Abstract
The classical approach to predicting the geographical extent of species invasions consists of training models in the native range and projecting them in distinct, potentially invasible areas. However, recent studies have demonstrated that this approach could be hampered by a change of the realized climatic niche, allowing invasive species to spread into habitats in the invaded ranges that are climatically distinct from those occupied in the native range. We propose an alternative approach that involves fitting models with pooled data from all ranges. We show that this pooled approach improves prediction of the extent of invasion of spotted knapweed (Centaurea maculosa) in North America on models based solely on the European native range. Furthermore, it performs equally well on models based on the invaded range, while ensuring the inclusion of areas with similar climate to the European niche, where the species is likely to spread further. We then compare projections from these models for 2080 under a severe climate warming scenario. Projections from the pooled models show fewer areas of intermediate climatic suitability than projections from the native or invaded range models, suggesting a better consensus among modelling techniques and reduced uncertainty.
Keywords: niche-based modelling, niche shift, climate change, spotted knapweed
1. Introduction
Biological invasions represent a growing threat to biodiversity (Pimentel et al. 2000). Once introduced species are established, they become difficult to eradicate (Genovesi 2005), thus, preventing future invasions is the most cost-effective form of management.
Niche-based models (NBM, Guisan & Thuiller 2005) are increasingly used to estimate risks of biological invasions (e.g. Thuiller et al. 2005). Two crucial assumptions are made when applying these models. First, the species is assumed to be in equilibrium with its environment in the range used to train the model, and second, the environmental niche of the species is assumed to be conserved across space and time (Wiens & Graham 2005; Pearman et al. 2008).
So far, NBM approaches have developed models by using observations either from the invaded range (e.g. Mau-Crimmins et al. 2006) or from the native range (e.g. Peterson & Vieglais 2001; Thuiller et al. 2005) and then predicted the potential extent of invasions. An obvious problem with the first approach is that in the invaded range, the invasion process may not be completed and thus, the invading species may not yet occupy all suitable environments (Wilson et al. 2007). Such a violation of the equilibrium assumption can subsequently bias the model, under-predicting the full potential for invasion. A problem with the second approach is that the ecological requirements of the species might have changed during the invasion process (Pearman et al. 2008), thus violating the assumption of niche conservatism.
We recently evidenced (Broennimann et al. 2007) a shift of the climatic niche of spotted knapweed (Centaurea maculosa Lam.), a weed introduced in the 1890s from Europe into western North America, where it now infests over 3×106 ha of disturbed and natural grassland habitats (see the electronic supplementary material, S2). A consequence of this difference was that models fitted in the native range could successfully predict areas of introduction in North America, but were unable to predict the full extent of invasion. Current NBM approaches may thus be inadequate for several invasive species. Moreover, distinct model training strategies may not only yield distinct predictions in the present, but also on future predictions in warmer climates (Araujo et al. 2005).
Here, we assess uncertainties in predictions of spotted knapweed invasion obtained with models trained in (i) the native range, (ii) the invaded range and (iii) both ranges, as similarly explored by Kriticos & Randall (2001). We further investigate the outcome of these training strategies with regard to the outcome of predictions under a severe warming scenario by 2080 (HadCM-SRES-A1FI; Nakicenovic & Swart 2000). Attempts to predict future distributions of invasive species in a warmer climate are few (e.g. Beerling 1993).
2. Material and methods
(a) Species occurrence data
We used the occurrence database described in Broennimann et al. (2007), complemented with occurrences from Canada (www.eflora.bc.ca), eastern USA and Russia (H. Müller-Schärer 2007, unpublished data), resulting in a robust cross-continental dataset of 373 occurrences for Europe and 2972 occurrences for North America. A corresponding number of pseudo-absences were sampled randomly (see Elith et al. 2006) across both native and invaded ranges.
(b) Climate data
We used the CRU05 climate data at 0.5° (New et al. 2000) to derive the following set of meaningful predictors (Guisan & Thuiller 2005): mean annual temperature (tAN), annual sum of precipitation (pAN), mean annual daily temperature range (dtrAN), minimum temperature (tMIN), maximum temperature (tMAX), total precipitation of the wettest quarter (pWETQ), total precipitation of the driest quarter (pDRYQ) and potential evaporation (pet; following Samani & Pessarakli 1986). The HadCM-A1FI climate scenario anomalies were retrieved from the CRU-TS2 dataset (Mitchell et al. 2004) and added to present climate maps to obtain future climate predictors for 2080.
(c) Statistical modelling
Models were fitted and evaluated using a standard split-sample strategy. Models were trained using three subsets (70%) of occurrence data, (i) from Europe (modEU), (ii) from North America (modNA) only and (iii) from both ranges (modEUNA). We used the same Biomod modelling framework (Thuiller 2003) as Broennimann et al. (2007), fitting for each species, generalized linear models (GLM), generalized additive models (GAM), boosted regression trees and random forest (RF). Models were validated in each case with the remaining 30% independent data (Thuiller 2003). The whole training-evaluation procedure was repeated 100 times. The predictive power of the models was tested using the area under the receiver operator characteristic function (area under the curve criteria, AUC; Thuiller 2003; Elith et al. 2006; electronic supplementary material, S1). Models were projected into future climates to generate species distribution maps for 2080.
3. Results
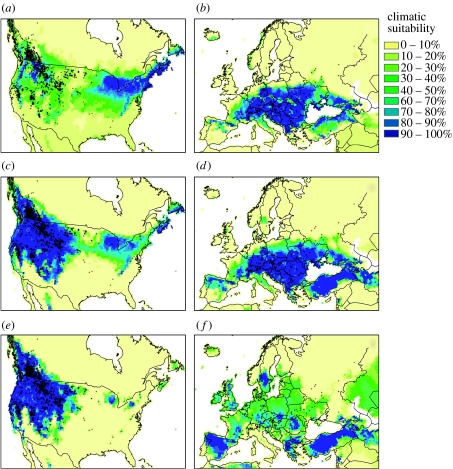
The modEU models (figure 1a,b) predict the distribution significantly better in EU than in NA (table 1). In NA, they correctly predict areas of introduction (Victoria, British Columbia) but not the full invasion extent. They also predict areas in eastern NA, where the plant is present but still infrequent. Similarly, the modNA models (figure 1e,f) predict the distribution significantly better in NA than in EU (Wilcoxon signed-rank tests; table 1). They predict the species in southern EU where it is absent and fail to predict most of its actual native distribution. The modEUNA models (figure 1c,d) predict in both ranges as accurately as modEU and modNA predict in EU and NA, respectively (table 1).
Figure 1.
Geographical predictions of spotted knapweed at the present time. (a,b) Models calibrated in Europe only (modEU), (c,d) in both North America and Europe (modEUNA) and (e,f) North America only (modNA) are projected to North America (a,c,e) and Europe (b,d,f). The climatic suitability (yellow to blue scale) indicates the percentage of models (all techniques pooled) predicting the species in the present. Dots represent known occurrences.
Table 1.
Model accuracy on evaluation dataset (30% independent data) using AUC. (Standard deviations (±) indicate the variability of model accuracy through 100 iterations. AUC values for models calibrated in Europe (EU), North America (NA) and both (EU+NA) are indicated for each modelling technique. The differences in model accuracy between ranges within and between models were tested with a paired Wilcoxon signed-rank test (n.s., not significant; *0.05<p<1×10−5; **p<1×10−5).)
| calibration in EU | ↔ | calibration in EU+NA | ↔ | calibration in NA | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| NA | ↔ | EU | EU | ↔ | NA | NA | ↔ | EU | |||
| GLM | 0.77±0.07 | ** | 0.87±0.03 | n.s. | 0.86±0.03 | ** | 0.96±0.01 | n.s. | 0.96±0.01 | ** | 0.73±0.04 |
| GAM | 0.89±0.05 | * | 0.88±0.03 | n.s. | 0.87±0.03 | ** | 0.96±0.01 | n.s. | 0.96±0.01 | ** | 0.70±0.04 |
| GBM | 0.85±0.02 | ** | 0.89±0.03 | n.s. | 0.88±0.03 | ** | 0.96±0.01 | n.s. | 0.97±0.01 | ** | 0.76±0.04 |
| RF | 0.84±0.03 | ** | 0.90±0.02 | n.s. | 0.90±0.03 | ** | 0.97±0.01 | n.s. | 0.98±0.01 | ** | 0.76±0.03 |
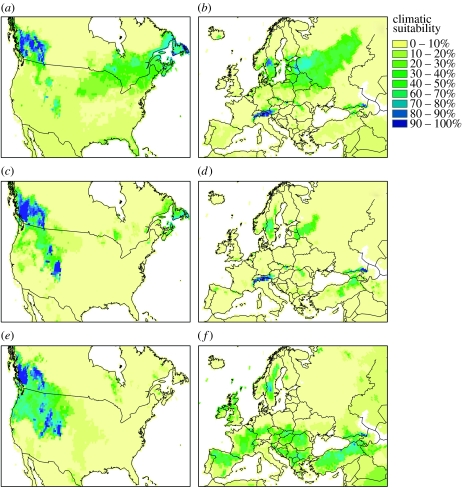
When projected for 2080, the three strategies yielded distinct predictions (figure 2). The modEU predictions were correlated 66% with the modEUNA predictions (66% in EU, 62% in NA) and only 40% with modNA predictions (45% in EU, 32% in NA). The modEUNA predictions were correlated 76% with the modNA predictions (79% in EU, 73% in NA). In EU, the modEU models predicted the species to persist in the Alps and to colonize Sweden, the Baltic countries, Caucasus and north-eastern Russia (figure 2b), whereas it predicted the colonization of British Columbia, Quebec and Labrador in the invaded range. In contrast, the modNA models predicted a decreased suitability in the EU and the migration of the species to British Columbia and the Central Rockies in NA. The models calibrated on both ranges indicated convergent patterns of distribution in EU and NA when compared with modEU and modNA alone (comparing figure 2d with b and c with e).
Figure 2.
Geographical predictions of spotted knapweed by 2080 under HadCM-A1FI warming climate scenario (description of the models and colour scale are the same as given in figure 1).
4. Discussion
Studying invasive species in their native and invaded ranges offers new opportunities for addressing theoretical challenges associated with niche-based modelling and the prediction of biological invasions. As we showed, a main challenge is the uncertainty of conservatism of the climatic niche between ranges, whether observed changes result from a modification of the realized niche and/or the fundamental niche (Pearman et al. 2008), and how these can further affect predictions of biological invasions.
Using spotted knapweed as a model species, we confirm that the current practice, consisting of training models in the native range, fails to predict the full extent of biological invasions (Broennimann et al. 2007). As a better alternative, we propose that models be trained using data from both native and invaded ranges, which ensures that the models can predict the future spread of the species in areas of the invaded range that are not yet invaded but where suitable conditions similar to the native range occur. We show that this approach yields accurate predictions across both ranges (figure 1, table 1). Furthermore, the prediction of the extent of invasion in North America improves on models based solely on the distribution in the native range and equally performs on models based on the invaded range, while ensuring the inclusion of areas with similar climate to the European niche, where the species is likely to spread further.
There are several factors that could contribute to the improvement demonstrated by our approach. In the native range, biotic constraints (e.g. competitors, herbivores) can exclude the species from part of the climatic envelope within which it could otherwise grow and reproduce (i.e. the fundamental niche is constrained to the realized niche). In the invaded range, however, many of these natural enemies are absent (e.g. specialist root-feeding herbivores; Müller 1989). Such biotic release may allow the species to occupy climatic situations (i.e. additional parts of the fundamental niche) from which it is excluded in the native range. Models trained in the native range will fail to account for these newly occupied climates. By taking both ranges into account in the training process, the niche implicitly fitted can be expanded or shifted to express a different part of the fundamental niche in the invaded range. Conversely, the pooled model should indicate, in Europe, areas where the species does not occur, potentially due to these negative biotic interactions. Interestingly, figure 1d reveals such areas in Turkey and southern and eastern Ukraine where a potential competitor—Centaurea diffusa—is present. One step further would be to build projections with a model based on worldwide occurrences. Further testing on a larger number of invasive species is also required to validate such a pooled approach.
Although less likely than a change of the realized niche, on which these models are implicitly based, one cannot exclude the possibility that the fundamental niche might also have evolved in the invaded range, due to evolutionary processes taking place after introduction (Dietz & Edwards 2006; Pearman et al. 2008). Disentangling changes in the realized niche versus fundamental niche is currently impossible based on empirical distribution data only. Further contributions from experimentalists and ecophysiologists are required.
Future predictions of invasions in a warmer climate based on the pooled approach exhibit similar patterns to the predictions based on models trained and projected within the same range (figure 2). However, the pooled approach significantly reduces areas of future predictions, forecasting a much reduced invasion extent for the species by 2080. Interestingly, the reduction mainly concerns areas with limited consensus between modelling techniques, thus reducing uncertainty. However, our projections cannot be compared to future species occurrence data. A strong test would be to predict current patterns of invasions from data on past invasions, but dated observations are currently missing for such analysis.
Acknowledgments
We thank Peter Rice and Townsend Peterson for providing occurrence data, Wilfried Thuiller and Robin Engler for providing improved versions of Biomod scripts and Dorothea Pio and Peter Pearman, Wilfried Thuiller and two anonymous referees for useful comments on the manuscript. This project was funded by the NCCR ‘Plant Survival’ (Swiss National Science Foundation).
Footnotes
One contribution of 12 to a Special Feature on ‘Global change and biodiversity: future challenges’.
Supplementary Material
Additional description of methods
Additional description of invasion history of spotted knapweed
Detailed discussion of the contributions of climatic predictors
Importance of the climatic variables
References
- Araújo M.B, Whittaker R.J, Ladle R.J, Erhard M. Reducing uncertainty in projections of extinction risk from climate change. Glob. Ecol. Biogeogr. 2005;14:529–538. doi:10.1111/j.1466-822X.2005.00182.x [Google Scholar]
- Beerling D.J. The impact of temperature on the northern distribution-limits of the introduced species Fallopia japonica and Impatiens glandulifera in north-west Europe. J. Biogeogr. 1993;20:45–53. doi:10.2307/2845738 [Google Scholar]
- Broennimann O, Treier U.A, Müller-Schärer H, Thuiller W, Peterson A.T, Guisan A. Evidence of climatic niche shift during biological invasion. Ecol. Lett. 2007;10:701–709. doi: 10.1111/j.1461-0248.2007.01060.x. doi:10.1111/j.1461-0248.2007.01060.x [DOI] [PubMed] [Google Scholar]
- Dietz H, Edwards P.J. Recognition that causal processes change during plant invasion helps explain conflicts in evidence. Ecology. 2006;87:1359–1367. doi: 10.1890/0012-9658(2006)87[1359:rtcpcd]2.0.co;2. doi:10.1890/0012-9658(2006)87[1359:RTCPCD]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Elith J, et al. Novel methods improve prediction of species' distributions from occurrence data. Ecography. 2006;29:129–151. doi:10.1111/j.2006.0906-7590.04596.x [Google Scholar]
- Genovesi P. Eradications of invasive alien species in Europe: a review. Biol. Invasions. 2005;7:127–133. doi:10.1007/s10530-004-9642-9 [Google Scholar]
- Guisan A, Thuiller W. Predicting species distribution: offering more than simple habitat models. Ecol. Lett. 2005;8:993–1009. doi: 10.1111/j.1461-0248.2005.00792.x. doi:10.1111/j.1461-0248.2005.00792.x [DOI] [PubMed] [Google Scholar]
- Kriticos D.J, Randall R.P. A comparison of systems to analyse potential weed distributions. In: Groves R.H, Panetta F.D, Virtue J.G, editors. Weed risk assessment. CSIRO Publishing; Collingwood, Australia: 2001. pp. 61–82. [Google Scholar]
- Mau-Crimmins T.M, Schussman H.R, Geiger E.L. Can the invaded range of a species be predicted sufficiently using only native-range data? Lehmann lovegrass (Eragrostis lehmanniana) in the southwestern United States. Ecol. Model. 2006;193:736–746. doi:10.1016/j.ecolmodel.2005.09.002 [Google Scholar]
- Mitchell, T. D., Carter, T. R., Jones, P. D., Hulme, M. & New, M. 2004 A comprehensive set of high-resolution grids of monthly climate for Europe and the globe: the observed record (1901–2000) and 16 scenarios (2001–2100). Tyndall Centre for Climate Change Research, University of East Anglia.
- Müller H. Structural analysis of the phytophagous insect guilds associated with the roots of Centaurea maculosa Lam., C. diffusa Lam., and C. vallesiaca Jordan in Europe: 1. Field observations. Oecologia. 1989;78:41–52. doi: 10.1007/BF00377196. doi:10.1007/BF00377196 [DOI] [PubMed] [Google Scholar]
- Nakicenovic, N. & Swart, R. 2000 Emissions scenarios. Special report of the Intergovernmental Panel on Climate Change, Cambridge University Press.
- New M, Hulme M, Jones P. Representing twentieth-century space–time climate variability. Part II: development of 1901–96 monthly grids of terrestrial surface climate. J. Clim. 2000;13:2217–2238. doi:10.1175/1520-0442(2000)013<2217:RTCSTC>2.0.CO;2 [Google Scholar]
- Pearman P.B, Guisan A, Broennimann O, Randin C.F. Niche dynamics in space and time. Trends Ecol. Evol. 2008;23:149–158. doi: 10.1016/j.tree.2007.11.005. doi:10.1016/j.tree.2007.11.005 [DOI] [PubMed] [Google Scholar]
- Peterson A.T, Vieglais D.A. Predicting species invasions using ecological niche modeling: new approaches from bioinformatics attack a pressing problem. Bioscience. 2001;51:363–371. doi:10.1641/0006-3568(2001)051[0363:PSIUEN]2.0.CO;2 [Google Scholar]
- Pimentel D, Lach L, Zuniga R, Morrison D. Environmental and economic costs of nonindigenous species in the United States. Bioscience. 2000;50:53–65. doi:10.1641/0006-3568(2000)050[0053:EAECON]2.3.CO;2 [Google Scholar]
- Samani Z.A, Pessarakli M. Estimating potential crop evapotranspiration with minimum data in Arizona. Trans. ASAE. 1986;29:522–524. [Google Scholar]
- Thuiller W. Biomod: optimizing predictions of species distributions and projecting potential future shifts under global change. Glob. Change Biol. 2003;9:1353–1362. doi: 10.1111/gcb.12728. doi:10.1046/j.1365-2486.2003.00666.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thuiller W, Richardson D.M, Pyšek P, Midgley G.F, Hughes G.O, Rouget M. Niche-based modelling as a tool for predicting the risk of alien plant invasions at a global scale. Glob. Change Biol. 2005;11:2234–2250. doi: 10.1111/j.1365-2486.2005.001018.x. doi:10.1111/j.1365-2486.2005.001018.x [DOI] [PubMed] [Google Scholar]
- Wiens J.J, Graham C.H. Niche conservatism: integrating evolution, ecology, and conservation biology. Annu. Rev. Ecol. Evol. Syst. 2005;36:519–539. doi:10.1146/annurev.ecolsys.36.102803.095431 [Google Scholar]
- Wilson J.R.U, Richardson D.M, Rouget M, Procheş Ş, Amis M.A, Henderson L, Thuiller W. Residence time and potential range: crucial considerations in modelling plant invasions. Diversity Distrib. 2007;13:11–22. doi:10.1111/j.1472-4642.2006.00302.x [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional description of methods
Additional description of invasion history of spotted knapweed
Detailed discussion of the contributions of climatic predictors
Importance of the climatic variables