Abstract
From invertebrates to mammals, cell-cycle progression during an asymmetric cell division is accompanied by precisely timed redistribution of cell-fate determinants so that they segregate asymmetrically to enable the two daughter cells to choose different fates. Interestingly, studies on how cell fates are specified in such divisions reveal that the same fate determinants can be reiteratively used to specify a variety of cell types through multiple rounds of cell divisions or to exert seemingly contradictory effects on cell proliferation and differentiation. Here I summarize the molecular mechanisms governing asymmetric cell division and review recent findings pointing to a novel mechanism for coupling intracellular signaling and cell-cycle progression. This mechanism uses changes in the morphology, subcellular distribution and molecular composition of cellular organelles like the Golgi apparatus and centrosomes, which also accompany the progression of cell cycle, to activate but also temporally constrain the activity of fate determinants during asymmetric cell divisions.
Findings from decades of research on how cell fates are specified during development show that a given fate determinant is often capable of specifying multiple cell types and affecting seemingly contradictory aspects of cellular differentiation. A major challenge in developmental biology is to understand how the activities of such fate determinants are regulated, in particular how the cellular context – the presence and absence of other regulators – affects their ability to exert their influences. This review is not intended as a comprehensive overview of the known mechanisms of cell-fate determination but focuses on those involved in intrinsically asymmetric divisions by precursor cells in Drosophila and mice, particularly during neurogenesis. I summarize the mechanisms of asymmetric cell division and discuss recent findings that point to the potential use of cellular organelles like the Golgi and centrosomes to couple cell-cycle progression and cell-fate determination to allow for the reiterative use of fate determinants to specify multiple cell types or influence different aspects of cellular differentiation.
Patterns of cell division during neurogenesis
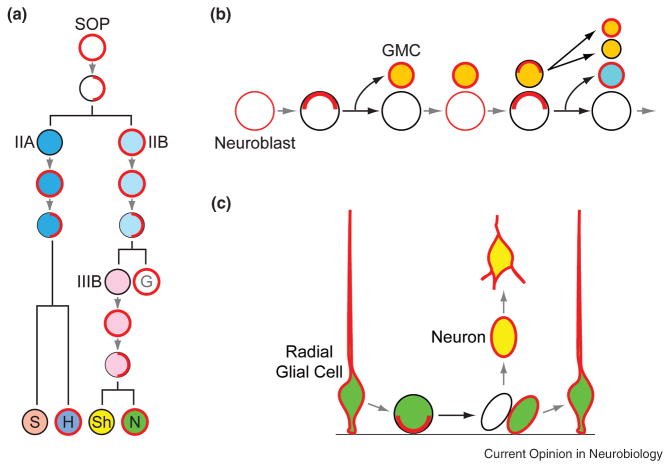
Intrinsically asymmetric cell division is a process that produces two daughter cells that are already different at birth. Such divisions are used extensively during Drosophila neurogenesis to produce the wide variety of neurons and glia necessary for building a functional nervous system. In the periphery nervous system (PNS), various sensory organs are produced through stereotypical divisions by single precursor cells [1]. An external sensory (ES) organ, for example, consists of four cells: a neuron, a sheath cell, a hair and a socket cell. The four cells are descendants of a single sensory organ precursor (SOP) cell through three rounds of asymmetric cell divisions (Figure 1a). The SOP cell first divides to produce a IIA and a IIB cell. The IIA cell then divides to generate a neuron and a sheath cell, whereas the IIB divides to generate a glial cell, which either moves away or dies, and a IIIB cell, which further divides to produce a neuron and a sheath cell. The precursor cells for other sensory organs, including the chordotonal organs, undergo similar patterns of divisions but generate different cell types.
Figure 1.
Reiterative use and diverse roles of the cell-fate determinant Numb during asymmetric cell divisions. (a) Drosophila SOP cells. (b) Drosophila neuroblasts. (c) Mammalian radial glial cells. G, glial cell; H, hair cell; N, neuron; S, socket cell; Sh, sheath cell.
In the Drosophila central nervous system (CNS), neurons and glia arise from precursors called neuroblasts. In Drosophila embryos, there are about 30 individually identifiable, segmentally reiterated neuroblasts, each divides repeatedly like a stem cell to generate a larger daughter cell that retains the neuroblast characteristics and bud off a smaller ganglion mother cell (GMC), which divides once, often asymmetrically, to generate two differentiated neurons and/or glia (Figure 1b). Each neuroblast has an invariant lineage tree, through which the origins of virtually all cell types in the embryonic CNS can be traced [2,3] (Figure 1B). Neurons and glia in the adult Drosophila brain are generated by neuroblasts in the larva. Larval neuroblasts generally divide in ways similar to that of their counterparts in the embryos, but their capacity to divide is considerably larger due to a need for producing much large numbers of neurons and glia. Moreover, unlike embryonic neuroblasts, which show similar division patterns and become smaller with each successive divisions, larval neuroblasts grow to their original size after each division and exhibit differences in division patterns. For example, the smaller daughter cell produced by neuroblasts of the dorsomedial lineage, instead of dividing only once like the GMC daughters in other lineages, also divide repeatedly [4].
While it remains unclear whether precursors in the developing mammalian nervous system generally undergo stereotypical divisions like their invertebrate counterparts, direct imaging experiments show that progenitor cells in the developing neocortex at least mostly divide asymmetrically. Radial glial cells (RGCs), which are the major population of cortical progenitor cells, undergo two types asymmetric divisions during cortical neurogenesis [5•,6•]. In type-I divisions, a RGC divides to produce another RGC (self-renew) and a daughter cell that becomes a neuron (Figure 1c). In type-II divisions, a RGC divides in a way resembling that of the Drosophila neuroblasts by generating two daughter cells that both re-enter cell cycle, but one remains as a RGC whereas the other divides only once to generate two neurons (Figure 1c).
Mechanisms of asymmetric cell division
Studies using C. elegans and Drosophila show that generating two intrinsically different daughter cells requires at least three steps: a cell must first polarize itself so that one side of the cell is different from the other, followed by localizing various cell-fate determinants to only one side of the cell and aligning the mitotic spindle along the axis of cell polarity so that the localized fate determinants are segregated primarily to one of the two daughter cells. The molecules involved and their respective roles in establishing cell polarity and segregating cell-fate determinants have been extensively reviewed recently [7–9] and are briefly summarized below.
Cell polarity is established and maintained through the polarized distribution of two evolutionarily conserved protein complexes. One complex includes the Par proteins (Par3 and Par6) and atypical protein kinase C (aPKC), whereas the other comprises a protein cassette related to heterotrimeric G protein signaling that includes Gαi and Partner of inscuteable (Pins), which is a receptor-independent regulator of Gαi, as well as Inscuteable (Insc) and Discs-large (Dlg). The polarity of embryonic neuroblasts, which delaminate from the neural ectoderm, is aligned along the apical-basal axis. The Par proteins, which are already apically localized in the neural ectodermal cells, recruit Insc and the G protein cassette to the apical cortex during neuroblast delamination. Subsequently, the Pax3/Par6/aPKC complex, through mechanisms that remain to be fully delineated but involve the tumor suppressor proteins Dlg and Lethal giant larvae (Lgl), helps to localize cell-fate determinants such as Numb (see more detailed discussion below), Prospero (Pros) and Brat to the opposite, basal cell cortex. The G protein related cassette, on the other hand, orients the mitotic spindle, at least in part through the Pins protein, which serves as a conformational switch linking Gαi and Mud. The latter is a Drosophila homologue of the microtubule and dynein binding protein NuMA, pointing to a model by which the apical complexes attract one of the spindle poles to orient the mitotic spindle along the apical-basal axis to ensure asymmetric segregation of the basally localized fate determinants [7–9].
In the Drosophila PNS, the polarity of SOP cells are aligned along the anterior-posterior body axis (planar polarity). SOP cells become polarized by responding to a planar polarity cue provided by the Wnt receptor Frizzled [10] and by localizing Gαi, Pins and Dlg to the anterior cell cortex [11,12]. The anterior Gαi/Pins/Dlg complex then directs the positioning of the Par3/Par6/aPKC complex to the posterior cortex, which in turns directs the accumulation of cell-fate determinants to the opposite (anterior) side of the cells. In other words, while the same players are involved in the critical steps of asymmetric cell division, different precursor cells use them differently. A good example is how the size difference between the two daughters is determined [11,13]. The two SOP daughters, IIA and IIB, are of similar size, whereas the GMC daughter of a neuroblast is considerably smaller than its sibling. Insc, an adaptor protein, helps neuroblasts to generate two differently sized daughter cells by binding to Pins through the GoLoco motif and recruiting Gαi/Pins complex to the Par3/Par6/aPKC complex [13]. Placing the two complexes on the same side of the neuroblasts results in an asymmetric mitotic spindle and, consequently, a cell cleavage plane that divides the cells unequally. Insc, however, is not expressed in SOP cells, and the Gαi/Pins/Dlg and Par3/Par6/aPKC complexes are localized to the opposite sides of the cell. This absence of Insc is important for SOP cells to generate a symmetric mitotic spindle and two similarly sized daughter cells, since Insc misexpression in SOP cells can lead to co-localization of the two complexes in one pole and asymmetry in daughter cell sizes [11].
Not surprisingly, the molecular components that regulate asymmetric cell division in C. elegans and Drosophila, including regulators of cell polarity like the Par proteins, aPKC, Insc, Pins and the components of G protein signaling as well as cell fate determinants like Numb, Pros and Brat, have highly conserved homologues in mammals. Like their invertebrate counterparts, the mammalian proteins show polarized distribution in mammalian neural progenitor cells, and perturbing their activities leads to defects in cell polarity, mitotic spindle orientation and daughter cell fates. The findings have been recently reviewed [7–9] and are not detailed here.
Reiterative use and diverse roles of cell-fate determinants
As summarized above, the key regulators of asymmetric cell division are evolutionarily conserved and used by precursors from C. elegans to mammals to establish cell polarity and orient mitotic spindle. Interestingly, although the precursor cells that divide asymmetrically, both in- and outside of the nervous system, often share no apparent class or lineage identities, many nevertheless use a common set of cell-fate determinants to distinguish their daughter cells. Numb, for example, is a cytosolic signaling protein that is widely expressed during embryogenesis and is segregated asymmetrically by a wide variety of neural and non-neural precursor cells to allow the two daughter cells to adopt distinct fates. In the CNS, neuroblasts are different from one another and bud off GMCs that produce morphologically and functionally distinct neurons and glia, and Numb can promote motor neuron over interneuron fates in some divisions but specify one interneuron fate over another in others [14,15]. Outside of the nervous system, asymmetric Numb segregation is required for specifying different founder progenitor cells for various muscles as well as different cell types in the Drosophila Malpighian tubules (kidney) [16,17]. It is believed that Numb does not specify a particular fate but simply enables the two daughter cells to choose differently between two fate options. It has been further suggested that Numb does this by inhibiting Notch activity to cause a bias in Notch mediated cell-cell communication, either between the two daughter cells or between the daughter cells and their environment, through its asymmetric presence [18,19].
That Numb simply acts as a binary switch also raises an interesting question. In the SOP lineage, for example, Numb is used reiteratively in successive asymmetric divisions [1,20] (Figure 1a). It is symmetrically distributed during interphase but becomes localized to only one half of the cell membrane after the SOP cell enters mitosis and is segregated primarily into the IIB cell. This asymmetric Numb presence is essential; in the absence of Numb, both daughter adopt the IIA fate, whereas its symmetric inheritance produces two IIB cells. Interestingly, the IIA cell, which divides to produce a hair and a socket cell, also uses Numb to distinguish its two daughter cells. Even though the IIA fate requires Numb to be absent initially, newly synthesized Numb quickly accumulates in IIA cells, which then segregate the protein asymmetrically to promote the hair cell fate. In other words, Numb appears to lose its ability to specify cell fates shortly after an asymmetric division but regains the ability to do so when the cells divide again.
The mammalian Numb homologues are encoded by two genes, m-numb (Numb) and numblike (Numbl) [21–24]. The two genes are functionally redundant during mouse neurogenesis and are expressed in both neural progenitor cells and neurons [24,25]. They are essential for neurogenesis but play two seemingly contradictory roles in this process. When neural progenitor cells divide asymmetrically to self-renew and produce a neuron, Numb proteins segregate asymmetrically to promote progenitor over neuronal fates [25,26]. After the division, however, they are required for the differentiation of newborn neurons [25,27]. In other words, the ability of mammalian Numb proteins to specify cell fates may also be limited to during and/or shortly after mitosis.
Golgi Fragmentation and Numb signaling
That cells whose fates depend on an initial Numb absence subsequently use Numb to specify the fates of their daughter cells or promote their own differentiation raises an interesting possibility that Numb activity in cell-fate specification is tightly regulated and coupled to cell-cycle progression. Our recent study points to a novel mechanism that uses the process of Golgi fragmentation and reconstitution during cell cycle to differentially regulate Numb signaling by changing the subcellular distribution of an essential partner of Numb, ACBD3, thereby allowing Numb signaling to be activated only at times when the two daughter cells are being generated and be deactivated shortly after they are born [28••].
Cell-cycle progression in mammalian cells is accompanied by dramatic changes in the morphology of the Golgi apparatus, an essential organelle that plays a variety of roles, including post-translational modification and trafficking of proteins. Mammalian Golgi membranes are organized into interconnected stacks of flattened cisternae during interphase and confined to the region surrounding the centrioles. When cells enter mitosis, however, they fragment the pericentriolar Golgi stacks and disperse the Golgi fragments throughout the cytosol to ensure that both daughter cells inherit this essential organelle, although there is debate as to whether the Golgi simply disappears and is regenerated after the division from the endoplasmic reticulum (ER) [29–31].
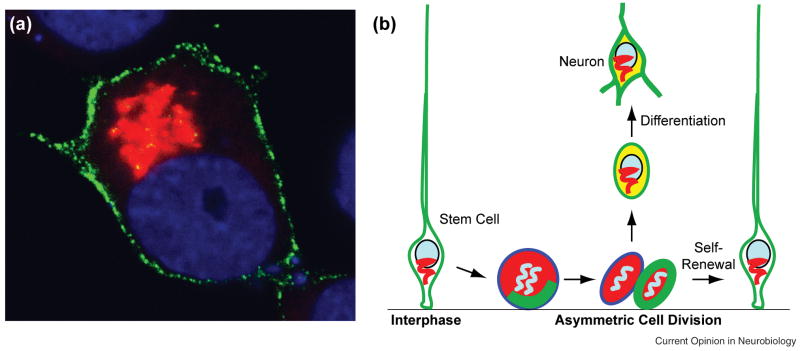
ACBD3 binds to Numb through a functionally essential Numb domain and can act synergistically with Numb to specify cell fates. Like mammalian Numb proteins, ACBD3 is widely expressed and present in both neural progenitor cells and neurons. However, unlike Numb and Numbl, which are cytosolic proteins, ACBD3 associates with the Golgi in postmitotic neurons and interphase progenitor cells (Figure 2a). After the Golgi fragments during mitosis, however, ACBD3 is released into the cytosol. In other words, the two partners, Numb and ACBD3, may only be able to interact with each other during mitosis, when Numb activity is needed to distinguish the daughter cells, and the ability of Numb to specify cell fates is quickly lost after the division as ACBD3 goes back to the Golgi, thereby allowing Numb to promote differentiation of newborn neurons by tapping into a different pathway or to wait for reactivation until the next division (Figure 2b). This model is supported by experiments using a myristoylated form of ACBD3, which remains in the cytosol throughout the cell cycle. As expected, this cytosolic form of ACBD3 can specify cell fates in the presence of Numb, and, more importantly, its misexpression in neural progenitor cells during mouse neurogenesis inhibits neuron production, likely by forcing both daughter cells to choose progenitor over neuronal fates [28].
Figure 2.
A Model for differentially regulating Numb signaling during mammalian neurogenesis through Golgi fragmentation and reconstitution. (a) The subcellular distribution of the exogenous, GFP-tagged mouse Numb protein (in green) and the endogenous ACBD3 (in red) in interphase NIE115 cells, a mouse neuroblastoma cell line. (b) Numb proteins promote self-renewal when neural stem (progenitor) cells divide asymmetrically to self-renew and produce a neuron, but this occurs only when ACBD3 (in red) is present in the cytosol after Golgi fragmentation during mitosis. Newly synthesized Numb proteins subsequently accumulate in newborn neurons but, with ACBD3 localized to the Golgi, tap into a different pathway to promote neuronal differentiation. Stem cells in many tissues may use this mechanism to balance self-renewal and differentiation.
It is worth noting that ACBD3 release into the cytosol likely is not simply a byproduct of Golgi fragmentation but rather a precisely regulated process. Golgi fragmentation is tightly coupled with cell-cycle progression. It occurs before cells enter prophase and is actually essential for entrance into mitosis [32]. ACBD3 associates with the cytoplasmic face of Golgi membranes and remains associated with fragmented Golgi fragments (blobs) until late metaphase, when it becomes mostly cytosolic [28]. Since the Golgi also further fragments and releases some of its contents between pro-metaphase and early anaphase [30], it will be interesting to determine whether and how the regulators of Golgi fragmentation and reconstitution affect ACBD3 distribution.
Cell-cycle regulators and centrosomes in asymmetric cell division
It is unclear whether and how this Golgi-based mechanism is conserved evolutionarily. In Drosophila embryos, the Golgi exists as widely dispersed units in the cytoplasm and does not further fragment during cell cycle [33]. ACBD3, however, has a highly conserved Drosophila homologue, and mammalian Numb and ACBD3 proteins can function synergistically to specify Numb-dependent fates in Drosophila. Therefore, at least some aspects of this novel mechanism are likely to be evolutionarily conserved. Moreover, since the steps of asymmetric cell division, including precisely timed redistribution of cell-fate determinants, are intimately associated with the phases of cell cycle, it is likely that the Golgi apparatus is not the only organelle used by cells to activate and also temporally constrain the activities of fate determinants. There is evidence that this may indeed be the case.
In Drosophila, Numb, Pros and Brat are symmetrically distributed in interphase cells but becomes asymmetrically localized by metaphase. Their asymmetric localization is mediated by two adaptor proteins, Partner of Numb (Pon), which binds to Numb, and Miranda, which bind to Pros and Brat [34–38]. Interestingly, whereas Miranda is essential for asymmetrically localizing Pros and Brat, Pon loss only delays Numb asymmetric localization, which nevertheless occurs by the time cells enter into anaphase and telophase [36]. The latter phenomenon, termed “telophase rescue”, which is observed in many mutants affecting asymmetric cell division, results from the presence of a pathway involving Kinesin heavy chain 73 (Khc-73), a plus-end-directed microtubule motor protein, and Dlg, the membrane-associated guanylate kinase (MAGUK) [39]. The Khc-73/Dlg pathway and the previously described Par3/Par6/aPKC/Insc pathway can both generate cell polarity by asymmetrically localizing Gαi/Pins but act independently and during different phases of the cell cycle. The Par3/Par6/aPKC/Insc pathway is initiated at late prophase by an unknown extrinsic cue, whereas the Khc-73/Dlg pathway is initiated later during prometaphase/metaphase by astral microtubules and is required for Gαi/Pins localization over the spindle pole (but only when Par and Insc proteins are absent).
Cell-cycle progression is driven by changes in the activity of cell-cycle regulators, and several such regulators, including Cdc2, Aurora-A and Polo, have been shown to be critical for asymmetric Numb localization [40–44]. Cdc2 is a Cyclin-dependent kinase and essential for entry into mitosis, whereas Aurora A and Polo are involved in many events during mitosis, including centrosome maturation, spindle formation and cytokinesis. Cdc2 is not required for initiating the formation of the apical complexes but is necessary for its maintenance and, consequently, the asymmetric localization of cell fate determinants [40]. Attenuating the function of Aurora-A [41••,44] or Polo [43••], while leaving many components of the polarity complexes unaffected, causes aPKC and Numb to distribute symmetrically in dividing neuroblasts and SOP cells. Most importantly, although Numb segregates symmetrically in cdc2, aurora-A and polo mutants, the phenotypes exhibited by these mutants are actually consistent with a loss of Numb function. Since Numb asymmetric localization is not essential for the ability to specify cell fates [24,45], these findings indicate that the cell-cycle regulators may also be required, directly or indirectly, for activating Numb signaling, thereby constraining Numb activity to during and shortly after mitosis. A recent study also points to another way of bringing two signaling partners together in a cell-cycle dependent manner. Bora, a nuclear protein, is required for the activation Aurora-A, which is present in the cytoplasm and also associates with the centrosomes. Interestingly, upon entry into mitosis, Bora becomes excluded from the nucleus in a Cdc2-dependent manner, pointing to a model that entrance into mitosis initiates the release of Bora into the cytoplasm to activate Aurora-A [42•]. Once Aurora-A is activated, if further triggers a phosphoryaltion cascade that at least is responsible for asymmetric Numb localization [41••]. In SOP cells, Lgl, Par-6 and aPKC form a complex at one pole. Aurora-A first phosphorylates Par-6, a regulatory subunit of aPKC, to activate aPKC, which in turn phosphorylates Lgl. A consequence of Lgl phosphorylation is the exchange of Lgl for Bazooka (the Drosophila Par-3 homologue) in the complex, and this changes the substrate specificity of aPKC, which then phosphorylates Numb and releases it from one side of cortex, thereby causing Numb to concentrate on the other side of the cell cortex.
In addition to the changing activities of cell-cycle regulators, progression through cell cycle is also accompanied by changes in the centrosome. Centrosomes are duplicated during S phase, and migration of the two centrosomes to the opposite poles of the cells during prophase and the subsequent formation of the mitotic spindle are essential for asymmetric cell division to occur by ensuring that the localized cell-fate determinants are segregated asymmetrically. Several recent studies, however, indicate that the importance of centrosomes during asymmetric cell division is not simply due to its roles in organizing the poles of the mitotic spindle; centrosomes may play a more direct role in regulating asymmetric cell division. Drosophila larval neuroblasts with extra centrosomes initially form multiple spindles. This, however, triggers the spindle assembly checkpoint, which in turn delays the progression of mitosis. Subsequently, the delay in mitosis enables the cells to either cluster the centrosomes at the two poles or inactivate those that are not clustered, and the spindles eventually become bipolar, allowing the cells to generate two daughters that maintain a stable diploid genome. Interestingly, despite the eventual presence of a bipolar mitotic spindle, some neuroblasts exhibit defects in polarized distribution of proteins involved in asymmetric cell division and divide symmetrically instead of asymmetrically, leading to tumor-like growth of neuroblasts as both daughter cells choose self-renewal over differentiation [46••]. Most intriguingly, the male germline stem cells (GSCs) in Drosophila, which divides asymmetrically to self-renew and produce a daughter cell that differentiates, segregate the two centrosomes non-randomly. Differential labeling of mother and daughter centrosomes reveal that the mother centrosome, which contains the oldest centriole, is consistently inherited by the daughter cell that will remain as a GSC [47••]. While the importance of this non-random segregation of the two centrosomes remains to be determined, it is conceivable that the two centrosomes may harbor different cell-fate determinants or their regulators that help to determine daughter cell fates [48].
Conclusions
The studies described above show that the mechanisms of asymmetric cell division –establishing and maintaining cell polarity, controlling mitotic spindle orientation and localizing cell-fate determinants – are highly conserved evolutionarily and such divisions are essential for development and for stem cells to balance self-renewal and differentiation from invertebrates to mammals. These studies also pose an interesting challenge – namely, to elucidate the molecular mechanisms that allow the same cell-fate determinants to be reiteratively used and play diverse roles. A hallmark of asymmetric cell division is that its regulation is precisely coordinated with the progression of cell cycle, which is also accompanied by a variety of other cellular changes, from the activities of cell-cycle regulators themselves to the morphology, distribution and molecular contents of organelles and other subcellular components. Numb signaling likely is not the only pathway taking advantage of Golgi fragmentation and reconstitution to couple its activation with cell-cycle progression. Therefore, further examination of how the activity of cell fate determinants are activated and constrained during asymmetric cell divisions may reveal novel mechanisms that integrate cellular signaling in a context dependent manner to regulate development and the behavior of stem cells.
Acknowledgments
I thank Panfeng Fang for the image in Figure 2a and apologize to colleagues whose works were not mentioned due to space limitation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest published in the last two years have been highlighted as:
• of special interest
• of outstanding interest
- 1.Gho M, Bellaiche Y, Schweisguth F. Revisiting the Drosophila microchaete lineage: a novel intrinsically asymmetric cell division generates a glial cell. Development. 1999;126:3573–3584. doi: 10.1242/dev.126.16.3573. [DOI] [PubMed] [Google Scholar]
- 2.Bossing T, Udolph G, Doe CQ, Technau GM. The embryonic central nervous system lineages of Drosophila melanogaster. I. Neuroblast lineages derived from the ventral half of the neuroectoderm. Dev Biol. 1996;179:41–64. doi: 10.1006/dbio.1996.0240. [DOI] [PubMed] [Google Scholar]
- 3.Schmid A, Chiba A, Doe CQ. Clonal analysis of Drosophila embryonic neuroblasts: neural cell types, axon projections and muscle targets. Development. 1999;126:4653–4689. doi: 10.1242/dev.126.21.4653. [DOI] [PubMed] [Google Scholar]
- 4.Bello BC, Izergina N, Caussinus E, Reichert H. Amplification of neural stem cell proliferation by intermediate progenitor cells in Drosophila brain development. Neural Develop. 2008;3:5. doi: 10.1186/1749-8104-3-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5•.Konno D, Shioi G, Shitamukai A, Mori A, Kiyonari H, Miyata T, Matsuzaki F. Neuroepithelial progenitors undergo LGN-dependent planar divisions to maintain self-renewability during mammalian neurogenesis. Nat Cell Biol. 2008;10:93–101. doi: 10.1038/ncb1673. [DOI] [PubMed] [Google Scholar]
- 6•.Noctor SC, Martinez-Cerdeno V, Kriegstein AR. Distinct behaviors of neural stem and progenitor cells underlie cortical neurogenesis. J Comp Neurol. 2008;508:28–44. doi: 10.1002/cne.21669. Along with Ref [5•], this paper provides a detailed analysis of the division patterns of neural progenitor cells and the relationship between the orientation of the mitotic spindle and the fates of the two daughter cells during mammalian neurogenesis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Doe CQ. Neural stem cells: balancing self-renewal with differentiation. Development. 2008;135:1575–1587. doi: 10.1242/dev.014977. [DOI] [PubMed] [Google Scholar]
- 8.Knoblich JA. Mechanisms of asymmetric stem cell division. Cell. 2008;132:583–597. doi: 10.1016/j.cell.2008.02.007. [DOI] [PubMed] [Google Scholar]
- 9.Zhong W, Chia W. Neurogenesis and asymmetric cell division. Curr Opin Neurobiol. 2008;18:4–11. doi: 10.1016/j.conb.2008.05.002. [DOI] [PubMed] [Google Scholar]
- 10.Gho M, Schweisguth F. Frizzled signalling controls orientation of asymmetric sense organ precursor cell divisions in Drosophila. Nature. 1998;393:178–181. doi: 10.1038/30265. [DOI] [PubMed] [Google Scholar]
- 11.Bellaiche Y, Radovic A, Woods DF, Hough CD, Parmentier ML, O’Kane CJ, Bryant PJ, Schweisguth F. The Partner of Inscuteable/Discs-large complex is required to establish planar polarity during asymmetric cell division in Drosophila. Cell. 2001;106:355–366. doi: 10.1016/s0092-8674(01)00444-5. [DOI] [PubMed] [Google Scholar]
- 12.Schaefer M, Petronczki M, Dorner D, Forte M, Knoblich JA. Heterotrimeric G proteins direct two modes of asymmetric cell division in the Drosophila nervous system. Cell. 2001;107:183–194. doi: 10.1016/s0092-8674(01)00521-9. [DOI] [PubMed] [Google Scholar]
- 13.Cai Y, Yu F, Lin S, Chia W, Yang X. Apical complex genes control mitotic spindle geometry and relative size of daughter cells in Drosophila neuroblast and pI asymmetric divisions. Cell. 2003;112:51–62. doi: 10.1016/s0092-8674(02)01170-4. [DOI] [PubMed] [Google Scholar]
- 14.Buescher M, Yeo SL, Udolph G, Zavortink M, Yang X, Tear G, Chia W. Binary sibling neuronal cell fate decisions in the Drosophila embryonic central nervous system are nonstochastic and require inscuteable-mediated asymmetry of ganglion mother cells. Genes Dev. 1998;12:1858–1870. doi: 10.1101/gad.12.12.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Spana EP, Kopczynski C, Goodman CS, Doe CQ. Asymmetric localization of numb autonomously determines sibling neuron identity in the Drosophila CNS. Development. 1995;121:3489–3494. doi: 10.1242/dev.121.11.3489. [DOI] [PubMed] [Google Scholar]
- 16.Ruiz Gomez M, Bate M. Segregation of myogenic lineages in Drosophila requires numb. Development. 1997;124:4857–4866. doi: 10.1242/dev.124.23.4857. [DOI] [PubMed] [Google Scholar]
- 17.Wan S, Cato AM, Skaer H. Multiple signalling pathways establish cell fate and cell number in Drosophila malpighian tubules. Dev Biol. 2000;217:153–165. doi: 10.1006/dbio.1999.9499. [DOI] [PubMed] [Google Scholar]
- 18.Guo M, Jan LY, Jan YN. Control of daughter cell fates during asymmetric division: interaction of Numb and Notch. Neuron. 1996;17:27–41. doi: 10.1016/s0896-6273(00)80278-0. [DOI] [PubMed] [Google Scholar]
- 19.Spana EP, Doe CQ. Numb antagonizes Notch signaling to specify sibling neuron cell fates. Neuron. 1996;17:21–26. doi: 10.1016/s0896-6273(00)80277-9. [DOI] [PubMed] [Google Scholar]
- 20.Rhyu MS, Jan LY, Jan YN. Asymmetric distribution of numb protein during division of the sensory organ precursor cell confers distinct fates to daughter cells. Cell. 1994;76:477–491. doi: 10.1016/0092-8674(94)90112-0. [DOI] [PubMed] [Google Scholar]
- 21.Salcini AE, Confalonieri S, Doria M, Santolini E, Tassi E, Minenkova O, Cesareni G, Pelicci PG, Di Fiore PP. Binding specificity and in vivo targets of the EH domain, a novel protein-protein interaction module. Genes Dev. 1997;11:2239–2249. doi: 10.1101/gad.11.17.2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Verdi JM, Schmandt R, Bashirullah A, Jacob S, Salvino R, Craig CG, Program AE, Lipshitz HD, McGlade CJ. Mammalian NUMB is an evolutionarily conserved signaling adapter protein that specifies cell fate. Curr Biol. 1996;6:1134–1145. doi: 10.1016/s0960-9822(02)70680-5. [DOI] [PubMed] [Google Scholar]
- 23.Zhong W, Feder JN, Jiang MM, Jan LY, Jan YN. Asymmetric localization of a mammalian numb homolog during mouse cortical neurogenesis. Neuron. 1996;17:43–53. doi: 10.1016/s0896-6273(00)80279-2. [DOI] [PubMed] [Google Scholar]
- 24.Zhong W, Jiang MM, Weinmaster G, Jan LY, Jan YN. Differential expression of mammalian Numb, Numblike and Notch1 suggests distinct roles during mouse cortical neurogenesis. Development. 1997;124:1887–1897. doi: 10.1242/dev.124.10.1887. [DOI] [PubMed] [Google Scholar]
- 25.Petersen PH, Zou K, Hwang JK, Jan YN, Zhong W. Progenitor cell maintenance requires numb and numblike during mouse neurogenesis. Nature. 2002;419:929–934. doi: 10.1038/nature01124. [DOI] [PubMed] [Google Scholar]
- 26.Petersen PH, Zou K, Krauss S, Zhong W. Continuing role for mouse Numb and Numbl in maintaining progenitor cells during cortical neurogenesis. Nat Neurosci. 2004;7:803–811. doi: 10.1038/nn1289. [DOI] [PubMed] [Google Scholar]
- 27.Huang EJ, Li H, Tang AA, Wiggins AK, Neve RL, Zhong W, Jan LY, Jan YN. Targeted deletion of numb and numblike in sensory neurons reveals their essential functions in axon arborization. Genes Dev. 2005;19:138–151. doi: 10.1101/gad.1246005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28••.Zhou Y, Atkins JB, Rompani SB, Bancescu DL, Petersen PH, Tang H, Zou K, Stewart SB, Zhong W. The mammalian Golgi regulates numb signaling in asymmetric cell division by releasing ACBD3 during mitosis. Cell. 2007;129:163–178. doi: 10.1016/j.cell.2007.02.037. This paper provides evidence for a novel mechanism in mammals that uses the process of Golgi fragmentation and reconstitution to precisely coordinate cell-cycle progression and cell-fate specification, thereby allowing a cell-fate determinant like Numb to play multiple roles during asymmetric cell divisions. [DOI] [PubMed] [Google Scholar]
- 29.Altan-Bonnet N, Sougrat R, Lippincott-Schwartz J. Molecular basis for Golgi maintenance and biogenesis. Curr Opin Cell Biol. 2004;16:364–372. doi: 10.1016/j.ceb.2004.06.011. [DOI] [PubMed] [Google Scholar]
- 30.Colanzi A, Corda D. Mitosis controls the Golgi and the Golgi controls mitosis. Curr Opin Cell Biol. 2007;19:386–393. doi: 10.1016/j.ceb.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 31.Shorter J, Warren G. Golgi architecture and inheritance. Annu Rev Cell Dev Biol. 2002;18:379–420. doi: 10.1146/annurev.cellbio.18.030602.133733. [DOI] [PubMed] [Google Scholar]
- 32.Sutterlin C, Hsu P, Mallabiabarrena A, Malhotra V. Fragmentation and dispersal of the pericentriolar Golgi complex is required for entry into mitosis in mammalian cells. Cell. 2002;109:359–369. doi: 10.1016/s0092-8674(02)00720-1. [DOI] [PubMed] [Google Scholar]
- 33.Stanley H, Botas J, Malhotra V. The mechanism of Golgi segregation during mitosis is cell type-specific. Proc Natl Acad Sci U S A. 1997;94:14467–14470. doi: 10.1073/pnas.94.26.14467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Betschinger J, Mechtler K, Knoblich JA. Asymmetric segregation of the tumor suppressor brat regulates self-renewal in Drosophila neural stem cells. Cell. 2006;124:1241–1253. doi: 10.1016/j.cell.2006.01.038. [DOI] [PubMed] [Google Scholar]
- 35.Lee CY, Wilkinson BD, Siegrist SE, Wharton RP, Doe CQ. Brat is a Miranda cargo protein that promotes neuronal differentiation and inhibits neuroblast self-renewal. Dev Cell. 2006;10:441–449. doi: 10.1016/j.devcel.2006.01.017. [DOI] [PubMed] [Google Scholar]
- 36.Lu B, Rothenberg M, Jan LY, Jan YN. Partner of Numb colocalizes with Numb during mitosis and directs Numb asymmetric localization in Drosophila neural and muscle progenitors. Cell. 1998;95:225–235. doi: 10.1016/s0092-8674(00)81753-5. [DOI] [PubMed] [Google Scholar]
- 37.Matsuzaki F, Ohshiro T, Ikeshima-Kataoka H, Izumi H. miranda localizes staufen and prospero asymmetrically in mitotic neuroblasts and epithelial cells in early Drosophila embryogenesis. Development. 1998;125:4089–4098. doi: 10.1242/dev.125.20.4089. [DOI] [PubMed] [Google Scholar]
- 38.Shen CP, Knoblich JA, Chan YM, Jiang MM, Jan LY, Jan YN. Miranda as a multidomain adapter linking apically localized Inscuteable and basally localized Staufen and Prospero during asymmetric cell division in Drosophila. Genes Dev. 1998;12:1837–1846. doi: 10.1101/gad.12.12.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Siegrist SE, Doe CQ. Microtubule-induced Pins/Galphai cortical polarity in Drosophila neuroblasts. Cell. 2005;123:1323–1335. doi: 10.1016/j.cell.2005.09.043. [DOI] [PubMed] [Google Scholar]
- 40.Tio M, Udolph G, Yang X, Chia W. cdc2 links the Drosophila cell cycle and asymmetric division machineries. Nature. 2001;409:1063–1067. doi: 10.1038/35059124. [DOI] [PubMed] [Google Scholar]
- 41••.Wirtz-Peitz F, Nishimura T, Knoblich JA. Linking cell cycle to asymmetric division: Aurora-A phosphorylates the Par complex to regulate Numb localization. Cell. 2008;135:161–173. doi: 10.1016/j.cell.2008.07.049. This paper shows that a phosphorylation cascade triggered by Aurora-A as cell cycle progresses is responsible for asymmetrically localizing Numb during asymmetric divisions by Drosophila neural precursor cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42•.Hutterer A, Berdnik D, Wirtz-Peitz F, Zigman M, Schleiffer A, Knoblich JA. Mitotic activation of the kinase Aurora-A requires its binding partner Bora. Dev Cell. 2006;11:147–157. doi: 10.1016/j.devcel.2006.06.002. This paper provides evidence that Bora activates Aurora-A through direct binding and a Cdc2-dependent mechanism that relocates Bora from the nucleus to the cytoplasm at the onset of mitosis. [DOI] [PubMed] [Google Scholar]
- 43••.Wang H, Ouyang Y, Somers WG, Chia W, Lu B. Polo inhibits progenitor self-renewal and regulates Numb asymmetry by phosphorylating Pon. Nature. 2007;449:96–100. doi: 10.1038/nature06056. This paper provides evidence that the cell-cycle regulator Polo kinase regulates asymmetric cell division by neuroblasts at least partly through phosphorylating Pon to affect the asymmetric localization and function of the cell-fate determinant Numb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang H, Somers GW, Bashirullah A, Heberlein U, Yu F, Chia W. Aurora-A acts as a tumor suppressor and regulates self-renewal of Drosophila neuroblasts. Genes Dev. 2006;20:3453–3463. doi: 10.1101/gad.1487506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Knoblich JA, Jan LY, Jan YN. The N terminus of the Drosophila Numb protein directs membrane association and actin-dependent asymmetric localization. Proc Natl Acad Sci U S A. 1997;94:13005–13010. doi: 10.1073/pnas.94.24.13005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46••.Basto R, Brunk K, Vinadogrova T, Peel N, Franz A, Khodjakov A, Raff JW. Centrosome amplification can initiate tumorigenesis in flies. Cell. 2008;133:1032–1042. doi: 10.1016/j.cell.2008.05.039. This paper provides evidence that centrosome amplification initiates tumorigenesis not by causing genome instability but through defects in asymmetric cell division. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47••.Yamashita YM, Mahowald AP, Perlin JR, Fuller MT. Asymmetric inheritance of mother versus daughter centrosome in stem cell division. Science. 2007;315:518–521. doi: 10.1126/science.1134910. This paper provides evidence that Drosophila male germline stem cells non-randomly segregate the mother and daughter centrosomes during self-renewing asymmetric cell divisions. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yamashita YM, Fuller MT. Asymmetric centrosome behavior and the mechanisms of stem cell division. J Cell Biol. 2008;180:261–266. doi: 10.1083/jcb.200707083. [DOI] [PMC free article] [PubMed] [Google Scholar]