Abstract
The trend for increasing biodiversity from the poles to the tropics is one of the best-known patterns in nature. This latitudinal biodiversity gradient has primarily been documented so far with extant species as the measure of biodiversity. Here, we evaluate the global pattern in biodiversity across latitudes based on the magnitude of genetic population divergence within plant species, using a robust spatial design to compare published allozyme datasets. Like the pattern of plant species richness across latitudes, we expected the divergence among populations of current plant species would have a similar pattern and direction. We found that lower latitudinal populations showed greater genetic differentiation within species after controlling for geographical distance. Our analyses are consistent with previous population-level studies in animals, suggesting a high possibility of tropical peaks in speciation rates associated with observed levels of species richness.
Keywords: genetic population divergence, latitudinal biodiversity gradient, Nei's genetic distance, speciation
1. Introduction
The latitudinal biodiversity gradient (LBG) is one of the best-known patterns in nature. This pattern of equatorial peak in biodiversity has been documented from a range of taxonomic groups in animals and plants, identified in the fossil records extending back to the Palaeozoic (325 Ma) and reported in a diverse array of environments (e.g. forests, grasslands, wetlands, fresh waters, deep seas) across a range of spatial scales (Rosenzweig 1995; Gaston & Blackburn 2000; Willig et al. 2003; Hillebrand 2004; Mittelbach et al. 2007). However, despite robust evidence of the LBG, we have little knowledge about latitudinal patterns in the diversity of genes or populations because most studies that have examined the LBG so far have focused on extant species richness (or higher taxa) as a unit of biodiversity (Gaston 2000).
Examining the latitudinal variation of intraspecific population divergence significantly advances our understanding of the generality of the LBG, because the level of population differentiation may correspond with incipient speciation and future species richness (Grant 1981; Avise 2000; Levin 2000). Despite rapid accumulation of population genetic and phylogeographic studies, there have been few comprehensive examinations of population divergences from a latitudinal perspective (but see Martin & McKay 2004; Allen et al. 2006). Here, we evaluated the LBG by examining genetic divergence among plant populations. We assumed that, as with the pattern of increasing species richness towards the equator (Currie & Paquin 1987; Davies et al. 2004; Allen & Gillooly 2006; Jablonski et al. 2006; Wright et al. 2006; but see Weir & Schluter 2007), the divergence among populations of extant species would have a similar pattern and direction (Martin & McKay 2004). Despite the considerable focus on hybridization and polyploidy as main factors driving plant speciation (Arnold 1997; Rieseberg & Wendel 2004), divergence among populations within a species may be considered a starting point of speciation (Coyne & Orr 2004). Here, we test the hypothesis that higher latitudinal populations within species show smaller genetic differentiation, using previous allozyme studies of plants. These data should illustrate whether the process of the LBG per se (when extinction and immigration/emigration rates are equal across latitudes) is happening on multiple scales (within as well as among species).
2. Material and methods
We reviewed individual studies in the supplemental sources cited in Cole (2003), which summarized allozyme genetic variation in plants. Although the data from other markers, such as sequence and AFLP (Amplified Fragment Length Polymorphism), for testing genetic variation are becoming more widely available, the largest appropriate dataset for plants is still from allozyme-based studies. We limited our meta-analysis to intraspecific studies with at least four populations, where tables for pairwise genetic distances among populations were given and where geographical information for all analysed populations was available. For studies containing both mainland and island populations, we included only mainland populations for analyses because islands often show extremes in genetic and phenotypic evolution. Additional studies were collected from searches of ‘Web of Science’ covering 1996–2005. Keyword strings included ‘genetic variation’, ‘genetic diversity’, ‘genetic divergence’, ‘genetic structure’, ‘phylogeography’, ‘gene flow’, ‘population genetics’ or ‘conservation genetics’, in conjunction with ‘allozyme’ or ‘isozyme’ in the title field. Forty-five datasets were compiled after 458 studies or taxa, revealed by the search, were checked for the above criteria.
We generally followed the methods of Martin & McKay (2004) to test the hypothesis that higher latitudinal populations within species show smaller pairwise genetic differentiation. We used either latitudinal data shown in each paper or geographical information, such as sampling location maps, to record latitude for each population and to calculate pairwise geographical distance between populations for each species. For a measure of pairwise genetic divergence, we used Nei's genetic distance as reported by the original authors in each paper. Nei's genetic distance (DN) is defined as , where pix is the frequency of allele i in population x; piy is the frequency of allele i in population y; and ln is logarithm to the base e (Nei 1987). Then, we estimated distance-controlled genetic differentiation (i.e. Nei's genetic distance/geographical distance in km) because geographical distance could create confounding effects on genetic differentiation. The populations within each species were separated into ‘high’ and ‘low’ latitudinal regions based on their location relative to the latitudinal midpoint of a species' sampled range (see fig. 1 in Martin & McKay 2004). This minimizes potential biases introduced by using the mean latitude of all sampled populations, particularly in cases in which sampled populations are not evenly distributed across latitudes. Owing to the limited numbers of sampled populations in many source papers, we could not further subdivide the dataset latitudinally without losing information.
To compare the magnitude of genetic differentiation between higher and lower latitudinal regions, we used analysis of variance with mean values of pairwise distance-controlled genetic differentiation as the dependent variable and with latitudinal regions as a treatment and species as a block. We used a linear regression with standardized differences between higher and lower latitudinal population differentiations to assay whether our results were affected by the latitudinal midpoints of the sampled taxa. Mean values of the distance-controlled genetic differentiation were log-transformed prior to analysis to normalize the distributions.
3. Results
In total, we analysed data on 409 populations, spanning from 65° N to 43° S latitude, of 45 taxa from Asia, Australia, Europe, North America and South America. These data are summarized in table S1 of the electronic supplementary material.
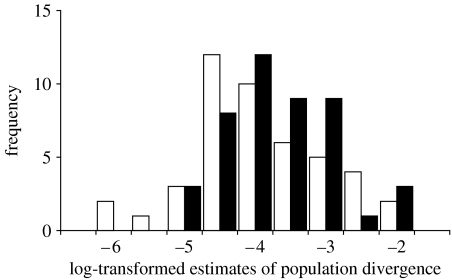
Our results indicated differences among distance-controlled genetic population differentiation for two latitudinal regions (table 1a). We found the expected pattern that lower latitudinal populations within species showed greater genetic differentiation in 31 out of the 45 taxa examined (overall 69%, Northern Hemisphere 68% and Southern Hemisphere 80%). The mean log-transformed distance-controlled estimates of population divergence were −3.40±0.79 and −3.65±0.94 for lower and higher latitudes, respectively (frequency distributions of population divergence estimates are depicted in figure 1). Although the effect size for latitude was not large (Cohen's d=0.29; 95% CI=0.06–0.56; Cohen 1988), this result was similar to latitudinal difference in vertebrate population divergence (Cohen's d=0.29; Martin & McKay 2004).
Table 1.
Effect of latitude and species on (a) distance-controlled genetic population differentiation and (b) geographical distance in plant species.
| d.f. | F | p | |
|---|---|---|---|
| (a) | |||
| latitude | 1 | 6.76 | 0.012 |
| species | 44 | 6.23 | <0.001 |
| (b) | |||
| latitude | 1 | 2.35 | 0.132 |
| species | 44 | 13.33 | <0.001 |
Figure 1.
Frequency distribution of log-transformed distance-controlled estimates of population divergence in lower (closed bars) and higher (open bars) latitudinal groups.
Mean geographical distance among lower latitudinal populations was not different from that among higher latitudinal populations (table 1b). The linear regression indicated no significant relationship between latitudinal midpoints of the examined taxa and the standardized differences between higher and lower latitudinal population differentiations (R2=0.0006, p>0.05). Therefore, it is unlikely that the observed pattern resulted from bias in geographical distances between the sampled populations or from latitudinal midpoints of examined taxa.
4. Discussion
We found that lower latitudinal populations within species showed greater genetic differentiation. Although the causal link among high species richness near the equator, high speciation rate and high population differentiation is not necessarily straightforward (Allen & Gillooly 2006; Jablonski et al. 2006; Wright et al. 2006; but see Weir & Schluter 2007), we would argue that genetic divergence among populations is positively related to speciation rates or species diversity (Turelli et al. 2001; Martin & McKay 2004; Kelly & Eernisse 2007). The greater genetic divergence should imply greater evolutionary independence, which may increase the chance that evolutionary processes can lead to speciation (Martin & McKay 2004). Thus, our results suggest that greater evolutionary independence in lower latitudes may promote speciation or future species diversity, reflecting currently observed latitudinal gradient in species richness.
Often, researchers have examined the LBG with evidence from species- or higher level studies based on comparative methods (e.g. Wright et al. 2006). Although species-level approaches have provided evidence for the generality of the LBG, they have several limitations. For example, current taxonomy or phylogeny includes more data in the temperate regions than in the tropics; thus important intraspecific but almost interspecific level variations in currently described species in the tropics may have been disregarded (Chek et al. 2003; Mittelbach et al. 2007). Such non-equivalent comparisons at species or higher level may be confounded by variation in taxonomic practice, ecology or different methods across taxa or original studies (Martin & McKay 2004). We used a robust spatial design to compare populations within species and treated populations independently when using data from distinct original studies. Therefore, our approach may have an advantage over species-level comparative methods because it controls for bias caused by potential disparities in such comparisons between tropical and temperate regions.
Although there has been an emphasis on natural selection (e.g. adaptation to different habitats) or other mechanisms (e.g. hybridization and polyploidy) as the main forces driving diversification or speciation in plants (Rieseberg & Wendel 2004), geographical divergence of populations within species is typically the first step in the origin of species and serves as an intuitive starting point for discussion of speciation (Grant 1981; Levin 2000; Coyne & Orr 2004). Plants are appropriate organisms for exploring the molecular effects of latitudinal variation in climate because they are sedentary ectotherms (Wright et al. 2006). Moreover, Barraclough & Savolainen (2001) revealed a link between the rate of neutral molecular change within populations and the evolution of species diversity in flowering plants, showing that rates of amino acid and structural ribosomal DNA substitutions, morphological change and diversification rates, all of them correlate with the neutral substitution rate based on a large taxonomically diverse sample of angiosperms. However, more work is necessary to isolate the effects of population divergence from other speciation mechanisms (e.g. polyploidy) and to estimate their relative contribution to the latitudinal gradient in plant species richness.
Our study was not intended to address causal mechanisms of this pattern. In this meta-analysis, we cannot distinguish whether higher divergence is due to lower gene flow or smaller population size, or whether this pattern is driven by faster molecular evolution or a longer time since divergence. Such causal mechanisms should be assessed using well-supported phylogeographic data by DNA sequences representing each population, in conjunction with population dynamics and ecological data, based on sufficient numbers of populations spanning each species' latitudinal range. For example, Martin & McKay (2004) tested a hypothesis that the pattern reflects recolonization history of higher latitudes after glaciation and found that subtraction of glaciated populations in their analysis did not affect the results. Unfortunately, our sample size after removing those populations was too small to analyse it. However, our results are also consistent with previous studies using vertebrate taxa (Martin & McKay 2004), suggesting that greater differentiation at lower latitudes is a common feature of plant and vertebrate populations and may be associated with increased likelihood of speciation. Although the latitudinal effect size is not large, the observed pattern is compatible with other studies across multiple scales: more isolated structure in tropical populations (Chek et al. 2003; Martin & McKay 2004); more subspecies in lower latitudes within species (P. R. Martin 2008, personal communication); increased frequency of smaller ranged endemics resulting from allopatric speciation in lower latitudes (Gentry 1986); and, finally, greater species richness towards the equator (Gaston 2000). In this vein, identifying the pattern of population divergence along latitudinal gradients is a critical step towards understanding the generality of the evolutionary dynamics underlying the LBG.
Acknowledgments
We thank M. Conroy, D. Promislow and three anonymous reviewers for their helpful comments on the manuscript.
Supplementary Material
Geographic and genetic data for plant species examined in this study
References
- Allen A.P, Gillooly J.F. Assessing latitudinal gradients in speciation rates and biodiversity at the global scale. Ecol. Lett. 2006;9:947–954. doi: 10.1111/j.1461-0248.2006.00946.x. doi:10.1111/j.1461-0248.2006.00946.x [DOI] [PubMed] [Google Scholar]
- Allen A.P, Gillooly J.F, Savage V.M, Brown J.H. Kinetic effects of temperature on rates of genetic divergence and speciation. Proc. Natl Acad. Sci. USA. 2006;103:9130–9135. doi: 10.1073/pnas.0603587103. doi:10.1073/pnas.0603587103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold M.L. Oxford University Press; Oxford, UK: 1997. Natural hybridization and evolution. [Google Scholar]
- Avise J.C. Harvard University Press; Cambridge, MA: 2000. Phylogeography: the history and formation of species. [Google Scholar]
- Barraclough T.G, Savolainen V. Evolutionary rates and species diversity in flowering plants. Evolution. 2001;55:677–683. doi: 10.1554/0014-3820(2001)055[0677:erasdi]2.0.co;2. doi:10.1111/j.0014-3820.2001.tb00803.x [DOI] [PubMed] [Google Scholar]
- Chek A.A, Austin J.D, Lougheed S.C. Why is there a tropical-temperate disparity in the genetic diversity and taxonomy of species? Evol. Ecol. Res. 2003;5:69–77. [Google Scholar]
- Cohen J. Lawrence Erlbaum; Hillsdale, NJ: 1988. Statistical power analysis for behavioral sciences. [Google Scholar]
- Cole C.T. Genetic variation in rare and common plants. Annu. Rev. Ecol. Evol. Syst. 2003;34:213–237. doi:10.1146/annurev.ecolsys.34.030102.151717 [Google Scholar]
- Coyne J.A, Orr H.A. Sinauer; Sunderland, MA: 2004. Speciation. [Google Scholar]
- Currie D.J, Paquin V. Large-scale biogeographical patterns of species richness of trees. Nature. 1987;329:326–327. doi:10.1038/329326a0 [Google Scholar]
- Davies T.J, Savolainen V, Chase M.W, Moat J, Barraclough T.G. Environmental energy and evolutionary rates in flowering plants. Proc. R. Soc. B. 2004;271:2195–2200. doi: 10.1098/rspb.2004.2849. doi:10.1098/rspb.2004.2849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaston K.J. Global patterns in biodiversity. Nature. 2000;405:220–227. doi: 10.1038/35012228. doi:10.1038/35012228 [DOI] [PubMed] [Google Scholar]
- Gaston K.J, Blackburn T.M. Blackwell; Malden, MA: 2000. Pattern and process in macroecology. [Google Scholar]
- Gentry A.H. Endemism in tropical versus temperate plant communities. In: Soule M.E, editor. Conservation biology: the science of scarcity and diversity. Sinauer; Sunderland, MA: 1986. pp. 153–181. [Google Scholar]
- Grant V. Columbia University Press; New York, NY: 1981. Plant speciation. [Google Scholar]
- Hillebrand H. On the generality of the latitudinal diversity gradient. Am. Nat. 2004;163:192–211. doi: 10.1086/381004. doi:10.1086/381004 [DOI] [PubMed] [Google Scholar]
- Jablonski D, Roy K, Valentine J.W. Out of the tropics: evolutionary dynamics of the latitudinal diversity gradient. Science. 2006;314:102–106. doi: 10.1126/science.1130880. doi:10.1126/science.1130880 [DOI] [PubMed] [Google Scholar]
- Kelly R.P, Eernisse D.J. Southern hospitality: a latitudinal gradient in gene flow in the marine environment. Evolution. 2007;61:700–707. doi: 10.1111/j.1558-5646.2007.00055.x. doi:10.1111/j.1558-5646.2007.00055.x [DOI] [PubMed] [Google Scholar]
- Levin D.A. Oxford University Press; London, UK: 2000. The origin, expansion and demise of plant species. [Google Scholar]
- Martin P.R, McKay J.K. Latitudinal variation in genetic divergence of populations and the potential for future speciation. Evolution. 2004;58:938–945. doi: 10.1111/j.0014-3820.2004.tb00428.x. doi:10.1554/03-611 [DOI] [PubMed] [Google Scholar]
- Mittelbach G.G, et al. Evolution and the latitudinal diversity gradient: speciation, extinction and biogeography. Ecol. Lett. 2007;10:315–331. doi: 10.1111/j.1461-0248.2007.01020.x. doi:10.1111/j.1461-0248.2007.01020.x [DOI] [PubMed] [Google Scholar]
- Nei M. Columbia University Press; New York, NY: 1987. Molecular evolutionary genetics. [Google Scholar]
- Rieseberg L.H, Wendel J. Plant speciation: rise of the poor cousins. New Phytol. 2004;161:3–8. doi:10.1111/j.1469-8137.2004.00957.x [Google Scholar]
- Rosenzweig M.L. Cambridge University Press; Cambridge, UK: 1995. Species diversity in space and time. [Google Scholar]
- Turelli M.N, Barton N.H, Coyne J.A. Theory and speciation. Trends Ecol. Evol. 2001;16:330–343. doi: 10.1016/s0169-5347(01)02177-2. doi:10.1016/S0169-5347(01)02177-2 [DOI] [PubMed] [Google Scholar]
- Weir J.T, Schluter D. The latitudinal gradient in recent speciation and extinction rates of birds and mammals. Science. 2007;315:1574–1576. doi: 10.1126/science.1135590. doi:10.1126/science.1135590 [DOI] [PubMed] [Google Scholar]
- Willig M.R, Kaufman D.M, Stevens R.D. Latitudinal gradients of biodiversity: patterns, process, and systhesis. Annu. Rev. Ecol. Evol. Syst. 2003;34:273–309. doi:10.1146/annurev.ecolsys.34.012103.144032 [Google Scholar]
- Wright S, Keeling J, Gillman L. The road from Santa Rosalia: a faster tempo of evolution in tropical climates. Proc. Natl Acad. Sci. USA. 2006;103:7718–7722. doi: 10.1073/pnas.0510383103. doi:10.1073/pnas.0510383103 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Geographic and genetic data for plant species examined in this study