Abstract
A classical paradigm in population genetics is that homozygosity or inbreeding affects individual fitness through increased disease susceptibility and mortality, and diminished breeding success. Using data from an insular population of mouflon (Ovis aries) founded by a single pair of individuals, we compare embryo number of ewes with different levels of inbreeding. Contrary to expectations, ewes with the highest levels of homozygosity showed the largest number of embryos. Using two different statistical approaches, we showed that this relationship is probably caused by heterozygosity at specific genes. The genetics of embryo number coupled with cyclic dynamics could play a central role in promoting genetic variation in this population.
Keywords: inbreeding, fecundity, genetic diversity
1. Introduction
Inbreeding is known to affect individual fitness (Frankham et al. 2002) by the expression of deleterious recessive alleles and/or the loss of heterozygote advantage in homozygous inbred individuals (Keller & Waller 2002). In animals, inbreeding has been found to reduce reproductive success characteristics such as fertility, litter or clutch size and juvenile survival (Keller & Waller 2002). In plants, inbreeding is also known to reduce seed production and germination (Husband & Schemske 1996).
Individual inbreeding is best quantified using pedigree information (Pemberton 2004). However, in natural populations, pedigrees are often not available or difficult to obtain, and pedigree-based indices of inbreeding are often replaced by molecular indices, such as standardized multi-locus heterozygosity (MLH; Coltman et al. 1999). The average heterozygosity of an individual, however, may not accurately represent genome-wide heterozygosity, and therefore, may not reflect its general level of inbreeding appropriately (Balloux et al. 2004). The relationship between heterozygosity measured by molecular markers and general level of inbreeding can be improved by using a very large number of loci (Balloux et al. 2004). Furthermore in small or recently bottlenecked populations, linkage disequilibrium (LD) increases the correlation between heterozygosity and inbreeding (Hansson & Westerberg 2002, see also Slate et al. 2004 for discussion).
Using data from an insular population of mouflon (Ovis aries) founded by a single pair of individuals (Chapuis et al. 1994), we compare embryo number of ewes with different levels of inbreeding estimated by MLH from 24 microsatellite loci. Approximately 34% of females produced twins in this population (Boussès & Réale 1998), which exceeds the twinning rate for European mouflon populations (2.5–20.7%; Garel et al. 2005). We then tested whether the embryo number was related to the overall level of inbreeding or local effects using a logistic regression approach and by calculating the H–H correlation (Balloux et al. 2004).
2. Material and Methods
(a) The population
The European mouflon has recently been classified as a feral form of the domestic sheep (O. aries) that separated ca 8000 years ago (Opinion 2027 (Case 3010) 2003). The study population was founded in 1957 by two individuals introduced on Haute Island, a small island (6.5 km2) of the Kerguelen subantarctic Archipelago (49°20′ S, 70°20′ E) from the Vincennes Zoo (Paris, France). The Vincennes Zoo population was created in 1934, probably with individuals from Hamburg and Hannover Zoos and from Corsica (Vincennes Zoo herbivore curator J. Villemain 2007, personal communication). The population grew exponentially to reach approximately 700 individuals in 1977. Since then, the population has fluctuated cyclically varying between 250 and 650 individuals and with winter crashes occurring at a periodicity of 3–5 years (Chapuis et al. 1994). In 1994, 54 females were culled and total mass of carcass (i.e. mass without the internal organs) measured. Forty-two ewes were pregnant and the number of developed embryos counted. Based on the number of definite incisors, all these females were age 2 and older, but we could not determine the age of females with precision. In other feral sheep, ewes between 2 and 6 years of age have identical fecundity (Clutton-Brock et al. 1996). Given the short lifespan observed here (Kaeuffer et al. 2007a), few ewes probably reached 6 years of age. We therefore estimated that 10 generations have elapsed from the introduction to 1994.
(b) Genetic analyses
We genotyped tissue samples collected from culled ewes at 24 microsatellite loci (ARO28, HEL10, MCM64, MCM152, BM3413, BM848, HUJ177, MAF64, MCM527, TGLA13, Ilsts059, TGLA176, RT1, AGLA226, Il2ra, MCM218, NRAMP, OarCP49, TEXAN4, DRBps, INRA26, oMHC1, TGLA387 and MAF33; see Kaeuffer et al. 2007a,b for details) to estimate multi-locus standardized heterozygosity (MLH; Coltman et al. 1999). Chromosomal positions were determined from the sheep genomic map (Maddox et al. 2001).
(c) Statistical analyses
To test for the relative effect of MLH and body mass on embryo number in pregnant ewes (i.e. one versus two embryos), we ran a generalized linear model (with a binomial distribution and a log link function), including MLH, body mass and their second-order interaction. We then determined whether embryo number was related to the overall level of inbreeding or to local effects by modelling the probability of producing two embryos as a function of heterozygosity at each locus. We used the false discovery rate method (FDR) to correct our results for multiple testing using the fdrtool function (Strimmer 2007) implemented in R (R Development Core Team 2007). We estimated the H–H correlation that is the average correlation between the mean H calculated from two random subsets of 12 markers, over a 1000 replicates (Balloux et al. 2004). A strong H–H correlation indicates that average H reflects whole genome heterozygosity well (Balloux et al. 2004).
3. Results
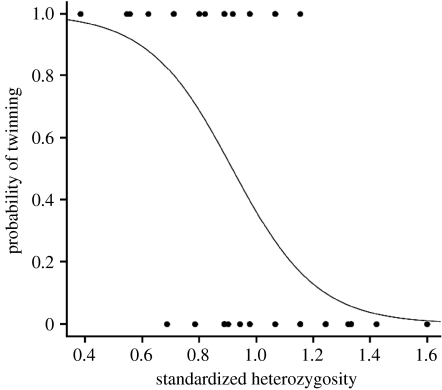
Average standardized heterozygosity was 0.99±0.040 (average±s.e.: 0.46±0.019 for non-standardized H). Females with a lower MLH and heavy females were more prone to have two embryos (likelihood ratio test; MLH, LRT=17.767, d.f.=1; p<0.001; body mass, LRT=6.126, d.f.=1, p=0.013; figure 1). The interaction between MLH and carcass mass was non-significant and removed from the model.
Figure 1.
Effect of standardized multi-locus standardized heterozygosity (MHL) on the probability of twinning in the Kerguelen mouflon population.
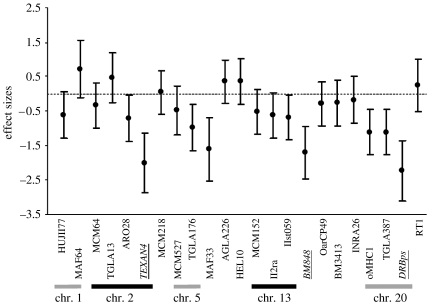
Three of the 24 loci used showed significant effects (likelihood ratio tests; TEXAN4: LRT=7.058, d.f.=1, p<0.01; DRBps: LRT=8.413, d.f.=1, p<0.05; BM848: LRT=5.898, d.f.=1, p=0.015; figure 2). After FDR correction, the effects of TEXAN4, BM848 and DRBPs were significant at a level of 6%. Heterozygosity at the three loci significantly affected embryo number when tested together in a generalized linear model (TEXAN4: LRT=12.387, d.f.=1, p<0.001; DRBps: LRT=10.625, d.f.=1, p=0.001; BM848: LRT=5.138, d.f.=1, p=0.023), showing that the link between each locus and embryo number was independent.
Figure 2.
Effect size (and their s.e.) of loci on gemelarity estimated with a logistic regression and linkage group of the loci used. Three loci showed a significant effect on embryo number (TEXAN4, BM848 and DRBps). Loci linked to TEXAN4 and DRBps show similar trends in their effect but are non-significant.
Two pairs of loci (NRAMP–ARO28 and TGLA387–oMHC1) linked to TEXAN4 and DRBps, respectively, and located within 3 centimorgans (Maddox et al. 2001), showed similar but non-significant effects on embryo number (figure 2). The NRAMP locus (not shown in the figure) was characterized by a particularly strong but non-significant effect size (effect=−9.544, s.e.=29.052, p=0.742) due to the absence of heterozygous individuals with twins for that locus.
After removing the loci with a significant effect on embryo number, the relationship between MLH and litter size was reduced but still significant (LRT=10.555, d.f.=1, p=0.001). We observed a relatively low H–H correlation (r=0.216±0.004; average±s.e.).
4. Discussion
Our study is the first to show a negative effect of MLH on embryo number in an unmanaged population. The positive effect of body mass has also been observed in feral Soay sheep (Clutton-Brock et al. 1996). The effects of MLH here contrast with conventional population and conservation genetic expectation of a negative effect of homozygosity on progeny number (Keller & Waller 2002). An increase in multiple births with inbreeding has been observed in captive chimpanzees, Pan troglodytes (Geissmann 1990). Inbreeding, however, was estimated from pedigree information and could not separate genome-wide and local effects.
We identified three chromosomal regions close to the loci TEXAN4 (chromosome 2; Maddox et al. 2001), DRBps (chromosome 20; Maddox et al. 2001) and BM848 (chromosome 10; Maddox et al. 2001), which may affect embryo number. This indicates that genes other than Booroola and Lacaune (chromosomes 6 and 11, respectively; Davis 2005) influence fecundity in O. aries. However, due to small samples sizes, we were unable to model the additive effects of specific alleles at these loci. The low H–H correlation suggests that MLH was weakly correlated with the individual level of inbreeding (approx. 10–30%; figure 3 in Balloux et al. 2004) and reinforces the idea that correlation between embryo number and MLH may result from local effects rather than general inbreeding level.
However, a significant and negative relationship between MLH and embryo number was still observed after removing the three loci with a significant effect. This may be because other marker loci are in LD with DRBps and TEXAN4 (figure 2). LD is expected between physically linked markers and also between unlinked markers in populations undergoing strong bottlenecks. We observed strong LD (mean rLD=0.146; range: 0–0.749; Hill & Robertson 1968), which was significant for some loci separated by more than 40 cM (i.e. Il2ra and MCM152), and also between the three loci and loci on other chromosomes (e.g. MAF33 linked to DRBps, see also Kaeuffer et al. 2007b). Thus, in addition to the specific effects of DRBps, BM848 and TEXAN4 on embryo number, we cannot rule out the contribution of genome-wide effects generated by LD to the correlation between embryo number and MLH.
We observed a high level of dizygotic twins (at least 9 out of the 19 pairs observed). This form of twinning is partially controlled by genes (Hoekstra et al. 2008), supporting the evidence for genetic effects on embryo number in the mouflon population. The exceptionally high twinning rate of the Kerguelen population (33.8%) and of the ancestral population (Boussès & Réale 1998) suggests that the mutations were present in the island founders. The strong founder effect at Kerguelen, coupled with the isolation of the population, has probably facilitated the increased frequency of recessive homozygous individuals at loci involved in twin production.
The Kerguelen mouflon exhibits cyclic population dynamics and an increase in heterozygosity over the time (Kaeuffer et al. 2007a). The genetics of litter size coupled with the cyclic dynamics could play a central role in promoting genetic variation (Hedrick 2007). Thus, homozygous twinning ewes may suffer higher costs of reproduction and increased mortality during population's crashes (Clutton-Brock et al. 1996), which may increase average heterozygosity in the population over time. This cyclic selection, acting only on females, could contribute to the maintenance of genetic polymorphism in wild populations (Reinhold 2000; Hedrick 2007).
Acknowledgments
We thank all fieldworkers who collected mouflon samples and data, A. Krupa and A. Llewellyn for their help with molecular analyses. This work was supported by the IPEV and the CNRS to D.P. and J.-L.C., the NSERC and the CFI to D.R., and the Royal Society to D.W.C. We thank T. Coulson, D. Garant, J.-M. Gaillard, A. Hendry and two anonymous reviewers for their helpful comments.
References
- Balloux F, Amos W, Coulson T. Does heterozygosity estimate inbreeding in real populations? Mol. Ecol. 2004;13:3021–3031. doi: 10.1111/j.1365-294X.2004.02318.x. doi:10.1111/j.1365-294X.2004.02318.x [DOI] [PubMed] [Google Scholar]
- Boussès P, Réale D. Biology of twinning and origin of an unusually high twinning rate in an insular mouflon population. Z. Saugetierkd. 1998;63:147–153. [Google Scholar]
- Chapuis J.L, Boussès P, Barnaud G. Alien mammals, impact and management in the French subantarctic islands. Biol. Conserv. 1994;67:97–104. doi:10.1016/0006-3207(94)90353-0 [Google Scholar]
- Clutton-Brock T.H, Stevenson I.R, Marrow P, MacColl A.D, Houston A.I, McNamara J.M. Population fluctuations, reproductive costs and life-history tactics in female Soay sheep. J. Anim. Ecol. 1996;65:675–689. doi:10.2307/5667 [Google Scholar]
- Coltman D.W, Pilkington J.G, Smith J.A, Pemberton J.M. Parasite-mediated selection against inbred Soay sheep in a free-living, island population. Evolution. 1999;53:1259–1267. doi: 10.1111/j.1558-5646.1999.tb04538.x. doi:10.2307/2640828 [DOI] [PubMed] [Google Scholar]
- Davis G.H. Major genes affecting ovulation rate in sheep. Genet. Sel. Evol. 2005;37:S11–S23. doi: 10.1186/1297-9686-37-S1-S11. doi:10.1051/gse:2004026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankham R, Ballou J.D, Briscoe D.A. Cambridge University Press; Cambridge, UK: 2002. Introduction to conservation genetics. [Google Scholar]
- Garel M, Cugnasse J.-M, Gaillard J.-M, Loison A, Gibert P, Douvre P, Dubray D. Reproductive output of female mouflon (Ovis gmelini musimon x Ovis sp.): a comparative analysis. J. Zool. 2005;266:65–71. doi:10.1017/S0952836905006667 [Google Scholar]
- Geissmann T. Familial incidence of multiple births in a colony of chimpanzees (Pan troglodytes) J. Med. Primatol. 1990;19:467–478. [PubMed] [Google Scholar]
- Hansson B, Westerberg L. On the correlation between heterozygosity and fitness in natural populations. Mol. Ecol. 2002;11:2467–2474. doi: 10.1046/j.1365-294x.2002.01644.x. doi:10.1046/j.1365-294X.2002.01644.x [DOI] [PubMed] [Google Scholar]
- Hedrick P.W. Cyclic fitness variation and polymorphism: cycling selection for litter size in arctic foxes. Heredity. 2007;98:339. doi: 10.1038/sj.hdy.6800980. doi:10.1038/sj.hdy.6800980 [DOI] [PubMed] [Google Scholar]
- Hill W.G, Robertson A. Linkage disequilibrium in finite populations. Theor. Appl. Genet. 1968;38:226–231. doi: 10.1007/BF01245622. doi:10.1007/BF01245622 [DOI] [PubMed] [Google Scholar]
- Hoekstra C, Zhao Z.Z, Lambalk C.B, Willemsen G, Martin N.G, Boomsma D.I, Montgomery G.W. Dizygotic twinning. Hum. Reprod. Update. 2008;14:37–47. doi: 10.1093/humupd/dmm036. doi:10.1093/humupd/dmm036 [DOI] [PubMed] [Google Scholar]
- Husband B.C, Schemske D.W. Evolution of the magnitude and timing of inbreeding depression in plants. Evolution. 1996;50:54–70. doi: 10.1111/j.1558-5646.1996.tb04472.x. doi:10.2307/2410780 [DOI] [PubMed] [Google Scholar]
- Kaeuffer R, Coltman D.W, Chapuis J.-L, Pontier D, Réale D. Unexpected heterozygosity in an island mouflon population founded by a single pair of individuals. Proc. R. Soc. B. 2007a;274:527–533. doi: 10.1098/rspb.2006.3743. doi:10.1098/rspb.2006.3743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeuffer R, Réale D, Coltman D.W, Pontier D. Detecting population structure using Structure software: effect of background linkage disequilibrium. Heredity. 2007b;99:374–380. doi: 10.1038/sj.hdy.6801010. doi:10.1038/sj.hdy.6801010 [DOI] [PubMed] [Google Scholar]
- Keller L.F, Waller D.M. Inbreeding effects in wild populations. Trends Ecol. Evol. 2002;17:230–241. doi:10.1016/S0169-5347(02)02489-8 [Google Scholar]
- Maddox J.F, et al. An enhanced linkage map of the sheep genome comprising more than 1000 loci. Genome Res. 2001;11:1275–1289. doi: 10.1101/gr.135001. doi:10.1101/gr.GR-1350R [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opinion 2027 (Case 3010) Usage of 17 specific names based on wild species which are pre-dated by or contemporary with those based on domestic animals (Lepidoptera, Osteichthyes, Mammalia): conserved. Bull. Zool. Nom. 2003;60:81–84. [Google Scholar]
- Pemberton J. Measuring inbreeding depression in the wild: the old ways are the best. Trends Ecol. Evol. 2004;19:613–615. doi: 10.1016/j.tree.2004.09.010. doi:10.1016/j.tree.2004.09.010 [DOI] [PubMed] [Google Scholar]
- R Development Core Team 2007 R: a language and environment for statistical computing Vienna, Austria: R Foundation for Statistical Computing.
- Reinhold K. Maintenance of a genetic polymorphism by fluctuating selection on sex-limited traits. J. Evol. Biol. 2000;13:1009–1014. doi:10.1046/j.1420-9101.2000.00229.x [Google Scholar]
- Slate J, David P, Dodds K.G, Veenvliet B.A, Glass B.C, Broad T.E, McEwan J.C. Understanding the relationship between the inbreeding coefficient and multilocus heterozygosity: theoretical expectations and empirical data. Heredity. 2004;93:255–265. doi: 10.1038/sj.hdy.6800485. doi:10.1038/sj.hdy.6800485 [DOI] [PubMed] [Google Scholar]
- Strimmer, K. 2007 fdrtool: estimation and control of (local) false discovery rates. R package, v. 1.1.4.