodes P56, is induced not only by type I IFNs but also by dsRNA, which exerts an effect through the TLR3 and RIG-I pathways. Because of the observed interaction between P56 and HPVE1, we wanted to examine whether treatments of cells with IFN or dsRNA had any effects on E1 functions. The major function of E1 is to support HPV DNA replication, which can be conveniently measured by a transient transfection assay. In this assay, C33A cells are transfected with expression vectors of E1 and E2, the two proteins necessary for supporting HPV DNA replication, and a plasmid containing the HPV Ori DNA. Because DNA synthesized in human cells is insensitive to Dpn-I due to its methylation status, replication of the HPV Ori region can be monitored by measuring the amounts of Dpn-I-insensitive Ori DNA. Using this assay, we tested the effects of IFN-β on HPV DNA replication. As expected, increasing doses of IFN induced increased levels of P56 (Figure 4A). In untreated cells, there was a substantial amount of replicated Ori DNA, which was not degraded by Dpn-I, although unreplicated Ori, E1 and E2 DNAs were completely degraded (Figure 4B). Quantification of the amounts of replicated Ori DNA showed that IFN-β strongly inhibited HPV DNA replication in a dose-dependent manner (Figure 4C). This effect was observed with both the high-risk strain HPV18 DNA and the low-risk strain HPV11 DNA (data not shown). To examine the effects of dsRNA on C33A cells, which do not express TLR3, exogenous expression of TLR3 was necessary. dsRNA efficiently induced P56 in two TLR3-expressing C33A cells, 44 and 14, but not in a sister clone, 29, that did not express TLR3 (Figure 4D); HPV18 Ori replication was strongly inhibited by dsRNA treatment of clones 44 and 14, but not 29 (Figure 4E). These results demonstrated that two agents, IFN and dsRNA, both of which could induce the synthesis of P56, inhibited HPV DNA replication, indicating that P56 might be responsible for inhibiting the action of HPV E1.
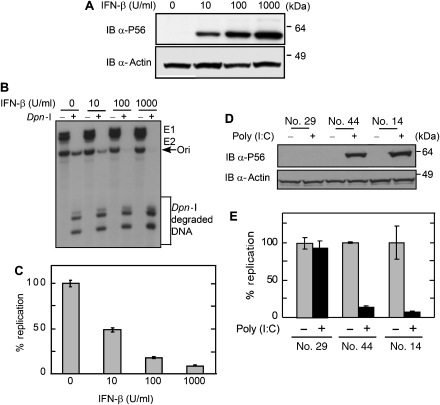
Figure 4.
Effect of IFN-β and poly (I)–poly (C) on HPV Ori DNA replication. (A) Dose-dependent induction of P56 by IFN: cells were treated with increasing doses of IFN, and cellular P56 levels were measured by immunoblotting. (B) Cellular replication assay. The plasmid pOri177 (which contain the HPV18 origin) and expression vectors of E1 and E2 were co-transfected into C33A cells and then cells were treated with different concentrations of IFN-β. Replication of Ori DNA was analysed by Southern blotting of Dpn-I-digested DNA. Arrows indicate the Ori DNA. (C) Dose-dependent inhibition of Ori DNA replication by IFN. Cells were transfected as above and treated with different concentrations of IFN. Replicated Ori DNA was quantitated and normalized. Data are represented as means of three independent experiments. (D) Establishment of TLR3-expressing C33A cells. In two cell clones (nos. 44 and 14) expressing TLR3, Poly (I:C) could induce P56. Another clone (no. 29) did not express TLR3 and P56 was not induced in it. Cell lysates were used for measuring the levels of P56 and actins by immunoblotting. (E) Inhibition of Ori DNA replication by dsRNA treatment. Replication assays were performed in poly (I)–poly (C)-treated and untreated C33A clones expressing or not expressing TLR3. Data are presented as means of three independent experiments.
P56-mediated inhibition of HPV DNA replication by IFN
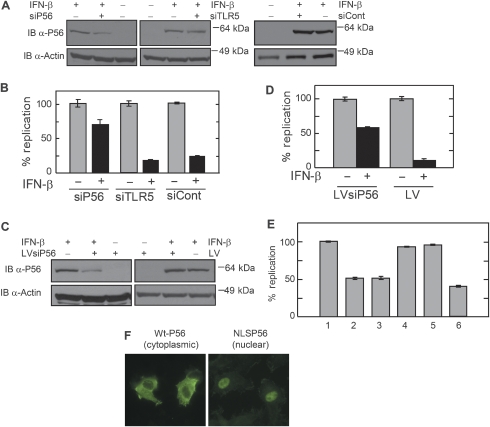
In the next series of experiments, we examined whether the observed inhibition of HPV DNA replication in IFN-treated cells was indeed mediated by P56. For this purpose, P56 expression in IFN-treated C33A cells was ablated by transfection of siRNA for P56. This treatment caused a partial lowering of the P56 level, whereas similar expression of a scrambled control siRNA or an unrelated siRNA did not cause any inhibition (Figure 5A). When HPV DNA replication was measured in these cells, lowering of the level of P56 caused substantial alleviation of the IFN's inhibitory effect (Figure 5B), indicating that P56 is a major, if not the sole, mediator of this effect. Similar conclusions were drawn from another experiment, which used a lentivirus expression vector for P56 short hairpin (shRNA); again less P56 expression relieved the inhibitory effects of IFN (Figure 5C and D). In a complementary experiment, exogenous P56 was expressed by infecting the C33A cells with lentiviral expression vectors for P56 and the extent of HPV DNA replication was measured. P56 expressed from the lentivirus did not cause any inhibition of E1 synthesis (Supplementary Figure S3). However, expression of WtP56 or its 1–179 deletion mutant caused about 50% inhibition of DNA replication (Figure 5E, lanes 2 and 3); no such inhibition was observed in cells expressing mutant P56 proteins that did not bind to E1 (Figure 5E, lanes 4 and 5). The above results strongly indicate that P56 can inhibit HPV DNA replication in vivo and it mediates the inhibitory action of IFN on HPV DNA replication. To examine whether this action of P56 was mediated by its ability to translocate E1 to the cytoplasm (Figure 1E), we generated a P56 mutant directed to the nucleus. This mutant, NLSP56, to which the nuclear localization signal of the simian virus 40 (SV40) T antigen was attached, was localized exclusively in the nucleus (Figure 5F) and it interacted with E1 (Supplementary Figure S4). The NLSP56 protein inhibited HPV DNA replication as strongly as the Wt protein (Figure 5E), suggesting that P56 could inhibit nuclear functions of E1.
Figure 5.
Effect of ablation in IFN-treated cells or ectopic expression of P56 on HPV Ori DNA replication. (A) Expression levels of P56. The levels were determined in IFN-treated and untreated C33A cells transfected with siRNA for P56, an unrelated siRNA for TLR5 or a scrambled control P56 siRNA. (B) Quantitation of HPV Ori DNA replication. Replication was measured in cells treated as in panel A. Data are presented as means of three independent experiments. (C) C33A cells were infected with lentivirus containing shRNAi for P56 (LVsiP56) or empty vector (LV) used for replication assays. Expression levels of P56 protein in IFN-treated cells were measured. (D) Quantitation of HPV DNA replication in the above cells. Data are presented as means of three independent experiments. (E) Inhibition of HPV DNA replication by the expression of P56 protein in C33A cells. Cells were infected with lentiviruses expressing wild-type or mutant P56 protein, transfected with E1, E2 and Ori plasmid, and DNA replication was measured. Data are represented as means of three independent experiments. Lane 1: no P56; lane2: WtP56; lane3: P56 (1–179); lane 4: M2P56; lane 5: M2P56 (1–179); lane 6: NLS P56. (F) WtP56 and NLS P56 were expressed in cells and their subcellular locations were determined by immunostaining.
Inhibitory effects of P56 and E1 functions in vitro
To confirm and further explore the effects of P56 on HPV E1, we resorted to in vitro assays for measuring the function of the viral protein. In an in vitro HPV Ori DNA replication system, recombinant E1, expressed either in Escherichia coli or in insect cells, was absolutely needed for de novo synthesis of the viral DNA (Figure 6A). In our hands, further addition of E2 enhanced the synthesis only marginally. Using this assay, we could demonstrate dose-dependent inhibition of HPV DNA replication by recombinant P56 (Figure 6B). In the presence of 2.5 μM WtP56, DNA replication was completely inhibited, whereas the same amount of M2P56 did not inhibit it at all. This conclusion was true for recombinant P56 purified for either bacteria or insect cells (Figure 6C). Similarly, inhibition was observed with the N-terminal fragment, 1–179, of P56, but not with the corresponding M2 protein (Figure 6D). These results demonstrated that interaction of E1 with P56 causes potent inhibition of the ability of the viral protein to support viral DNA replication. To support DNA replication, E1 requires its helicase activity to unwind DNA at the Ori region and its ATPase activity. Full-length P56 completely blocked the helicase activity of E1 (Figure 7A, lane 3). The same was true for P56 1–179 (Figure 7B); Wt (lane 4), but not M2 (lane 5), P56 1–179 blocked the helicase activity as strongly as full-length P56 (lane 6). In contrast to the above results, P56 could not inhibit the ATPase activity of E1 as measured by free phosphate release from ATP (Figure 7C). The above results clearly demonstrated that binding of P56 to E1 blocked its ability to unwind DNA, but not hydrolyse ATP.
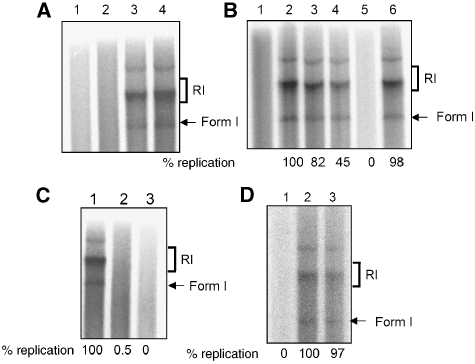
Figure 6.
Effect of P56 on HPV11 Ori replication in vitro. (A) In vitro DNA replication assay. Purified E1 and E2 proteins were added to the replication assay system and radiolabelled newly synthesized DNA was analysed by gel electrophoresis. Replication intermediates, RI and form I, are indicated on the side and the relative replication levels, as quantified, are shown at the bottom. Lane 1: no addition; lane 2: E2; lane 3: E1; lane 4: E1 and E2. (B) Effect of P56. Replication assay was performed in the presence of increasing amounts of bacterially expressed P56. Lane 1: no E1; lane2: E1; lane 3: E1 and 0.5 μM P56; lane 4: E1 and 1.0 μM P56; lane 5: E1 and 2.5 μM P56; lane 6: E1 and 2.5 μM M2P56. Replicated DNA was analysed and quantitated. (C) Effect of E1 expressed in insect cells. Lane 1, no P56; lane 2, 2.5 μM of bacterially expressed P56; lane 3, 2.5 μM of P56 expressed in insect cells. (D) Effect of P56 1–179. GST-linked 1–179 amino-acid fragment of Wt or M2 P56 proteins was purified from bacteria and added to the reaction. Lane1: GST-WtP56 (1–179); lane 2: GST; lane 3: GST-M2P56 (1–179).
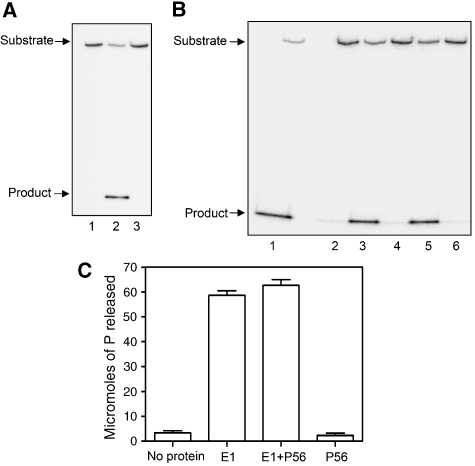
Figure 7.
Effect of P56 on E1 enzyme activities. (A) Inhibition of DNA-unwinding activity of E1 by P56. E1 helicase assay was performed using E1 expressed and purified from insect cells. The position of the substrates and products is indicated by arrows. Lane 1: no E1; lane 2: E1; lane 3: E1 and P56. (B) Inhibition of unwinding activity by P56 1–179. GST-linked P56 1–179 and its mutants were purified from bacteria and added to the reaction. Lane 1: E1; lane 2: no E1; lane 3: E1 and GST; lane 4: E1 and GST–P56 (1–179); lane 5: E1 and GST–M2P56 (1–179); lane 6: E1 and WtP56. (C) Effects of P56 on ATPase activity of E1. Hydrolysis of ATP by E1 was assayed; means of three independent experiments are shown. Lane 1: no protein; lane 2: E1; lane 3: E1 and P56; lane 4: P56.
Discussion
HPV is a major human pathogen causing cancer and hyperproliferative lesions in epithelial cells (Stanley et al, 2007). Although type I IFN has been used clinically to treat genital warts and other HPV-caused lesions, the underlying antiviral mechanism remained unclear; this study provides the first clear evidence of a specific IFN-induced protein blocking HPV DNA replication. It remains possible that other ISG products can interfere with other steps of HPV life cycle, but our results strongly indicate that P56 is the major component of the anti-HPV action of IFN. Such a direct functional correlation between one IFN-induced protein and a specific virus is rare in the IFN literature; usually several such proteins exert an effect at many steps of replication of a virus. Moreover, the biochemical mechanism of P56 action against HPV is clear and straightforward, which is hardly the case for other ISG products, except Mx, which strongly inhibits the replications of influenza and other related viruses (Haller et al, 1998).
Our experimental results explain many observations in the literature reporting progressive losses of HPV episomal DNA on IFN treatment of virus-infected tissues or cells (Turek et al, 1982; Herdman et al, 2006). Maintenance of episomal virus DNA in infected tissues can lead to genomic integration of the viral DNA, especially for high-risk HPV strains, causing neoplastic progression (Pett and Coleman, 2007). In tissue culture, IFN treatment of cells harbouring bovine papillomavirus or HPV31 DNA episomes causes a gradual loss of the viral DNA, suggesting a specific mode of action of IFN against HPV and a rationale for treating non-cancerous HPV lesions in patients with IFNs (Chang et al, 2002). The results presented here demonstrate that the underlying cause is an IFN-induced inhibition of HPV DNA replication, which is mediated by the interaction of P56 with E1, a viral protein that is essential for DNA replication.
Because of the paucity of human cell lines that are amenable to de novo HPV infection, it has been difficult to study different aspects of virus replication in the same cells and the effects of exogenous agents on them. To circumvent this problem, different parts of viral metabolism have been often studied in isolation and the assay, used here to examine HPV DNA replication, has been very useful to identify the Ori sequences and the E1 and E2 protein domains that are necessary for efficient DNA replication. Using this assay, we could demonstrate strong effects of type I IFN and TLR3 signalling on HPV DNA replication (Figure 4). The assay condition itself did not cause any IFN synthesis (Supplementary Figure S5) and exogenous IFN treatment did not cause any cell death or perturbation of cell cycle regulation that could account for the observed inhibition of viral DNA synthesis (Supplementary Table S1). Manipulations of P56 expression strongly indicated that the inhibitory action was mediated by P56 (Figure 5). Ectopic expression of P56 itself caused strong inhibition of viral DNA replication, although it was not as pronounced as in cells treated with IFN (Figure 5E). The levels of P56 expression in these experiments were comparable to those in cells treated with IFN, and the effects were not due to an inhibition of E1 synthesis (Supplementary Figure S3). The latter consideration is important because P56 is known to cause translational inhibition (Guo et al, 2000a). The actions of the two mutants of P56, M2 P56 and P56 1–179, further confirmed that the antiviral effect of P56 is distinct. The first mutant was incapable of inhibiting DNA replication (Figure 5E) because it could not bind to E1 (Supplementary Figure S2); although it inhibited protein synthesis as strongly as WtP56 (Figure 3E). On the other hand, the second mutant, P56 1–179 did not bind to eIF-3 or inhibit protein synthesis (Guo and Sen, 2000), but it could still inhibit DNA replication by binding to E1 (Figures 2 and 5E). When P56 was artificially directed to the nucleus (Figure 5F), it could still inhibit DNA replication, indicating that the cytoplasmic translocation of E1 by WtP56 may not contribute much to the phenomenon. We can speculate several reasons for the observed incompleteness of the inhibition of DNA replication by ectopically expressed P56. It is possible that in IFN-treated cells, P56 works in conjunction with another IFN-induced protein or that it is modified by IFN treatment to a more potent form; however, given our in vitro results these are unlikely explanations. The more likely explanation is that P56 is the major, but not the only, IFN-induced protein that inhibits HPV DNA replication. Ablation of P56 expression in IFN-treated cells solidified our conclusions further. Although, P56 expression was only partially ablated (Figure 5A and C), there was substantial alleviation of the inhibition of DNA replication in IFN-treated cells.
The in vitro analysis of the action of P56 on E1 functions produced unequivocal results confirming its strong inhibitory effects. New HPV DNA synthesis was inhibited by recombinant P56, expressed in either bacteria or insect cells, in a dose-dependent manner (Figure 6B) and the inhibition was observed only with full-length P56 or a fragment that bound to E1, but not with the corresponding mutants that did not bind to E1 (Figure 6). The same was true for the DNA helicase activity of E1 (Figure 7). Surprisingly, another activity of E1, the ATPase activity, was not affected by P56 at all (Figure 7C). The latter result demonstrated that the effect of P56 on E1 was selective, thus reassuring that it was not causing gross distortions of the E1 structure. Future studies should analyse detailed steps of the E1 helicase activity and identify the one that is inhibited by P56. For example, P56 may affect E1 oligomerization, DNA binding or the helicase activity itself (Sedman et al, 1997; Sun et al, 1998; Schuck and Stenlund, 2005).
The detailed analysis of P56–E1 interactions provided important information. The P56–E1 interaction is direct, the two purified recombinant proteins bound to each other (Figure 1D). It is also specific, other members of the human P56 family did not interact with E1 (Figure 1C). On the other hand, P56 recognizes E1 from many strains of HPV E1; we tested HPV18, HPV11 and HPV31 E1 and P56 interacted with and inhibited functions of all of them. It is the extreme N-terminal region of P56, between residues 29 and 152, which interacted with E1. This region contains three TPR motifs (Sarkar and Sen, 2004), of which TPR2 is the most critical (Figure 3A); substitution of three residues, in TPR2, which is known to perturb the TPR structure (Lamb et al, 1995), eliminated E1 interaction. On the other hand, similar mutations in TPR1 and TPR3 or both did not block E1 interaction (Figure 3B). Results from the co-immunoprecipitation and pull-down assays were corroborated by immunofluorescence results demonstrating cytoplasmic translocation of E1 on its interaction with P56 (Figure 3C).
The reciprocal mapping of the E1 domain that interacted with P56 was less complete. The original yeast two-hybrid assay revealed that it was the C-terminal region of E1, containing residues 360–657, that interacted with P56. This conclusion was confirmed by their co-immunoprecipitation from mammalian cell extracts. However, further deletions, from either end, eliminated P56 interaction. Future analysis should introduce internal mutations to alter specific structural and functional motifs present in this region of E1. This region is required for the ATPase and the helicase activities of E1 but not for its DNA binding or nuclear localization (White et al, 2001); the three-dimensional structure of most of this region is known (Abbate et al, 2004; Auster and Joshua-Tor, 2004). It is interesting to note that binding of P56 to this region blocked one enzymatic function of E1 (helicase), but not the other (ATPase). Moreover, the same binding caused the over-riding function of the nuclear localization signals present in the N-terminal region of the protein (Deng et al, 2004). Future structural analysis of a complex between the P56 N-terminal region with E1 will be illuminating in explaining the observed functional consequences.
Our results demonstrated two general modes of action of P56 on E1 functions. In cells, P56 binding caused cytoplasmic translocation of E1, thus removing it from its site of action in the nucleus. In vitro, direct binding of the two proteins impaired the helicase activity of E1 and hence its ability to support DNA replication in vitro. Our experiment with the NLSP56 mutant indicates that the functional inhibition observed in vitro was also the primary mechanism in vivo. In an infected cell, either effect should cause inhibition of HPV episomal DNA replication and hence, with progressive cell divisions, dilution and eventual loss of the viral DNA. Although in most cases such a loss of HPV episomal DNA should be beneficial to the host, a recent report indicates that the effect may not be so predictable (Herdman et al, 2006). In this study, the authors used HPV16-infected cervical keratinocytes to examine the fate of viral DNA after IFN treatment. They observed that such treatments enriched the population with cells in which the viral DNA was integrated, an effect with undesirable clinical consequences. It will be interesting to examine whether ectopic expression of P56, in the absence of hundreds of other IFN-induced proteins, will have the same effect. Because cell delivery of peptides is highly efficient, it will be worthwhile to derive an E1-binding peptide from the N-terminal region of P56 that can block the actions of E1. Such a peptide may be a better antiviral agent than IFN because of its targeted action avoiding the undesirable side effects of IFN treatment.
Materials and methods
Cells and viruses
C33A (human cervical carcinoma), HEK293 (human embryonic kidney) and HT1080 (human fibrosarcoma) cell lines were obtained from ATCC. P2.1 cell line has been described earlier (Leaman et al, 1998). All cell lines were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum and antibiotics. Spodoptera frugiperda (Sf21) cells were obtained from ATCC and were maintained in Graces insect cell culture medium with 10% fetal bovine serum at 28°C.
Generation of C33A/TLR3 cell line
Flag-TLR3 plasmid (5 μg) was co-transfected into C33A cells with 0.5 μg of pBABE Puro plasmid using Fugene 6 (Roche) according to the manufacturer's protocol. Cells were selected in DMEM containing 800 μg/ml G418 and 1 μg/ml puromycin. Individual colonies were screened for P56 induction by 100 μg/ml dsRNA [poly (I: C)] (Pharmacia Biotech) for 6 h. Cell lysates (50 μg) were used for western blot analysis using anti-P56 antibody. Stable cell lines were propagated and maintained in complete DMEM supplemented with 800 μg/ml G418.
Yeast two-hybrid assay
Full-length P56 expressed as GAL-4-binding domain fusion protein, BD-P56, was used as the bait. A total of 1 × 107 transformants from a human HeLa cell matchmaker library (Clontech) were screened in the yeast strain Y190 (Clontech) as described before (Guo et al, 2000a). Sequence analysis of the cDNA revealed that it encoded a protein identical to the C terminus of HPV18 E1 (aa 359–657).
Plasmid constructs
Full-length and different deletion mutants of HPV18 E1 were generated by PCR amplification using HPV18 E1 as a template and cloned into pcDNA3 vector (Invitrogen) (Sverdrup and Khan, 1994). Family members of P56 gene were cloned by PCR and subcloned into pFLAG-CMV2 vector (Sigma). pcDNA3-P56, pET15b-P56 and Flag-PKR plasmids were described earlier (Guo et al, 2000b; Peters et al, 2002; Hui et al, 2005). P56 deletion mutants were generated by PCR using pcDNA3-P56 as a template and cloned into pcDNA3. N-terminal region of P56 (aa 1–179) containing point mutations described below were generated by PCR and subcloned into pcDNA3; M1 contained A71T, L75S and A78T in TPR1 (aa 1–85) (M1P56 (1–179 aa)). M2 contained A114T, L118R and V121E in TPR2 (aa 95–128) (M2P56 (1–179 aa)). M3 mutant contained A160T, F164S and V167E in the TPR3 (aa 141–174) (M3P56 (1–179 aa)). M1, 3 is a combination of M1 and M3 mutations above in TPR1–3 (aa 1–179) (M1, 3P56 (1–179 aa)). M2P56, containing the M2 mutations in the context of full-length P56, was subcloned into pcDNA3 vector. M2 in the context of full-length or N-terminal domain (aa 1–179) was also cloned in pET15b vector (Novagen), and N-terminal domain of P56 (aa 1–179) was subcloned into pGEX4T1 vector (as GST-fused protein) (Amersham Biosciences). The proteins were expressed and purified according to the manufacturer's protocols. All constructs were confirmed by DNA sequencing.
shRNA constructs and lentivirus infection
A PCR-based strategy (PCR SHAGging) for generating RNA polymerase III (U6 snRNA promoter)-driven construct expressing 29-bp shRNA was used as described before (Di Nardo et al, 2005). Two shRNA expression cassettes were generated that targeted P56 mRNA at nt 12–41 (shRNA1) and nt 1443–1472 (shRNA2), respectively; each cassette was driven by the U6 snRNA promoter. The shRNA1 (siP56) was subcloned into pcDNA3 and used in transient replication assay. Similarly, a scrambled shRNA construct (siControl) against P56 mRNA at nt 12–41 was generated; a shRNA for TLR5 (siTLR5) was also used as a control. The two cassettes were also subcloned in tandem into lentivirus vector pLV-noCMV containing G418 resistance gene (LVsiP56). Lentivirus plasmid encoding siRNA for luciferase was used as a control. To express P56 protein in the nucleus, the SV40 nuclear localization signal sequence (5′-CCGAAGAAGAAAAGGAAGAAGGTG-3′) was fused to 5′ P56 ORF by PCR and cloned into the pLVpuro Flag sequence containing vector (NLSP56) at XhoI and BamHI sites. Recombinant virus was used to infect C33A cells using 4 μg/ml of Polybrene; a pLV-CMV-EGFP plasmid was used as a control for efficiency of infection. The cells were selected in 800 μg/ml G418 and used in replication assay.
Antibodies
Rabbit polyclonal antibody against human P56 protein has been described earlier (Guo et al, 2000b) and used at 1:2000. c-Myc 9E10 monoclonal antibody (used at 1:1000) and rabbit antibody HisG-18 (used at 1:660) were purchased from Santa Cruz Biotechnology. Polyclonal antibody against MBP (NEB) was used at 1:10 000. The anti-FLAG-M2 monoclonal antibody and anti-FLAG-M2 HRP-conjugated antibody (Sigma) were used at a 1:2000 dilution. Antibody to actin was from Sigma (1:1000).
Immunoprecipitation and western blot
HT1080 cells were transfected with 4 μg of appropriate plasmids by FUGENE 6, and cell lysates were prepared as described (Leonard and Sen, 1997). Immunoprecipitation conditions were as described before (Terenzi et al, 2005).
In vitro interaction of HPV E1 with P56 protein
Purified His–E1 was mixed with purified Flag–P56 (18 pmol of each) and incubated with anti-FLAG-M2 agarose beads for 2 h at 4°C. The bound proteins were washed with RIPA buffer and analysed by 10% SDS–PAGE and western blot with anti-His antibody.
In vitro translation assay
Luciferase mRNA (0.5 μg; Promega) was translated in the presence of P56 or M2P56 proteins in 25 μl reactions using the rabbit reticulocyte lysate system (Promega). After 2 h at 30°C, newly synthesized 35S-labelled protein was analysed by loading 5 μl of reaction on 10% SDS–PAGE and quantitated by Phosphorimager using the molecular dynamics ImageQuant software.
ATPase assay
ATPase activity of E1 was assayed spectrophotometrically by measuring the absorbance between 620 and 660 nm resulting from coloured reaction of malachite green with free phosphate. In a 25 μl reaction mixture, containing 50 mM Tris–Cl, pH 8.0, 10 mM MgCl2, 1 mM DTT, 500 μM ATP was incubated with purified E1 protein. After 2 h at 37°C, 100 μl malachite green solution (Upstate) was added to the reaction and allowed to remain at room temperature for 15 min. The absorbance was measured in a spectrophotometer at a wavelength mentioned above.
Helicase assay
Helicase activity was detected by the release of a 32P-labelled oligonucleotide annealed to M13mp18 single-stranded (ss) DNA (White et al, 2001) . The partial double-stranded DNA was purified through a G-25 spun column (Pharmacia Biotech) to remove unannealed oligonucleotide. The helicase reaction mixture contained 5 nM substrate in 20 mM Tris–Cl, 2 mM DTT, 0.1 mg/ml BSA, 5 mM MgCl2, 5% glycerol, 5 mM ATP, and the solution was adjusted to pH 7.5. The reaction was incubated at 37°C for 2 h and stopped with 0.4% SDS, 20 mM EDTA, 5% glycerol and 0.03% bromophenol blue. The substrate and products were electrophoretically separated on a 6% non-denaturing polyacrylamide gel that was dried under vacuum.
Methods for transient DNA replication assay, preparation of cell extracts for DNA replication, immunofluoresence and expression and purification of recombinant proteins from insect cells and bacteria are described in the Supplementary data.
Supplementary Material
Supplementary Information
Acknowledgments
We thank Jinjiao Guo for initiating this project and Louise Chow for carrying out the initial in vitro DNA replication assays. We are indebted to Saleem Khan, Louise Chow, Joseph DiDonato, Andrei Gudkov and Peter Chumakov for sharing important reagents. This investigation was supported by National Institutes of Health grants CA068782 and CA062220.
References
- Abbate EA, Berger JM, Botchan MR (2004) The X-ray structure of the papillomavirus helicase in complex with its molecular matchmaker E2. Genes Dev 18: 1981–1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano K, Merrick WC, Hershey JW (1997) The translation initiation factor eIF3-p48 subunit is encoded by int-6, a site of frequent integration by the mouse mammary tumor virus genome. J Biol Chem 272: 23477–23480 [DOI] [PubMed] [Google Scholar]
- Auster AS, Joshua-Tor L (2004) The DNA-binding domain of human papillomavirus type 18 E1. Crystal structure, dimerization, and DNA binding. J Biol Chem 279: 3733–3742 [DOI] [PubMed] [Google Scholar]
- Chang YE, Laimins LA (2001) Interferon-inducible genes are major targets of human papillomavirus type 31: insights from microarray analysis. Dis Markers 17: 139–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YE, Pena L, Sen GC, Park JK, Laimins LA (2002) Long-term effect of interferon on keratinocytes that maintain human papillomavirus type 31. J Virol 76: 8864–8874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Stenlund A (2002) Sequential and ordered assembly of E1 initiator complexes on the papillomavirus origin of DNA replication generates progressive structural changes related to melting. Mol Cell Biol 22: 7712–7720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow LT, Broker TR (1997) Small DNA tumor viruses. In Viral Pathogenesis, Nathanson N (ed), pp 267–302. Philadelphia: Lippincott-Raven [Google Scholar]
- Deng W, Lin BY, Jin G, Wheeler CG, Ma T, Harper JW, Broker TR, Chow LT (2004) Cyclin/CDK regulates the nucleocytoplasmic localization of the human papillomavirus E1 DNA helicase. J Virol 78: 13954–13965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Der SD, Zhou A, Williams BR, Silverman RH (1998) Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligonucleotide arrays. Proc Natl Acad Sci USA 95: 15623–15628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Nardo A, Cicchetti G, Falet H, Hartwig JH, Stossel TP, Kwiatkowski DJ (2005) Arp2/3 complex-deficient mouse fibroblasts are viable and have normal leading-edge actin structure and function. Proc Natl Acad Sci USA 102: 16263–16268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dollard SC, Wilson JL, Demeter LM, Bonnez W, Reichman RC, Broker TR, Chow LT (1992) Production of human papillomavirus and modulation of the infectious program in epithelial raft cultures. OFF. Genes Dev 6: 1131–1142 [DOI] [PubMed] [Google Scholar]
- Elco CP, Guenther JM, Williams BR, Sen GC (2005) Analysis of genes induced by Sendai virus infection of mutant cell lines reveals essential roles of interferon regulatory factor 3, NF-kappaB, and interferon but not toll-like receptor 3. J Virol 79: 3920–3929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiss G, Jin G, Guo J, Bumgarner R, Katze MG, Sen GC (2001) A comprehensive view of regulation of gene expression by double-stranded RNA-mediated cell signaling. J Biol Chem 276: 30178–30182 [DOI] [PubMed] [Google Scholar]
- Grandvaux N, Servant MJ, tenOever B, Sen GC, Balachandran S, Barber GN, Lin R, Hiscott J (2002) Transcriptional profiling of interferon regulatory factor 3 target genes: direct involvement in the regulation of interferon-stimulated genes. J Virol 76: 5532–5539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, Hui DJ, Merrick WC, Sen GC (2000a) A new pathway of translational regulation mediated by eukaryotic initiation factor 3. EMBO J 19: 6891–6899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, Peters KL, Sen GC (2000b) Induction of the human protein P56 by interferon, double-stranded RNA, or virus infection. Virology 267: 209–219 [DOI] [PubMed] [Google Scholar]
- Guo J, Sen GC (2000) Characterization of the interaction between the interferon-induced protein P56 and the Int6 protein encoded by a locus of insertion of the mouse mammary tumor virus. J Virol 74: 1892–1899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haller O, Frese M, Kochs G (1998) Mx proteins: mediators of innate resistance to RNA viruses. Rev Sci Tech 17: 220–230 [DOI] [PubMed] [Google Scholar]
- Herdman MT, Pett MR, Roberts I, Alazawi WO, Teschendorff AE, Zhang XY, Stanley MA, Coleman N (2006) Interferon-beta treatment of cervical keratinocytes naturally infected with human papillomavirus 16 episomes promotes rapid reduction in episome numbers and emergence of latent integrants. Carcinogenesis 27: 2341–2353 [DOI] [PubMed] [Google Scholar]
- Hirt B (1967) Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol 26: 365–369 [DOI] [PubMed] [Google Scholar]
- Howley PM, Lowy DR (2001) Papillomaviruses and their replication. In Fields Virology, Knipe DM, Howley PM (eds), pp 2197–2229. Philadelphia: Lippincott, Williams & Wilkins [Google Scholar]
- Hughes FJ, Romanos MA (1993) E1 protein of human papillomavirus is a DNA helicase/ATPase. Nucleic Acids Res 21: 5817–5823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui DJ, Bhasker CR, Merrick WC, Sen GC (2003) Viral stress-inducible protein p56 inhibits translation by blocking the interaction of eIF3 with the ternary complex eIF2.GTP.Met-tRNAi. J Biol Chem 278: 39477–39482 [DOI] [PubMed] [Google Scholar]
- Hui DJ, Terenzi F, Merrick WC, Sen GC (2005) Mouse p56 blocks a distinct function of eukaryotic initiation factor 3 in translation initiation. J Biol Chem 280: 3433–3440 [DOI] [PubMed] [Google Scholar]
- Khan MA, Tolleson WH, Gangemi JD, Pirisi L (1993) Inhibition of growth, transformation, and expression of human papillomavirus type 16 E7 in human keratinocytes by alpha interferons. J Virol 67: 3396–3403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koromilas AE, Li S, Matlashewski G (2001) Control of interferon signaling in human papillomavirus infection. Cytokine Growth Factor Rev 12: 157–170 [DOI] [PubMed] [Google Scholar]
- Kuo SR, Liu JS, Broker TR, Chow LT (1994) Cell-free replication of the human papillomavirus DNA with homologous viral E1 and E2 proteins and human cell extracts. J Biol Chem 269: 24058–24065 [PubMed] [Google Scholar]
- Lamb JR, Tugendreich S, Hieter P (1995) Tetratrico peptide repeat interactions: to TPR or not to TPR? Trends Biochem Sci 20: 257–259 [DOI] [PubMed] [Google Scholar]
- Leaman DW, Salvekar A, Patel R, Sen GC, Stark GR (1998) A mutant cell line defective in response to double-stranded RNA and in regulating basal expression of interferon-stimulated genes. Proc Natl Acad Sci USA 95: 9442–9447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard GT, Sen GC (1997) Restoration of interferon responses of adenovirus E1A-expressing HT1080 cell lines by overexpression of p48 protein. J Virol 71: 5095–5101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JJ, Kelly TJ (1984) Simian virus 40 DNA replication in vitro. Proc Natl Acad Sci USA 81: 6973–6977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin OS, Keeffe EB (2001) Current treatment strategies for chronic hepatitis B and C. Annu Rev Med 52: 29–49 [DOI] [PubMed] [Google Scholar]
- Lowy DR, Howley PM (2001) Pappilomaviruses. In Fields Virology, Knipe DM, Howley PM (eds), pp 2231–2264. Philadelphia: Lippincot-Raven [Google Scholar]
- Lowy DR, Kirnbauer R, Schiller JT (1994) Genital human papillomavirus infection. Proc Natl Acad Sci USA 91: 2436–2440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchetti A, Buttitta F, Miyazaki S, Gallahan D, Smith GH, Callahan R (1995) Int-6, a highly conserved, widely expressed gene, is mutated by mouse mammary tumor virus in mammary preneoplasia. J Virol 69: 1932–1938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters GA, Khoo D, Mohr I, Sen GC (2002) Inhibition of PACT-mediated activation of PKR by the herpes simplex virus type 1 Us11 protein. J Virol 76: 11054–11064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pett M, Coleman N (2007) Integration of high-risk human papillomavirus: a key event in cervical carcinogenesis? J Pathol 212: 356–367 [DOI] [PubMed] [Google Scholar]
- Samuel CE (2001) Antiviral actions of interferons. Clin Microbiol Rev 14: 778–809, table of contents [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar SN, Sen GC (2004) Novel functions of proteins encoded by viral stress-inducible genes. Pharmacol Ther 103: 245–259 [DOI] [PubMed] [Google Scholar]
- Schuck S, Stenlund A (2005) Role of papillomavirus E1 initiator dimerization in DNA replication. J Virol 79: 8661–8664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedman T, Sedman J, Stenlund A (1997) Binding of the E1 and E2 proteins to the origin of replication of bovine papillomavirus. J Virol 71: 2887–2896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen GC (2001) Viruses and interferons. Annu Rev Microbiol 55: 255–281 [DOI] [PubMed] [Google Scholar]
- Sen GC, Peters GA (2007) Viral stress-inducible genes. Adv Virus Res 70: 233–263 [DOI] [PubMed] [Google Scholar]
- Stanley MA, Pett MR, Coleman N (2007) HPV: from infection to cancer. Biochem Soc Trans 35 (Part 6): 1456–1460 [DOI] [PubMed] [Google Scholar]
- Stark GR, Kerr IM, Williams BR, Silverman RH, Schreiber RD (1998) How cells respond to interferons. Annu Rev Biochem 67: 227–264 [DOI] [PubMed] [Google Scholar]
- Sun Y, Han H, McCance DJ (1998) Active domains of human papillomavirus type 11 E1 protein for origin replication. J Gen Virol 79 (Part 7): 1651–1658 [DOI] [PubMed] [Google Scholar]
- Sverdrup F, Khan SA (1994) Replication of human papillomavirus (HPV) DNAs supported by the HPV type 18 E1 and E2 proteins. J Virol 68: 505–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terenzi F, Pal S, Sen GC (2005) Induction and mode of action of the viral stress-inducible murine proteins, P56 and P54. Virology 340: 116–124 [DOI] [PubMed] [Google Scholar]
- Turek LP, Byrne JC, Lowy DR, Dvoretzky I, Friedman RM, Howley PM (1982) Interferon induces morphologic reversion with elimination of extrachromosomal viral genomes in bovine papillomavirus-transformed mouse cells. Proc Natl Acad Sci USA 79: 7914–7918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wacher C, Muller M, Hofer MJ, Getts DR, Zabaras R, Ousman SS, Terenzi F, Sen GC, King NJ, Campbell IL (2007) Coordinated regulation and widespread cellular expression of interferon-stimulated genes (ISG) ISG-49, ISG-54, and ISG-56 in the central nervous system after infection with distinct viruses. J Virol 81: 860–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Pflugheber J, Sumpter R Jr, Sodora DL, Hui D, Sen GC, Gale M Jr (2003) Alpha interferon induces distinct translational control programs to suppress hepatitis C virus RNA replication. J Virol 77: 3898–3912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White PW, Pelletier A, Brault K, Titolo S, Welchner E, Thauvette L, Fazekas M, Cordingley MG, Archambault J (2001) Characterization of recombinant HPV6 and 11 E1 helicases: effect of ATP on the interaction of E1 with E2 and mapping of a minimal helicase domain. J Biol Chem 276: 22426–22438 [DOI] [PubMed] [Google Scholar]
- Whitley RJ (2001) Herpes simplex viruses. In Field Virology, Knipe DM, Howley PM (eds), pp 2461–2509. Philadelphia: Lippincott, Williams & Wilkins [Google Scholar]
- zur Hausen H (1996) Papillomavirus infections—a major cause of human cancers. Biochim Biophys Acta 1288: F55–F78 [DOI] [PubMed] [Google Scholar]
- zur Hausen H, de Villiers EM (1994) Human papillomaviruses. Annu Rev Microbiol 48: 427–447 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information