Abstract
Two amino acid substitutions (L1014F and M918T) in the voltage-gated sodium channel confer target-site resistance to pyrethroid insecticides in the peach potato aphid, Myzus persicae. Pyrethroid-resistant and -susceptible M. persicae clones with various combinations of these mutations were crossed under laboratory conditions, and the genotypes of aphid progeny were analysed by direct DNA sequencing of the IIS4–S6 region of the sodium channel gene. Segregation patterns showed that in aphids heterozygous for both L1014F and M918T, both mutations were present in the same resistance allele. Despite these mutations appearing largely recessive in other pest species, such aphids exhibited strong resistance to pyrethroids in leaf-dip bioassays. These results have important implications for the spread and management of pyrethroid resistance in field populations.
Keywords: Myzus persicae, knock-down resistance, sodium channel, pyrethroids, inheritance, dominance
1. Introduction
The peach potato aphid, Myzus persicae (Sulzer; Hemiptera: Aphididae), is a pest of major economic importance on a wide range of crops throughout the world. Pyrethroid insecticides have been widely used to control agricultural pests and disease vectors over the past 40 years (Casida & Quistad 1998). However, their intensive use has led to resistance in many insect species including M. persicae (Soderlund & Knipple 2003). Pyrethroid-resistant clones of M. persicae have a leucine-to-phenylalanine mutation (L1014F) within the IIS6 transmembrane segment of the para-type sodium channel protein (Martinez-Torres et al. 1999), which is the major target site of pyrethroids (Soderlund & Knipple 2003). Resistance conferred by L1014F is termed knock-down resistance or kdr. An additional mutation, a methionine-to-threonine replacement (M918T) in the short cytoplasmic domain between segments IIS4 and IIS5 of the channel protein, has also been detected together with the L1014F mutation in some M. persicae clones (Anstead et al. 2004; Eleftherianos et al. 2008). This confers an enhanced form of pyrethroid resistance termed super-kdr.
Any attempt to understand the evolution of insecticide resistance in M. persicae must take into account its complex life cycle. This species can exhibit either holocycly (a sexual phase in winter) or anholocycly (continuous parthenogenesis), depending on the environmental conditions it encounters and the genetic characteristics of the aphids (Blackman 1974). The different life cycles of M. persicae have important implications for the development of resistance and the reassortment of resistance genes into genetic backgrounds that favour their long-term persistence (Devonshire et al. 1998).
Management of insecticide resistance benefits from a thorough knowledge of its genetic basis and the mechanisms involved. The mode of inheritance influences resistance detection, monitoring, modelling and risk assessment (Tabashnik et al. 1997). In order to investigate inheritance and dominance of insecticide resistance mechanisms in M. persicae, it is necessary to perform controlled laboratory crosses between clonal lineages established from field populations. Such crosses are inherently very challenging and require methods for inducing sexual forms and maintaining viable eggs through an obligatory winter diapause. The objective of the current study was to investigate the inheritance of pyrethroid-resistant and -susceptible phenotypes in M. persicae by crossing aphids from clones with different combinations of the L1014F and M918T mutations, and testing the response of progeny in pyrethroid bioassays.
2. Material and methods
(a) M. persicae: rearing methods and crossing experiments
Aphids were maintained on individual excised leaves of Chinese cabbage seedlings (Brassica napus L. var. chinensis cv. ‘Tip-Top’ or ‘Wong-Bok’) under 21°C and 16 L : 8 D to assure continuous asexual reproduction. Details of the parental M. persicae clones used in this study are given in table 1. All possessed the same (R3) level of enhanced carboxylesterase to avoid complications caused by the minor contribution that this often coexisting resistance mechanism makes to pyrethroid resistance (Martinez-Torres et al. 1999). Procedures for establishing and performing crosses between aphid clones were adapted from those described by Blackman et al. (1996). The major modification was that diapause development of eggs involved incubation at a transitional temperature of 10°C prior to being placed under freezing and thawing conditions. Viable progeny were used to generate progeny clones for genetic characterization and bioassays.
Table 1.
Origins and resistance genotypes of M. persicae clones used for laboratory crosses and insecticide bioassays.
| clone | origin | esterase | kdr | super-kdr |
|---|---|---|---|---|
| 800F | Italy, 1978 | R3 | SS | SS |
| 108T | product of cross | R3 | SR | SS |
| 794J | England, 1994 | R3 | RR | SS |
| 2169G | England, 1997 | R3 | SR | SR |
(b) Determination of the genotype of aphid progeny
Genomic DNA was extracted from single insects using methods described within the Nucleon Phytopure DNA extraction kit (Nucleon Biosciences). The IIS4–S6 region of the aphid para-type sodium channel gene, containing the L1014F and M918T pyrethroid resistance-associated mutations, was amplified from the genomic DNA of aphid progeny by two rounds of PCR, as described by Anstead et al. (2004). Sequencing was performed with the ABI Prism BigDye terminator cycle sequencing ready reaction kit, according to the supplier's standard procedure using primers Aph15 and Aph21 (Anstead et al. 2004). The samples were analysed using an Applied Biosystems 310 automated DNA sequencer. Sequence data were aligned and analysed using Vector NTI (Informax, Inc.).
(c) Determination of resistance phenotypes
The pyrethroid-resistant phenotype of aphid progeny was assessed using leaf-dip bioassays with apterous adult aphids (Barber et al. 1999). A single representative clone of each kdr and super-kdr genotypic combination was tested with at least six concentrations of lambda-cyhalothrin (‘Hallmark’, 50 g l−1 EC; Syngenta) ranging from 0.01 to 1000 ppm. Clone 800F (which lacked both resistance mutations; table 1) was used as the standard susceptible clone in all bioassays. Mortality was assessed 48 hours after treatment. Results were subjected to probit analysis (POLO LeOra Software), with a lack of overlap between 95% CI of calculated EC50 values being used as the criterion for significant differences in response (Robertson & Preisler 2007).
3. Results
(a) Success of crosses
Although up to 50% egg hatch was obtained in some cases, egg mortality was a recurring problem and many crosses failed completely, yielding only a few viable eggs or survival of the fundatrices was extremely low. No reasons could be found to account for the differential egg mortality under closely controlled conditions. This is a long-standing problem and a major constraint on genetic analyses of M. persicae (Blackman et al. 1996). A total of 60 M. persicae progeny clones were produced from the crosses, involving 3 of the 4 parental clones (table 2). Several attempts were made to cross 800F♂×794J♀ and 2169G♂×794J♀ aphids, but none of the hundreds of eggs laid were viable. Other crosses (e.g. 108T♂×794J♀, 108T♂×108T♀) proved impossible because aphids did not produce males in sufficient numbers when subjected to reduced photoperiod and temperature.
Table 2.
Inheritance of L1014F and M918T sodium channel point mutations and kdr/super-kdr genotypes of progeny clones produced from the laboratory crossing experiments between M. persicae clones (L and F refer to the L1014F mutation and M and T to the M918T mutation).
| cross | 800F♂×800F♀ | 800F♂×2169G♀ | 800F♂×108T♀ | 2169G♂×2169G♀ | 2169G♂×108T♀ |
|---|---|---|---|---|---|
| parent genotypes | LM/LM×LM/LM | LM/LM×FT/LM | LM/LM×FM/LM | FT/LM×FT/LM | FT/LM×FM/LM |
| expected progeny genotypesa | LM/LM | LM/LM, FT/LM | LM/LM, FM/LM | LM/LM, FT/LM, FT/FT | LM/LM, FM/LM, FT/LM, FT/FM |
| expected ratio | all LM/LM | 1 : 1 | 1 : 1 | 1 : 2 : 1 | 1 : 1 : 1 : 1 |
| observed ratio | all LM/LM | 13 : 4 | 5 : 7 | 0 : 2 : 0 | 1 : 2 : 2 : 2 |
| no. of progeny | 22 | 17 | 12 | 2 | 7 |
On the assumption that both resistance mutations are on the same allele (figure 1).
(b) Combinations of resistance mutations in M. persicae offspring
Clone 800F had previously been determined as homozygous for L1014 and M918 (LM/LM using the terminology in table 2; Martinez-Torres et al. 1999). Further confirmation of its genotype was obtained by selfing 800F individuals, which produced 22 progeny clones with the LM/LM genotype. Crossing 800F♂×2169G♀ yielded four progeny clones that contained both mutations in the heterozygous form (FT/LM), a genotype that was only probable if both mutations were present in the same allele in the female parent (figure 1a). The remaining 13 progeny clones from this cross were LM/LM, again consistent with both mutations occurring in the same allele. The 800F♂×108T♀ cross generated 12 progeny clones, 5 of which were characterized as FM/LM (as in the female parent) and the other 7 being LM/LM as in the male parent. Selfing of clone 2169G unfortunately generated only two progeny clones, both of which proved to be heterozygous for both mutations (FT/LM) like the parental clone. Two other genotypes expected from this cross (LM/LM and FT/FT) were not observed. Crossing 2169G♂×108T♀ generated seven progeny clones representing all four of the genotypes expected if both mutations in the 2169G parent were present in the same allele (figure 1b; table 2).
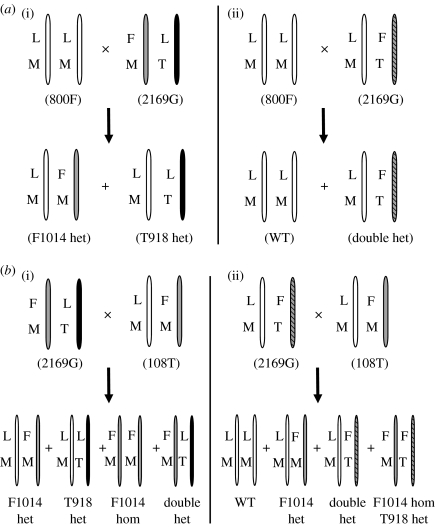
Figure 1.
Representation of crosses between M. persicae clones with different kdr/skdr genotypes showing the two possible scenarios for segregation of resistance mutations in the progeny. (a) 800 F (wild type, WT)×2169G (double het) and (b) 2169G (double het)×108T (F1014 het). (i) F1014 and T918 reside in different alleles for 2169G and (ii) F1014 and T918 reside in the same allele for 2169G. L, L1014 (WT); F, F1014 (kdr mutation); M, M918 (WT); T, T918 (super-kdr mutation).
(c) Response of aphid progeny to lambda-cyhalothrin
As expected, clones produced from the 800F♂×800F♀ cross proved fully susceptible to lambda-cyhalothrin (table 3). EC50 values for the two genotypes derived from 800F♂×2169G♀ were significantly different (p<0.05), with FT/LM progeny being 517-fold resistant compared with 800F aphids. Similarly, resistance factors for clones derived from 800F♂×108T♀ ranged from 1.13 for LM/LM progeny to 6 for FT/LM progeny (i.e. heterozygous for the L1014F mutation alone). FT/LM progeny produced from selfing 2169G♂ showed similarly strong resistance (465-fold) to aphids of this genotype from the 800F♂×2169G♀ cross. Finally, resistance in aphids of the four genotypes generated from 2169G♂×108T♀ varied widely from 1.15- to 702-fold. FT/LM and FT/FM aphids both showed strong pyrethroid resistance but their EC50 values were not significantly different (p>0.05).
Table 3.
Results of pyrethroid leaf-dip bioassays on M. persicae progeny clones representing all observed progeny genotypes. (EC50, effective concentration to give 50% dead or with irreversible symptoms of poisoning. Values followed by the same letter do not differ significantly (p<0.05). 95% CI, 95% confidence interval; RF (resistance factor)=EC50 for clone/EC50 for 800F. L and F refer to the L1014F mutation and M and T to the M918T mutation.)
| cross | progeny genotypes | EC50 (ppm) | 95% CI | slope | RF |
|---|---|---|---|---|---|
| 800F♂×800F♀ | LM/LM | 0.73a | 0.23–1.73 | 0.65 | 1.10 |
| 800F♂×2169G♀ | LM/LM | 0.88ab | 0.37–1.84 | 0.81 | 1.30 |
| FT4/LM | 341c | 125–624 | 0.61 | 517 | |
| 800F♂×108T♀ | LM/LM | 0.75a | 0.27–1.67 | 0.73 | 1.13 |
| FM/LM | 4.20b | 1.74–9.48 | 0.67 | 6.0 | |
| 2169G♂×2169G♀ | FT/LM | 307c | 97.3–1985 | 0.51 | 465 |
| 2169G♂×108T♀ | LM/LM | 0.76ab | 0.25–1.76 | 0.67 | 1.15 |
| FM/LM | 3.36ab | 0.91–9.47 | 0.57 | 5.0 | |
| FT/LM | 315c | 95.2–2375 | 0.48 | 477 | |
| FT/FM | 463c | 124–5173 | 0.45 | 702 |
4. Discussion
Despite the long history of research on resistance in M. persicae, little has been done to investigate its inheritance due to the technical difficulties with establishing controlled crosses in the laboratory. Blackman et al. (1996) is the only previous study of this kind, which focused on the inheritance of amplified carboxylesterase genes to relate data from the crosses to the physical location of amplicons in the aphid genome. The current work was established to improve understanding of the kdr (L1014F) and the more recently detected super-kdr (M918T) mutations and their practical implications. We were especially interested in determining the dominance characteristics of these mutations and generating novel genotypic combinations from clones isolated directly from field samples. Although crossing experiments failed to generate progeny homozygous for F1014 and T918 (FT/FT), cross 2169G♂×108T♀ produced aphids homozygous for F1014 and heterozygous for M918T (FT/FM); this kdr/skdr genotypic combination had not been found in the field so far.
The most important finding is that the skdr mutation is located in the same allele as the kdr mutation in the double heterozygote (FT/LM) clone (2169G). This was demonstrated by the genotypic combinations of aphid clones produced from certain crosses (figure 1). Firstly, cross 800F♂×2169G♀ would generate progeny with only LM/FM and LT/LM genotypes if the kdr and skdr mutations resided in different alleles (figure 1a(i)). However, progeny from this cross were LM/LM and FT/LM as predicted, if both mutations reside in the same allele (figure 1a(ii)). Similarly, if kdr and skdr mutations occurred in different alleles, progeny from the 2169G♂×108T♀ cross would have a different suite of four genotypes (figure 1b(i)) to that predicted (and observed) by a scenario in which both mutations reside in the same allele (figure 1b(ii)).
Clone 2169G and other M. persicae clones heterozygous for L1014F and M918T have been shown to exhibit higher resistance to pyrethroids than the ones homozygous for F1014 alone (Anstead et al. 2004; Eleftherianos et al. 2008). All double heterozygote (FT/LM) aphids tested in this study also showed strong pyrethroid resistance, despite knock-down resistance mutations usually being considered to be recessive in their expression (Soderlund & Knipple 2003). Had the kdr and super-kdr mutations proved to be on different chromosomes in double heterozygotes, dominance could arise from each allele encoding a sodium channel protein with an amino acid substitution conferring insensitivity to pyrethroids. This is clearly not the case and the reason why individuals with a single resistance allele display strong resistance remains unclear. However, this phenomenon has profound implications for resistance management since even heterozygotes can be selected rapidly by pyrethroids to economically damaging frequencies, and persists indefinitely through clonal propagation in areas such as the UK where sexual crossing is seemingly absent or extremely rare.
Finally, aphids heterozygous for L1014F and M918T have been shown to exhibit lower levels of disturbance after exposure to measured amounts of synthetic alarm pheromone than aphids homozygous for F1014 alone (Eleftherianos et al. 2002). This may have potential important implications for the survival of M. persicae expressing both resistance mutations, as the alteration in the behaviour of these forms may result in greater vulnerability to predation and parasitism.
Acknowledgments
We thank Diana Cox and Barbara Hackett for their technical assistance. I.E. was supported by a grant from IKY (Greek State Scholarships Foundation). Rothamsted Research receives grant-aided support from the Biotechnology and Biological Sciences Research Council (BBSRC) of the UK.
References
- Anstead J.A, Williamson M.S, Eleftherianos I, Denholm I. High-throughput detection of knockdown resistance in Myzus persicae using allelic discriminating quantitative PCR. Insect Biochem. Mol. Biol. 2004;34:871–877. doi: 10.1016/j.ibmb.2004.06.002. [DOI] [PubMed] [Google Scholar]
- Barber M.D, Moores G.D, Tatchell G.M, Vice W.E, Denholm I. Insecticide resistance in the currant-lettuce aphid, Nasonovia ribisnigri (Hemiptera: Aphididae) in the UK. Bull. Entomol. Res. 1999;89:17–23. doi:10.1017/S0007485399000036 [Google Scholar]
- Blackman R.L. Life cycle variation of Myzus persicae (Sulz.) (Hom., Aphididae) in different parts of the world, in relation to genotype and environment. Bull. Entomol. Res. 1974;63:595–607. [Google Scholar]
- Blackman R.L, Spence J.M, Field L.M, Javed N, Devine G.J, Devonshire A.L. Inheritance of the amplified esterase genes responsible for insecticide resistance in Myzus persicae (Homoptera: Aphididae) Heredity. 1996;77:154–167. doi:10.1038/hdy.1996.120 [Google Scholar]
- Casida J.E, Quistad G.B. Golden age of insecticide research: past, present, or future? Annu. Rev. Entomol. 1998;43:1–16. doi: 10.1146/annurev.ento.43.1.1. doi:10.1146/annurev.ento.43.1.1 [DOI] [PubMed] [Google Scholar]
- Devonshire A.L, Field L.M, Foster S.P, Moores G.D, Williamson M.S, Blackman R.L. The evolution of insecticide resistance in the peach-potato aphid, Myzus persicae. Phil. Trans. R. Soc. B. 1998;353:1677–1684. doi:10.1098/rstb.1998.0318 [Google Scholar]
- Eleftherianos I, Foster S.P, Williamson M.S, Denholm I. Behavioural consequences of pyrethroid resistance in the peach potato aphid, Myzus persicae (Sulzer) Proc. Brighton Crop Protect. Conf. 2002;1:745–749. [Google Scholar]
- Eleftherianos I, Foster S.P, Williamson M.S, Denholm I. Characterization of the M918T sodium channel gene mutation associated with strong resistance to pyrethroid insecticides in the peach-potato aphid, Myzus persicae (Sulzer) Bull. Entomol. Res. 2008;98:183–191. doi: 10.1017/S0007485307005524. doi:10.1017/S0007485307005524 [DOI] [PubMed] [Google Scholar]
- Martinez-Torres D, Foster S.P, Field L.M, Devonshire A.L, Williamson M.S. A sodium channel point mutation is associated with resistance to DDT and pyrethroid insecticides in the peach-potato aphid, Myzus persicae (Sulzer) (Hemiptera: Aphididae) Insect Mol. Biol. 1999;8:339–346. doi: 10.1046/j.1365-2583.1999.83121.x. doi:10.1046/j.1365-2583.1999.83121.x [DOI] [PubMed] [Google Scholar]
- Robertson J.L, Preisler H.K. 2nd edn. CRC; Boca Raton, FL: 2007. Bioassays with arthropods. [Google Scholar]
- Soderlund D.M, Knipple D.C. The molecular biology of knockdown resistance to pyrethroid insecticides. Insect Biochem. Mol. Biol. 2003;33:563–577. doi: 10.1016/s0965-1748(03)00023-7. doi:10.1016/S0965-1748(03)00023-7 [DOI] [PubMed] [Google Scholar]
- Tabashnik B.E, Liu Y.B, Malvar T, Heckel D.G, Masson L, Ballester V, Granero F, Ménsua J.L, Ferré J. Global variation in the genetic and biochemical basis of diamondback moth resistance to Bacillus thuringiensis. Proc. Natl Acad. Sci. USA. 1997;94:12 780–12 785. doi: 10.1073/pnas.94.24.12780. doi:10.1073/pnas.94.24.12780 [DOI] [PMC free article] [PubMed] [Google Scholar]