Abstract
Inbreeding may lead to morphological malformations in a wide variety of taxa. We used genetic markers to evaluate whether malformed urodeles were more inbred and/or had less genetic diversity than normal salamanders. We captured 687 adult and 1259 larval tiger salamanders (Ambystoma tigrinum tigrinum), assessed each individual for gross malformations, and surveyed genetic variation among malformed and normal individuals using both cytoplasmic and nuclear markers. The most common malformations in both adults and larvae were brachydactyly, ectrodactyly and polyphalangy. The overall frequency of adults with malformations was 0.078 compared to 0.081 in larval samples. Genetic diversity was high in both normal and malformed salamanders, and there were no significant difference in measures of inbreeding (f and F), allele frequencies, mean individual heterozygosity or mean internal relatedness. Environmental contaminants or other extrinsic factors may lead to genome alternations that ultimately cause malformations, but our data indicate that inbreeding is not a causal mechanism.
Keywords: tiger salamander, Ambystoma tigrinum, genetic variation, genetic diversity
1. Introduction
Inbreeding, the union of consanguineous gametes, can lead to a reduction in fitness caused by the expression of recessive deleterious alleles in homozygotes (i.e. inbreeding depression). The quantification of inbreeding depression is notoriously difficult, especially in wild populations (Keller & Waller 2002). Often, inbreeding depression is associated with qualitative morphological malformations, such as cryptorchidism in Florida panthers (Mansfield & Land 2002), lordosis in fish (Afonso et al. 2000) and skeletal dwarfing in lizards (Olsson et al. 1996). This is not to say that all malformations are the result of inbreeding, but in many different taxa there are strong associations between specific malformations and an increased incidence of consanguinity.
Malformations play an unknown role in worldwide amphibian declines, and there is growing trepidation about whether they may be linked to factors such as UVB radiation and parasite infection (Blaustein et al. 1997; Johnson et al. 2001). Inbreeding and other measures of genetic diversity have largely been ignored in studies of amphibian malformations, but may be important because habitat fragmentation and philopatric tendencies may increase the probability for consanguineous matings and the concomitant loss of genetic diversity. The specific goal of this study was to determine whether malformed individuals had reduced genetic diversity relative to normal individuals.
2. Material and methods
(a) Sample collection and malformation assessment
Individual salamanders were captured from a large wetland complex (Williams 2007; Bos et al. 2008) in Tippecanoe County, Indiana, USA using minnow traps (larvae) or standard drift fence and pitfall traps (adults). Upon initial capture, each salamander was thoroughly inspected for gross malformations (e.g. missing limbs or digits, extra limbs or digits, or pronounced asymmetries) and tail or toe tissue was collected for genetic analyses.
(b) DNA extraction and amplification procedures
DNA extractions and microsatellite genotyping were performed as described in Williams (2007). To minimize microsatellite genotyping errors, 40% of all individuals were rerun at four or more loci to ensure that systematic genotyping errors had not occurred. We also sequenced a 635 bp portion of the NADH dehydrogenase subunit 2 mitochondrial gene (ND2) following Bos et al. (2008).
(c) Genetic variability
Genetic samples were partitioned into the following four separate groups: (i) normal adults, (ii) malformed adults, (iii) normal larvae, and (iv) malformed larvae. We used genetic data analysis (Lewis & Zaykin 1999) to calculate microsatellite diversity and genetic structure. For each group, inbreeding coefficients (f, F) and differences in allele frequencies (θ) were calculated as in Weir & Cockerham (1984). We also calculated individual multilocus heterozygosity (IH) and internal relatedness (IR; Amos et al. 2001) for each individual. We arcsine transformed individual heterozygosity values and tested for differences in the means between the normal versus malformed adults and between the normal versus the malformed larvae using one-tailed t-tests. We also compared the mean IR between the two groups using one-tailed t-tests. We used Api-Calc (Ayres & Overall 2004) to calculate the probability of identity and the ‘identity function’ in Cervus v. 3.0 (Marshall et al. 1998) to ensure that none of our salamanders lost their physical mark and were inadvertently counted twice. Chi-squared tests were used to test for a sex bias in the frequency of malformations among adults (larvae cannot be sexed) as well as for a bias in the frequency of malformations in the front limbs versus the hind limbs in both larvae and adults.
Mitochondrial sequences were aligned using BioEdit v. 7.0.1 (Hall 1999) and sorted using Genelex v. 6 (Peakall & Smouse 2006). Arlequin v. 3.01 (Schneider et al. 2000) was used to estimate haplotype and nucleotide diversities; analysis of molecular variance (AMOVA) was used to test for differences in allele frequencies among the four groups.
3. Results
(a) Sample collection and malformation assessment
We report only gross malformations representing substantial deviations from the normal body plan (figure 1). Approximately 2000 salamanders (687 adults and 1259 larvae) were captured and evaluated for malformations. Among the 687 adults, 54 (7.9%) were malformed (table 1). Of these 54 adults, 46 (85%) had missing (ectrodactyly), extra (polyphalangy) or dwarfed digits (brachydactyly). We also observed three adults with two or more digits fused together (syndactyly) and two adults with extra limbs (polymelia). There was no significant difference in the occurrence of malformations between adult males and females (Χ12=1.23, p=0.27). The frequency of malformations found in fore and hind limbs were similar in adults (Χ12=0.49, p=0.48), whereas larvae had more malformations in hind limbs (Χ12=23.6, p<0.0001). Among the 102 malformed larvae, 94 (92%) had malformations involving ectrodactyly, polyphalangy and brachydactyly (table 1). The most profound malformations (e.g. missing limbs, incomplete limbs and missing eyes) were only found in larvae and represented less than 8% of the larval total. In general, the most common malformations (i.e. brachydactyly, ectrodactyly and polyphalangy) were observed in both adults and larvae, with remarkably similar rates of incidence (7.8 and 8.1%, respectively; table 1).
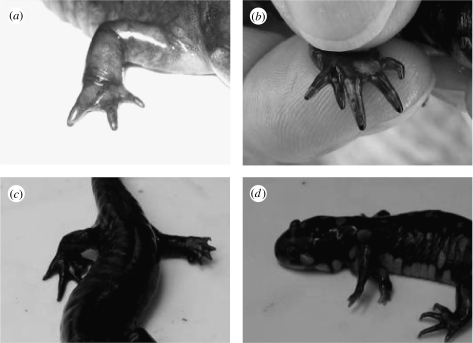
Figure 1.
Representative malformations observed in adult Ambystoma tigrinum. (a) Missing digit on the right fore limb, (b) extra digit on the right fore limb, (c) all five digits dwarfed on the left hind limb and (d) extra fore limb.
Table 1.
Type and frequency of malformations observed in Ambystoma tigrinum from 2003 to 2007. Percentage of each malformation type is given in parentheses. n, sample size.
| adults (n=687) | larvae (n=1259) | ||
|---|---|---|---|
| malformation | description | frequency | frequency |
| brachydactyly | dwarfed toe | 11 (1.6%) | 17 (1.4%) |
| ectrodactyly | missing toe | 19 (2.8%) | 5 (0.4%) |
| polyphalangy | excessive number of digits | 16 (2.3%) | 72 (5.7%) |
| syndactyly | fused digits | 3 (0.4%) | 0 |
| bifurcated tail | forked tail | 3 (0.4%) | 0 |
| polymelia | excessive number of limbs | 2 (0.3%) | 0 |
| amelia | missing limb | 0 | 3 (0.2%) |
| ectromelia | incomplete limb | 0 | 2 (0.2%) |
| phocomelia | absence of proximal portion of limb | 0 | 1 (0.1%) |
| hemimelia | short bone | 0 | 1 (0.1%) |
| anophthalmia | missing eye | 0 | 1 (0.1%) |
| total | 54 (7.8%) | 102 (8.1%) |
(b) Genetic variability
We generated microsatellite genotypes (n=6 loci) for 51 malformed adults, 50 normal adults, 30 malformed larvae and 33 normal larvae for a total of 164 individuals. Among the four groups, there were 2–27 alleles per locus (mean 10.4) and a mean observed heterozygosity of 0.68. Internal quality controls (i.e. approx. 40% of the samples were run twice at each locus) indicate that our genotyping error rate was less than 3%. The combined probability of identity (PID) was 2.3×10−7 and no two individuals shared identical multilocus genotypes across all six loci. Thus, our physical marks were not responsible for malformations in subsequent years.
There was no evidence that malformed individuals were more inbred than normal individuals, as gauged by different lines of evidence. There was no significant difference in measures of inbreeding (f and F) or differences in allele frequencies (θ) between pairwise comparisons of normal versus malformed individuals (table 2). Similarly, mean IH and mean IR values were not significantly different between normal and malformed individuals (table 3).
Table 2.
Estimates of inbreeding (f, F) and differences in allele/haplotype frequencies (θ and Fst) using both nuclear and cytoplasmic markers in eastern tiger salamanders from Indiana.
| microsatellite | ND2 | |||||||
|---|---|---|---|---|---|---|---|---|
| group | f | p-value | f | p-value | θ | p-value | Fst | p-value |
| normal adults | 0.051 | >0.05 | 0.019 | >0.05 | −0.003 | >0.05 | −0.003 | >0.05 |
| malformed adults | −0.003 | |||||||
| normal larvae | 0.061 | >0.05 | 0.040 | >0.05 | −0.000 | >0.05 | 0.014 | >0.05 |
| malformed larvae | 0.017 | |||||||
Table 3.
Mean multilocus IH and mean IR values of normal and malformed adult and larval A. t. tigrinum. n, sample size; d.f., degrees of freedom.
| comparison | measure | n | normal | n | malformed | t-value | d.f. | p-value |
|---|---|---|---|---|---|---|---|---|
| adult | IH | 50 | 0.655 | 51 | 0.699 | 1.57 | 99 | 0.119 |
| IR | 50 | 0.048 | 51 | −0.008 | 1.51 | 99 | 0.134 | |
| larvae | IH | 33 | 0.667 | 30 | 0.686 | 0.82 | 61 | 0.418 |
| IR | 33 | 0.049 | 30 | 0.042 | 0.8 | 61 | 0.455 |
Mitochondrial DNA diversity was also similar in normal and malformed individuals. A total of nine ND2 haplotypes were identified among the four groups of Ambystoma tigrinum tigrinum. Haplotype frequencies were remarkably consistent among all four groups. Likewise, overall haplotype diversity (range 0.8000–0.8841) and nucleotide diversity (range 0.0083–0.0096) were similar among the comparisons. AMOVA indicated that most haplotype variation was distributed within groups, whereas little variation was observed among the groups (table 2).
4. Discussion
Our data from approximately 2000 salamanders reveal new patterns of malformations in the wild, and our genetic data effectively rule out inbreeding and/or lack of genetic diversity as causal factors. We documented, in both larval and adult tiger salamanders, an incidence of malformations (approx. 8%) nearly twice as high as previously reported in adult newts (Meyer-Rochow & Asahima 1988) and similar to many anuran species (Johnson et al. 2001). Despite the differences in locations of malformations (hind and forelimb malformations were equally frequent in adults whereas larvae had significantly more hind limb malformations), our results reveal few differences in the frequency of malformations among life-history stages. This is an interesting pattern, as malformations should negatively impact survival (Sessions & Ruth 1990). In our population, abnormal adults were recorded nearly as often as abnormal larvae (and with similar malformations), suggesting that malformed larvae do not suffer substantially higher mortality than normal conspecifics.
We found substantial variation in the incidence of malformations across sampling years. For larvae, we detected malformations in 75 of 646 individuals (11.6%) sampled in 2006, but in only 27 of 613 individuals (4.4%) sampled in 2007. For adults, we detected malformations in only 1 of 109 individuals sampled in 2003, 14 of 220 in 2004, 6 of 44 in 2005 and 32 of 314 in 2006. Similar variation in the malformation rate over time is also seen in newts (Taricha torosa) and many anurans (Johnson et al. 2001).
The large effective size of our population (Ne) and the very low recapture rate (Bos et al. 2008) suggest that identity by descent is not a confounding issue. In our study population, DNA sequence diversity is high at a key nuclear gene under strong selection (Bos & DeWoody 2005). The microsatellite and mtDNA data presented herein indicate that levels of neutral genetic variation are also very high, roughly twice that found in most terrestrial animals (DeWoody & Avise 2000). Furthermore, allelic diversity and heterozygosity are similar between groups of normal and malformed individuals. This is important because even under a random union of gametes model, some matings are consanguineous and produce inbred offspring. We find no evidence that such inbred individuals are more likely to be morphologically abnormal.
Recently, genetic parentage data have begun to reshape our view of ambystomatid biology. The lack of strong site fidelity (Tennessen & Zamudio 2003) coupled with highly polyandrous mating systems, where both sexes benefit from multiple mating (Gopurenko et al. 2007) produces a large Ne with little inbreeding (Bos et al. 2008). Our mtDNA data buttress the microsatellite data; there are no differences in diversity between normal and malformed individuals and thus provide no support for the idea that an individual's genetic background is related to the incidence of malformations. We do not mean to imply that genes play no role in malformations, but our molecular data provide the first ‘scan’ for genome-wide effects. In total, they suggest that other biotic and abiotic factors (e.g. pathogens, UV radiation, regeneration following trauma) are responsible for salamander malformations.
Acknowledgments
Animals were collected under permits issued by the Indiana Department of Natural Resources and the Purdue University Animal Care and Use Committee.
We thank B. Williams and B. Reinhart for their field assistance and the DeWoody laboratory for comments on the earlier drafts of this manuscript. Financial support was provided by Purdue University and the US National Science Foundation (DEB-0514815 to J.A.D.).
References
- Afonso J.M, Montero D, Robaina L, Astrga N, Izquierdo M.S, Gines R. Association of a lordosis–scoliosis–kyphosis deformity in gilthead seabream (Sparus aurata) with family structure. Fish Phys. Biochem. 2000;22:159–163. doi:10.1023/A:1007811702624 [Google Scholar]
- Amos W, Worthington Wilmer J, Fullard K, Burg T.M, Croxall J.P, Bloch D, Coulson T. The influence of parental relatedness on reproductive success. Proc. R. Soc. B. 2001;268:2021–2027. doi: 10.1098/rspb.2001.1751. doi:10.1098/rspb.2001.1751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayres K.L, Overall A.D.J. Api-Calc 1.0: a computer program for calculating the average probability of identity allowing for substructure, inbreeding and the presence of close relatives. Mol. Ecol. Notes. 2004;4:315–318. doi:10.1111/j.1471-8286.2004.00616.x [Google Scholar]
- Blaustein A.R, Kiesecker J.M, Chivers D.P, Anthony R.G. Ambient UV-B radiation causes deformities in amphibian embryos. Proc. Natl Acad. Sci. USA. 1997;94:13 735–13 737. doi: 10.1073/pnas.94.25.13735. doi:10.1073/pnas.94.25.13735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bos D.H, DeWoody J.A. Molecular characterization of major histocompatibility complex class II alleles in wild tiger salamanders (Ambystoma tigrinum) Immunogenetics. 2005;57:775–781. doi: 10.1007/s00251-005-0038-5. doi:10.1007/s00251-005-0038-5 [DOI] [PubMed] [Google Scholar]
- Bos D.H, Gopurenko D, Williams R.N, DeWoody J.A. Inferring population history and demography using microsatellites, mitochondrial DNA, and major histocompatibility complex (MHC) genes. Evolution. 2008;62:1458–1468. doi: 10.1111/j.1558-5646.2008.00364.x. doi:10.1111/j.1558-5646.2008.00364.x [DOI] [PubMed] [Google Scholar]
- DeWoody J.A, Avise J.C. Microsatellite variation in marine, freshwater and anadromous fishes compared with other animals. J. Fish Biol. 2000;56:461–473. doi:10.1111/j.1095-8649.2000.tb00748.x [Google Scholar]
- Gopurenko D, Williams R.N, DeWoody J.A. Reproductive and mating success in the small-mouthed salamander (Ambystoma texanum) estimated via microsatellite parentage analysis. Evol. Biol. 2007;34:130–139. doi:10.1007/s11692-007-9009-0 [Google Scholar]
- Hall T.A. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999;41:95–98. [Google Scholar]
- Johnson P.T.J, Lunde K.B, Haight R.W, Bowerman J, Blaustein A.R. Ribeiroia ondatrae (Trematoda: Digenea) infection induces severe limb malformation in western toads (Bufo boreas) Can. J. Zool. 2001;79:370–379. doi:10.1139/cjz-79-3-370 [Google Scholar]
- Keller L.F, Waller D.M. Inbreeding effects in wild populations. Trends Ecol. Evol. 2002;17:230–241. doi:10.1016/S0169-5347(02)02489-8 [Google Scholar]
- Lewis, P. O. & Zaykin, D. 1999 Genetic data analysis: computer program for the analysis of allelic data, v. 1.0 (d12). Storrs, CT: Department of Ecology and Evolutionary Biology, The University of Connecticut.
- Mansfield K.G, Land E.D. Cryptorchidism in Florida panthers: prevalence, features, and influence of genetic restoration. J. Wildl. Dis. 2002;38:693–698. doi: 10.7589/0090-3558-38.4.693. [DOI] [PubMed] [Google Scholar]
- Marshall T.C, Slate J, Kruuk L.E.B, Pemberton J.M. Statistical confidence for likelihood-based paternity inference in natural populations. Mol. Ecol. 1998;7:639–655. doi: 10.1046/j.1365-294x.1998.00374.x. doi:10.1046/j.1365-294x.1998.00374.x [DOI] [PubMed] [Google Scholar]
- Meyer-Rochow V, Asahima M. Naturally occurring morphological abnormalities in wild populations of the Japanese newt Cynops pyrrhogaster (salamandridae: Urodela: Amphibia) Zool. Anz. 1988;221:70–80. [Google Scholar]
- Olsson M, Gullberg A, Tegelstrom H. Malformed offspring, sibling matings, and selection against inbreeding in the sand lizard (Lacerta agilis) J. Evol. Biol. 1996;9:229–242. doi:10.1046/j.1420-9101.1996.9020229.x [Google Scholar]
- Peakall R, Smouse P.E. Genelex 6: genetic analysis in Excel. Population genetics software for teaching and research. Mol. Ecol. Notes. 2006;6:288–295. doi: 10.1093/bioinformatics/bts460. doi:10.1111/j.1471-8286.2005.01155.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider, S., Roessli, D. & Excoffier, L. 2000 Arlequin: a software for population genetics data analysis (v. 3.01). Geneva, Switzerland: University of Geneva, Department of Anthropology.
- Sessions S.K, Ruth S.B. Explanation for naturally occurring supernumary limbs in amphibians. J. Exp. Zool. 1990;254:38–47. doi: 10.1002/jez.1402540107. doi:10.1002/jez.1402540107 [DOI] [PubMed] [Google Scholar]
- Tennessen J.A, Zamudio K.R. Early male reproductive advantage, multiple paternity and sperm storage in an amphibian aggregate breeder. Mol. Ecol. 2003;12:1567–1576. doi: 10.1046/j.1365-294x.2003.01830.x. doi:10.1046/j.1365-294X.2003.01830.x [DOI] [PubMed] [Google Scholar]
- Weir B.S, Cockerham C.C. Estimating F-statistics for analysis of populations structure. Evolution. 1984;38:1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. doi:10.2307/2408641 [DOI] [PubMed] [Google Scholar]
- Williams, R. N. 2007 Population biology, genetics, and sexual selection in wild Ambystomatid salamanders, 120 pp. PhD dissertation, Purdue University.