Abstract
DNA markers have enormous potential to improve the efficiency and precision of conventional plant breeding via marker-assisted selection (MAS). The large number of quantitative trait loci (QTLs) mapping studies for diverse crops species have provided an abundance of DNA marker–trait associations. In this review, we present an overview of the advantages of MAS and its most widely used applications in plant breeding, providing examples from cereal crops. We also consider reasons why MAS has had only a small impact on plant breeding so far and suggest ways in which the potential of MAS can be realized. Finally, we discuss reasons why the greater adoption of MAS in the future is inevitable, although the extent of its use will depend on available resources, especially for orphan crops, and may be delayed in less-developed countries. Achieving a substantial impact on crop improvement by MAS represents the great challenge for agricultural scientists in the next few decades.
Keywords: marker-assisted selection, plant breeding, QTL mapping, marker-assisted backcrossing, pyramiding, early generation selection
1. Introduction
Plant breeding—in combination with developments in agricultural technology such as agrochemicals—has made remarkable progress in increasing crop yields for over a century. However, plant breeders must constantly respond to many changes. First, agricultural practices change, which creates the need for developing genotypes with specific agronomic characteristics. Second, target environments and the organisms within them are constantly changing. For example, fungal and insect pests continually evolve and overcome host–plant resistance. New land areas are regularly being used for farming, exposing plants to altered growing conditions. Finally, consumer preferences and requirements change. Plant breeders therefore face the endless task of continually developing new crop varieties (Evans 1997).
The outlook for global crop production in the twenty-first century has been analysed by many researchers and does not look bright (Pinstrup-Andersen et al. 1999). A rising global population will require increased crop production and some research suggests that the rate of increase in crop yields is currently declining (Pingali & Heisey 1999). This required increase in crop production will need to occur in the context of mounting water scarcity, decreasing area and environmental degradation of arable land (partly caused by agriculture), increasing pollution, inevitable emergence of new races and biotypes of pathogens and pests, and possible adverse effects of climate change. Thus, the task of increasing crop yields represents an unprecedented challenge for plant breeders and agricultural scientists.
Plant breeding will play a key role in this coordinated effort for increased food production. Given the context of current yield trends, predicted population growth and pressure on the environment, traits relating to yield stability and sustainability should be a major focus of plant breeding efforts. These traits include durable disease resistance, abiotic stress tolerance and nutrient- and water-use efficiency (Mackill et al. 1999; Slafer et al. 2005; Trethowan et al. 2005). Furthermore, there is a need to develop varieties for cultivation in marginal land areas, especially in developing countries, and give greater emphasis to improving minor or ‘orphan’ crops (Naylor et al. 2004).
Despite optimism about continued yield improvement from conventional breeding, new technologies such as biotechnology will be needed to maximize the probability of success (Ortiz 1998; Ruttan 1999; Huang et al. 2002). One area of biotechnology, DNA marker technology, derived from research in molecular genetics and genomics, offers great promise for plant breeding. Owing to genetic linkage, DNA markers can be used to detect the presence of allelic variation in the genes underlying these traits. By using DNA markers to assist in plant breeding, efficiency and precision could be greatly increased. The use of DNA markers in plant breeding is called marker-assisted selection (MAS) and is a component of the new discipline of ‘molecular breeding’.
2. Overview of DNA markers, QTL mapping, and marker-assisted selection
(a) Features of cereal breeding
The fundamental basis of plant breeding is the selection of specific plants with desirable traits. Selection typically involves evaluating a breeding population for one or more traits in field or glasshouse trials (e.g. agronomic traits, disease resistance or stress tolerance), or with chemical tests (e.g. grain quality). The goal of plant breeding is to assemble more desirable combinations of genes in new varieties.
Standard breeding techniques for inbreeding cereal crops have been outlined in various textbooks (e.g. Allard 1999). In the commonly used pedigree breeding method, selecting desirable plants begins in early generations for traits of higher heritability. However, for traits of low heritability, selection is often postponed until the lines become more homozygous in later generations (F5 or F6). Selection of superior plants involves visual assessment for agronomic traits or resistance to stresses, as well as laboratory tests for quality or other traits. When the breeding lines become homozygous (F5 or later), they can be harvested in bulk and evaluated in replicated field trials. The entire process involves considerable time (5–10 years for elite lines to be identified) and expense.
The size and composition of a plant population is an important consideration for a breeding programme. The larger the number of genes segregating in a population, the larger the population size required in order to identify specific gene combinations. Typical breeding programmes usually grow hundreds or even thousands of populations, and many thousands or millions of individual plants (Witcombe & Virk 2001). Given the extent and complexity of selection required in breeding programmes, and the number and size of populations, one can easily appreciate the usefulness of new tools that may assist breeders in plant selection. The scale of breeding programmes also underlines the challenges of incorporating a relatively expensive technology such as MAS.
(b) Main types of DNA markers used in MAS
There are five main considerations for the use of DNA markers in MAS: reliability; quantity and quality of DNA required; technical procedure for marker assay; level of polymorphism; and cost (Mackill & Ni 2000; Mohler & Singrun 2004).
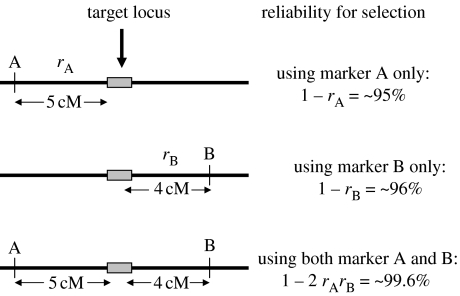
Reliability. Markers should be tightly linked to target loci, preferably less than 5 cM genetic distance. The use of flanking markers or intragenic markers will greatly increase the reliability of the markers to predict phenotype (figure 1).
Figure 1.
Reliability of selection using single and flanking markers (adapted from Tanksley (1983), assuming no crossover interference). The recombination frequency between the target locus and marker A is approximately 5% (5 cM). Therefore, recombination may occur between the target locus and marker in approximately 5% of the progeny. The recombination frequency between the target locus and marker B is approximately 4% (4 cM). The chance of recombination occurring between both marker A and marker B (i.e. double crossover) is much lower than for single markers (approx. 0.4%). Therefore, the reliability of selection is much greater when flanking markers are used. Adapted from formulae from Liu (1998, p. 310).
DNA quantity and quality. Some marker techniques require large amounts and high quality of DNA, which may sometimes be difficult to obtain in practice, and this adds to the cost of the procedures.
Technical procedure. The level of simplicity and the time required for the technique are critical considerations. High-throughput simple and quick methods are highly desirable.
Level of polymorphism. Ideally, the marker should be highly polymorphic in breeding material (i.e. it should discriminate between different genotypes), especially in core breeding material.
Cost. The marker assay must be cost-effective in order for MAS to be feasible.
The most widely used markers in major cereals are called simple sequence repeats (SSRs) or microsatellites (Gupta et al. 1999; Gupta & Varshney 2000). They are highly reliable (i.e. reproducible), co-dominant in inheritance, relatively simple and cheap to use and generally highly polymorphic. The only disadvantages of SSRs are that they typically require polyacrylamide gel electrophoresis and generally give information only about a single locus per assay, although multiplexing of several markers is possible. These problems have been overcome in many cases by selecting SSR markers that have large enough size differences for detection in agarose gels, as well as multiplexing several markers in a single reaction. SSR markers also require a substantial investment of time and money to develop, and adequate numbers for high-density mapping are not available in some orphan crop species. Sequence tagged site (STS), sequence characterized amplified region (SCAR) or single nucleotide polymorphism (SNP) markers that are derived from specific DNA sequences of markers (e.g. restriction fragment length polymorphisms: RFLPs) that are linked to a gene or quantitative trait locus (QTL) are also extremely useful for MAS (Shan et al. 1999; Sanchez et al. 2000; Sharp et al. 2001).
(c) QTL mapping and MAS
The detection of genes or QTLs controlling traits is possible due to genetic linkage analysis, which is based on the principle of genetic recombination during meiosis (Tanksley 1993). This permits the construction of linkage maps composed of genetic markers for a specific population. Segregating populations such as F2, F3 or backcross (BC) populations are frequently used. However, populations that can be maintained and produced permanently, such as recombinant inbreds and doubled haploids, are preferable because they allow replicated and repeated experiments. These types of populations may not be applicable to outbreeding cereals where inbreeding depression can cause non-random changes in gene frequency and loss of vigour of the lines. Using statistical methods such as single-marker analysis or interval mapping to detect associations between DNA markers and phenotypic data, genes or QTLs can be detected in relation to a linkage map (Kearsey 1998). The identification of QTLs using DNA markers was a major breakthrough in the characterization of quantitative traits (Paterson et al. 1988).
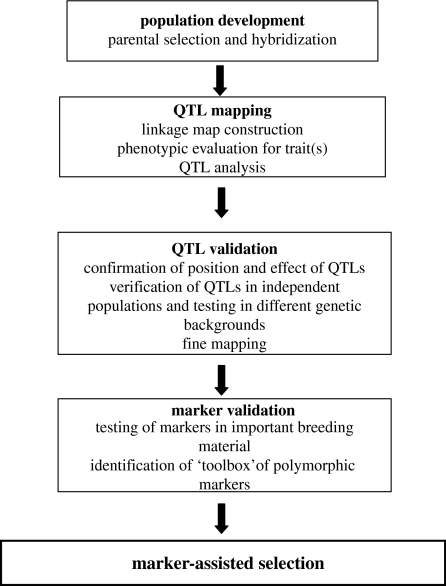
Reports have been numerous of DNA markers linked to genes or QTLs (Mohan et al. 1997; Francia et al. 2005). An overview of marker development is presented in figure 2. Previously, it was assumed that most markers associated with QTLs from preliminary mapping studies were directly useful in MAS. However, in recent years it has become widely accepted that QTL confirmation, QTL validation and/or fine (or high resolution) mapping may be required (Langridge et al. 2001). Although there are examples of highly accurate preliminary QTL mapping data as determined by subsequent QTL mapping research (Price 2006), ideally a confirmation step is preferable because QTL positions and effects can be inaccurate due to factors such as sampling bias (Melchinger et al. 1998). QTL validation generally refers to the verification that a QTL is effective in different genetic backgrounds (Langridge et al. 2001). Additional marker-testing steps may involve identifying a ‘toolbox’ or ‘suite’ of markers within a 10 cM ‘window’ spanning and flanking a QTL (due to a limited polymorphism of individual markers in different genotypes) and converting markers into a form that requires simpler methods of detection.
Figure 2.
Marker development ‘pipeline’.
Once tightly linked markers that reliably predict a trait phenotype have been identified, they may be used for MAS. The fundamental advantages of MAS over conventional phenotypic selection are as follows.
It may be simpler than phenotypic screening, which can save time, resources and effort. Classical examples of traits that are difficult and laborious to measure are cereal cyst nematode and root lesion nematode resistance in wheat (Eastwood et al. 1991; Eagles et al. 2001; Zwart et al. 2004). Other examples are quality traits which generally require expensive screening procedures.
Selection can be carried out at the seedling stage. This may be useful for many traits, but especially for traits that are expressed at later developmental stages. Therefore, undesirable plant genotypes can be quickly eliminated. This may have tremendous benefits in rice breeding because typical rice production practices involve sowing pre-germinated seeds and transplanting seedlings into rice paddies, making it easy to transplant only selected seedlings to the main field.
Single plants can be selected. Using conventional screening methods for many traits, plant families or plots are grown because single-plant selection is unreliable due to environmental factors. With MAS, individual plants can be selected based on their genotype. For most traits, homozygous and heterozygous plants cannot be distinguished by conventional phenotypic screening.
These advantages can be exploited by breeders to accelerate the breeding process (Ribaut & Hoisington 1998; Morris et al. 2003). Target genotypes can be more effectively selected, which may enable certain traits to be ‘fast-tracked’, resulting in quicker line development and variety release. Markers can also be used as a replacement for phenotyping, which allows selection in off-season nurseries making it more cost-effective to grow more generations per year (Ribaut & Hoisington 1998). Another benefit from using MAS is that the total number of lines that need to be tested can be reduced. Since many lines can be discarded after MAS early in a breeding scheme, this permits more efficient use of glasshouse and/or field space—which is often limited—because only important breeding material is maintained.
Considering the potential advantages of MAS over conventional breeding, one rarely discussed point is that markers will not necessarily be useful or more effective for every trait, despite the substantial investment in time, money and resources required for their development. For many traits, effective phenotypic screening methods already exist and these will often be less expensive for selection in large populations. However, when whole-genome scans are being used, even these traits can be selected for if the genetic control is understood.
3. Applications of MAS in plant breeding
The advantages described above may have a profound impact on plant breeding in the future and may alter the plant breeding paradigm (Koebner & Summers 2003). In this section, we describe the main uses of DNA markers in plant breeding, with an emphasis on important MAS schemes. We have classified these schemes into five broad areas: marker-assisted evaluation of breeding material; marker-assisted backcrossing; pyramiding; early generation selection; and combined MAS, although there may be overlap between these categories. Generally, for line development, DNA markers have been integrated in conventional schemes or used to substitute for conventional phenotypic selection.
(a) Marker-assisted evaluation of breeding material
Prior to crossing (hybridization) and line development, there are several applications in which DNA marker data may be useful for breeding, such as cultivar identity, assessment of genetic diversity and parent selection, and confirmation of hybrids. Traditionally, these tasks have been done based on visual selection and analysing data based on morphological characteristics.
(i) Cultivar identity/assessment of ‘purity’
In practice, seed of different strains is often mixed due to the difficulties of handling large numbers of seed samples used within and between crop breeding programmes. Markers can be used to confirm the true identity of individual plants. The maintenance of high levels of genetic purity is essential in cereal hybrid production in order to exploit heterosis. In hybrid rice, SSR and STS markers were used to confirm purity, which was considerably simpler than the standard ‘grow-out tests’ that involve growing the plant to maturity and assessing morphological and floral characteristics (Yashitola et al. 2002).
(ii) Assessment of genetic diversity and parental selection
Breeding programmes depend on a high level of genetic diversity for achieving progress from selection. Broadening the genetic base of core breeding material requires the identification of diverse strains for hybridization with elite cultivars (Xu et al. 2004; Reif et al. 2005). Numerous studies investigating the assessment of genetic diversity within breeding material for practically all crops have been reported. DNA markers have been an indispensable tool for characterizing genetic resources and providing breeders with more detailed information to assist in selecting parents. In some cases, information regarding a specific locus (e.g. a specific resistance gene or QTL) within breeding material is highly desirable. For example, the comparison of marker haplotypes has enabled different sources of resistance to Fusarium head blight, which is a major disease of wheat worldwide, to be predicted (Liu & Anderson 2003; McCartney et al. 2004).
(iii) Study of heterosis
For hybrid crop production, especially in maize and sorghum, DNA markers have been used to define heterotic groups that can be used to exploit heterosis (hybrid vigour). The development of inbred lines for use in producing superior hybrids is a very time-consuming and expensive procedure. Unfortunately, it is not yet possible to predict the exact level of heterosis based on DNA marker data although there have been reports of assigning parental lines to the proper heterotic groups (Lee et al. 1989; Reif et al. 2003). The potential of using smaller subsets of DNA marker data in combination with phenotypic data to select heterotic hybrids has also been proposed (Jordan et al. 2003).
(iv) Identification of genomic regions under selection
The identification of shifts in allele frequencies within the genome can be important information for breeders since it alerts them to monitor specific alleles or haplotypes and can be used to design appropriate breeding strategies (Steele et al. 2004). Other applications of the identification of genomic regions under selection are for QTL mapping: the regions under selection can be targeted for QTL analysis or used to validate previously detected marker–trait associations (Jordan et al. 2004). Ultimately, data on genomic regions under selection can be used for the development of new varieties with specific allele combinations using MAS schemes such as marker-assisted backcrossing or early generation selection (described below; Ribaut et al. 2001; Steele et al. 2004).
(b) Marker-assisted backcrossing
Backcrossing has been a widely used technique in plant breeding for almost a century. Backcrossing is a plant breeding method most commonly used to incorporate one or a few genes into an adapted or elite variety. In most cases, the parent used for backcrossing has a large number of desirable attributes but is deficient in only a few characteristics (Allard 1999). The method was first described in 1922 and was widely used between the 1930s and 1960s (Stoskopf et al. 1993).
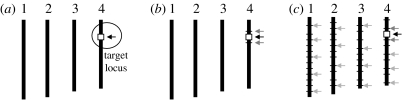
The use of DNA markers in backcrossing greatly increases the efficiency of selection. Three general levels of marker-assisted backcrossing (MAB) can be described (Holland 2004; figure 3). In the first level, markers can be used in combination with or to replace screening for the target gene or QTL. This is referred to as ‘foreground selection’ (Hospital & Charcosset 1997). This may be particularly useful for traits that have laborious or time-consuming phenotypic screening procedures. It can also be used to select for reproductive-stage traits in the seedling stage, allowing the best plants to be identified for backcrossing. Furthermore, recessive alleles can be selected, which is difficult to do using conventional methods.
Figure 3.
Levels of selection during marker-assisted backcrossing. A hypothetical target locus is indicated on chromosome 4. (a) Foreground selection, (b) recombinant selection and (c) background selection.
The second level involves selecting BC progeny with the target gene and recombination events between the target locus and linked flanking markers—we refer to this as ‘recombinant selection’. The purpose of recombinant selection is to reduce the size of the donor chromosome segment containing the target locus (i.e. size of the introgression). This is important because the rate of decrease of this donor fragment is slower than for unlinked regions and many undesirable genes that negatively affect crop performance may be linked to the target gene from the donor parent—this is referred to as ‘linkage drag’ (Hospital 2005). Using conventional breeding methods, the donor segment can remain very large even with many BC generations (e.g. more than 10; Ribaut & Hoisington 1998; Salina et al. 2003). By using markers that flank a target gene (e.g. less than 5 cM on either side), linkage drag can be minimized. Since double recombination events occurring on both sides of a target locus are extremely rare, recombinant selection is usually performed using at least two BC generations (Frisch et al. 1999b).
The third level of MAB involves selecting BC progeny with the greatest proportion of recurrent parent (RP) genome, using markers that are unlinked to the target locus—we refer to this as ‘background selection’. In the literature, background selection refers to the use of tightly linked flanking markers for recombinant selection and unlinked markers to select for the RP (Hospital & Charcosset 1997; Frisch et al. 1999b). Background markers are markers that are unlinked to the target gene/QTL on all other chromosomes, in other words, markers that can be used to select against the donor genome. This is extremely useful because the RP recovery can be greatly accelerated. With conventional backcrossing, it takes a minimum of six BC generations to recover the RP and there may still be several donor chromosome fragments unlinked to the target gene. Using markers, it can be achieved by BC4, BC3 or even BC2 (Visscher et al. 1996; Hospital & Charcosset 1997; Frisch et al. 1999a,b), thus saving two to four BC generations. The use of background selection during MAB to accelerate the development of an RP with an additional (or a few) genes has been referred to as ‘complete line conversion’ (Ribaut et al. 2002).
Some examples of MAB in cereals are presented in table 1. MAB will probably become an increasingly more popular approach, largely for the same reasons that conventional backcrossing has been widely used (Mackill 2006). For practical reasons, farmers in developed and developing countries generally prefer to grow their ‘tried and tested’ varieties. Farmers have already determined the optimum sowing rates and date, fertilizer application rates and number and timing of irrigations for these varieties (Borlaug 1957). There may also be reluctance from millers or the marketing industry to dramatically change a variety since they have established protocols for testing flour characteristics. Furthermore, even with the latest developments in genetic engineering technology and plant tissue culture, some specific genotypes are still more amenable to transformation than others. Therefore, MAB must be used in order to trace the introgression of the transgene into elite cultivars during backcrossing.
Table 1.
Examples of marker-assisted backcrossing in cereals.
| species | trait(s) | gene/QTLs | foreground selection | background selection | reference |
|---|---|---|---|---|---|
| barley | barley yellow dwarf virus | Yd2 | STS | not performed | Jefferies et al. (2003) |
| barley | leaf rust | Rphq6 | AFLP | AFLP | van Berloo et al. (2001) |
| barley | stripe rust | QTLs on 4H and 5H | RFLP | not performed | Toojinda et al. (1998) |
| barley | yield | QTLs on 2HL and 3HL | RFLP | RFLP | Schmierer et al. (2004) |
| maize | corn borer resistance | QTLs on chromosomes 7, 9 and 10 | RFLP | RFLP | Willcox et al. (2002) |
| maize | earliness and yield | QTLs on chromosomes 5, 8 and 10 | RFLP | RFLP | Bouchez et al. (2002) |
| rice | bacterial blight | Xa21 | STSa | RFLP | Chen et al. (2000) |
| rice | bacterial blight | Xa21 | STSa | AFLP | Chen et al. (2001) |
| rice | bacterial blight | xa5, xa13 and Xa21 | STS, CAPS | not performed | Sanchez et al. (2000) |
| rice | bacterial blight | xa5, xa13 and Xa21 | STS | not performed | Singh et al. (2001) |
| rice | bacterial blight+quality | xa13, Xa21 | STS and SSR | AFLP | Joseph et al. (2004) |
| rice | blast | Pi1 | SSR | ISSRb | Liu et al. (2003) |
| rice | deep roots | QTLs on chromosomes 1, 2, 7 and 9 | RFLP and SSR | SSR | Shen et al. (2001) |
| rice | quality | waxy | RFLPa | AFLP | Zhou et al. (2003a) |
| rice | root traits and aroma | QTLs on chromosomes 2, 7, 8, 9 and 11 | RFLP and SSR | RFLP and SSR | Steele et al. (2006) |
| rice | submergence tolerance | Sub1 QTL | phenotyping and SSRa | SSR | Mackill et al. (2006) |
| rice | submergence tolerance, disease resistance, quality | Subchr9 QTL, Xa21, Bph and blast QTLs and quality loci | SSR and STS | not performed | Toojinda et al. (2005) |
| wheat | powdery mildew | 22 Pm genes | phenotyping | AFLP | Zhou et al. (2005) |
Indicates recombinant selection performed to minimize linkage drag around target locus.
ISSR and inter SSRs.
(c) Marker-assisted pyramiding
Pyramiding is the process of combining several genes together into a single genotype. Pyramiding may be possible through conventional breeding but it is usually not easy to identify the plants containing more than one gene. Using conventional phenotypic selection, individual plants must be evaluated for all traits tested. Therefore, it may be very difficult to assess plants from certain population types (e.g. F2) or for traits with destructive bioassays. DNA markers can greatly facilitate selection because DNA marker assays are non-destructive and markers for multiple specific genes can be tested using a single DNA sample without phenotyping.
The most widespread application for pyramiding has been for combining multiple disease resistance genes (i.e. combining qualitative resistance genes together into a single genotype). The motive for this has been the development of ‘durable’ or stable disease resistance since pathogens frequently overcome single-gene host resistance over time due to the emergence of new plant pathogen races. Some evidence suggests that the combination of multiple genes (effective against specific races of a pathogen) can provide durable (broad spectrum) resistance (Kloppers & Pretorius 1997; Shanti et al. 2001; Singh et al. 2001). The ability of a pathogen to overcome two or more effective genes by mutation is considered much lower compared with the ‘conquering’ of resistance controlled by a single gene. In the past, it has been difficult to pyramid multiple resistance genes because they generally show the same phenotype, necessitating a progeny test to determine which plants possess more than one gene. With linked DNA markers, the number of resistance genes in any plant can be easily determined. The incorporation of quantitative resistance controlled by QTLs offers another promising strategy to develop durable disease resistance. Castro et al. (2003) referred to quantitative resistance as an insurance policy in case of the breakdown of qualitative resistance. A notable example of the combination of quantitative resistance was the pyramiding of a single stripe rust gene and two QTLs (Castro et al. 2003).
Pyramiding may involve combining genes from more than two parents. For example, Hittalmani et al. (2000) and Castro et al. (2003) combined genes originating from three parents for rice blast and stripe rust in barley, respectively. MAS pyramiding was also proposed as an effective approach to produce three-way F1 cereal hybrids with durable resistance (Witcombe & Hash 2000). Strategies for MAS pyramiding of linked target genes have also been evaluated (Servin et al. 2004). For many linked target loci, pyramiding over successive generations is preferable in terms of minimizing marker genotyping.
In theory, MAS could be used to pyramid genes from multiple parents (i.e. populations derived from multiple crosses). Some examples of MAS pyramiding in cereals are presented in table 2. In the future, MAS pyramiding could also facilitate the combination of QTLs for abiotic stress tolerances, especially QTLs effective at different growth stages. Another use could be to combine single QTLs that interact with other QTLs (i.e. epistatic QTLs). This was experimentally validated for two interacting resistance QTLs for rice yellow mottle virus (Ahmadi et al. 2001).
Table 2.
Examples of gene or QTL pyramiding in cereals.
| species | trait(s) | genes from parent 1 | genes from parent 2 | selection stage | DNA marker(s) used | reference |
|---|---|---|---|---|---|---|
| barley | barley yellow mosaic virus | rym1 | rym5 | F2 | RFLP, CAPS | Okada et al. (2004) |
| barley | barley yellow mosaic virus | rym4, rym9, rym11 | rym4, rym9, rym11 | F1-derived doubled haploids | RAPD, SSR | Werner et al. (2005) |
| barley | stripe rust | RspxRspx | QTLs 4, 7 QTL 5 | F1-derived doubled haploids | SSR | Castro et al. (2003) |
| rice | bacterial blight | xa5, xa13 | Xa4, Xa21 | F2 | RFLP, STS | Huang et al. (1997) |
| rice | bacterial blight, yellow stem borer, sheath blight | Xa21, Bt | RC7 chitinase gene, Bt | F2 | STS | Datta et al. (2002) |
| rice | blast disease | Pi1, Piz-5 | Pi1, Pita | F2 | RFLP, STS | Hittalmani et al. (2000) |
| rice | brown plant hopper | Bph1 | Bph2 | F4 | STS | Sharma et al. (2004) |
| rice | insect resistance and bacterial blight | Xa21 | Bt | F2 | STS | Jiang et al. (2004) |
| wheat | powdery mildew | Pm2 | Pm4a | F2 | RFLP | Liu et al. (2000) |
(d) Early generation marker-assisted selection
Although markers can be used at any stage during a typical plant breeding programme, MAS is a great advantage in early generations because plants with undesirable gene combinations can be eliminated. This allows breeders to focus attention on a lesser number of high-priority lines in subsequent generations. When the linkage between the marker and the selected QTL is not very tight, the greatest efficiency of MAS is in early generations due to the increasing probability of recombination between the marker and QTL. The major disadvantage of applying MAS at early generations is the cost of genotyping a larger number of plants.
One strategy proposed by Ribaut & Betran (1999) involving MAS at an early generation was called single large-scale MAS (SLS–MAS). The authors proposed that a single MAS step could be performed on F2 or F3 populations derived from elite parents. This approach used flanking markers (less than 5 cM, on both sides of a target locus) for up to three QTLs in a single MAS step. Ideally, these QTLs should account for the largest proportion of phenotypic variance and be stable in different environments.
The population sizes may soon become quite small due to the high selection pressure, thus providing an opportunity for genetic drift to occur at non-target loci, so it is recommended that large population sizes be used (Ribaut & Betran 1999). This problem can also be minimized by using F3 rather than F2 populations, because the selected proportion of an F3 population is larger compared with that of an F2 population (i.e. for a single target locus, 38% of the F3 population will be selected compared with 25% of the F2). Ribaut & Betran (1999) also proposed that, theoretically, linkage drag could be minimized by using additional flanking markers surrounding the target QTLs, much in the same way as in MAB.
For self-pollinated crops, an important aim may be to fix alleles in their homozygous state as early as possible. For example, in bulk and single-seed descent breeding methods, screening is often performed at the F5 or F6 generations when most loci are homozygous. Using co-dominant DNA markers, it is possible to fix specific alleles in their homozygous state as early as the F2 generation. However, this may require large population sizes; thus, in practical terms, a small number of loci may be fixed at each generation (Koebner & Summers 2003). An alternative strategy is to ‘enrich’ rather than fix alleles—by selecting homozygotes and heterozygotes for a target locus—within a population in order to reduce the size of the breeding populations required (Bonnett et al. 2005).
(e) Combined marker-assisted selection
There are several instances when phenotypic screening can be strategically combined with MAS. In the first instance, ‘combined MAS’ (coined by Moreau et al. 2004) may have advantages over phenotypic screening or MAS alone in order to maximize genetic gain (Lande & Thompson 1990). This approach could be adopted when additional QTLs controlling a trait remain unidentified or when a large number of QTLs need to be manipulated. Simulation studies indicate that this approach is more efficient than phenotypic screening alone, especially when large population sizes are used and trait heritability is low (Hospital et al. 1997). Bohn et al. (2001) investigated the prospect of MAS for improving insect resistance in tropical maize and found that MAS alone was less efficient than conventional phenotypic selection. However, there was a slight increase in relative efficiency when MAS and phenotypic screening were combined. In an example in wheat, MAS combined with phenotypic screening was more effective than phenotypic screening alone for a major QTL on chromosome 3BS for Fusarium head blight resistance (Zhou et al. 2003b). In practice, all MAS schemes will be used in the context of the overall breeding programme, and this will involve phenotypic selection at various stages. This will be necessary to confirm the results of MAS as well as select for traits or genes for which the map location is unknown.
In some (possibly many) situations, there is a low level of recombination between a marker and QTL, unless markers flanking the QTL are used (Sanchez et al. 2000; Sharp et al. 2001). In other words, a marker assay may not predict phenotype with 100% reliability. However, plant selection using such markers may still be useful for breeders in order to select a subset of plants using the markers to reduce the number of plants that need to be phenotypically evaluated. This may be particularly advantageous when the cost of marker genotyping is cheaper than phenotypic screening, such as for quality traits (Han et al. 1997). This was referred to as ‘tandem selection’ by Han et al. (1997) and ‘stepwise selection’ by Langridge & Chalmers (2005).
In addition to complementing conventional breeding methods, mapping QTLs for important traits may have an indirect benefit in a conventional breeding programme. In many cases, this occurs when traits which were thought to be under the complex genetic control are found to be under the influence of one or a few major QTLs. For example, in pearl millet downy mildew resistance was found to be under the control of genes of major effect (Jones et al. 1995). Likewise, submergence tolerance of rice was found to be under the control of the major QTL Sub1, which helped simplify the breeding for this trait (Mackill et al. 2006).
4. Reasons to explain the low impact of marker-assisted selection
(a) Still at the early stages of DNA marker technology development
Although DNA markers were first developed in the late 1980s, more user-friendly PCR-based markers such as SSRs were not developed until the mid- to late 1990s. Although currently large numbers of SSRs are publicly available for major cereals, this number was initially very low. It is only during the last 5–7 years that these markers could have been widely used, and tangible results may not yet have been produced. Inspection of the publication dates for the examples in tables 1 and 2 supports this. If this is the case, there should be a notable increase in the number of published papers describing MAS in the next 10 years and beyond.
(b) Marker-assisted selection results may not be published
Although QTL mapping has many potential practical outcomes, it is considered to be a basic research process, and results are typically published in scientific journals. However, for plant breeding, the final ‘product’ is a new variety. Although these varieties are registered, explicit details regarding the use of DNA markers during breeding may not be provided. Another reason for the limited number of published reports could be that private seed companies typically do not disclose details of methodology due to competition with other seed companies. In general, the problem of publishing also extends to QTL validation and QTL mapping. New QTLs are frequently reported in scientific journals, but reconfirmation of these QTLs in other germplasm and identification of more useful markers are usually not considered novel enough to warrant new publications. This is unfortunate because it is exactly this type of information that is needed for MAS. Some of this information can be found in symposia abstracts or web sites, but often this information is not very informative. An excellent example of successful MAS is the development of an improved version of the pearl millet hybrid HHB 67 with resistance to downy mildew, described at http://www.dfid-psp.org/AtAGlance/HotTopic.html.
(c) Reliability and accuracy of quantitative trait loci mapping studies
The accuracy of the QTL mapping study is critical to the success of MAS. This is particularly important when QTL mapping is undertaken for more complex traits, such as yield, that are controlled by many QTLs with small effects compared with simple traits. Many factors may affect the accuracy of a QTL mapping study such as the level of replication used to generate phenotypic data and population size (Kearsey & Farquhar 1998; Young 1999). Simulation and experimental studies have indicated that the power of QTL detection is low with the typical populations (less than 200) that are used (Beavis 1998; Kearsey & Farquhar 1998). As a result, confidence intervals for regions containing QTLs may be large, even for QTLs with large effects. Furthermore, sampling bias can lead to a large bias in estimates of QTL effects, especially in relatively small population sizes (Melchinger et al. 1998). These factors have important implications for MAS, since the basis for selecting markers depends on the accurate determination of the position and effect of a QTL.
(d) Insufficient linkage between marker and gene/ quantitative trait locus
In some cases, recombination occurs between the marker and gene/QTL due to loose linkage (Sharp et al. 2001; Thomas 2003). This may occur even if genetic distances from a preliminary QTL mapping study indicated tight linkage, because data from a single QTL mapping experiment may not be accurate (Sharp et al. 2001). The process of marker validation is required to determine the reliability of a marker to predict phenotype and this points out the advantages of using flanking markers.
(e) Limited markers and limited polymorphism of markers in breeding material
Ideally, markers should be ‘diagnostic’ for traits in a wide range of breeding material. In other words, markers should clearly discriminate between varieties that do and do not express the trait. Unfortunately, in practice, DNA markers are not always diagnostic. For example, a wheat SSR marker was diagnostic for the Sr2 gene (controlling stem rust resistance) for all except four susceptible Australian cultivars, in which the same marker allele was detected as for the source of resistance (Spielmeyer et al. 2003). This would preclude the use of this SSR marker for the introgression of resistance in the four susceptible cultivars, requiring that additional markers be developed. Even with the large numbers of available markers in some crops, there can be specific chromosome regions containing an important gene or QTL for which it is difficult to find polymorphic markers.
(f) Effects of genetic background
It has been observed that QTLs identified in a particular mapping population may not be effective in different backgrounds (Liao et al. 2001). For example, Steele et al. (2006) found that only one of four root-length QTLs were effective when transferred by backcrossing into a new rice variety. In some cases, this is due to the small effect of an allele transferred into elite varieties (Charcosset & Moreau 2004). Often for QTL mapping experiments, parents that represent the extreme ends of a trait phenotype are selected. This increases the chance of detecting QTLs because QTL mapping is based on statistically different means of marker groups. The main disadvantage with this approach is that one (or even both) parent(s) may possess QTL alleles that are similar or even identical to the elite germplasm used in breeding programmes. In this case, the effect of a QTL may be insignificant when used for introgression into elite varieties. In other cases, the effect of a QTL may differ in different genetic backgrounds due to interactions with other loci or epistasis (Holland 2001; Li 2000).
(g) Quantitative trait loci×environment effects
While the effects of many QTLs appear to be consistent across environments, the magnitude of effect and even direction of QTLs may vary depending on environmental conditions due to QTL×environment interactions (Hayes et al. 1993; Romagosa et al. 1999; Bouchez et al. 2002; Li et al. 2003). This often occurs for QTLs with smaller effects. The extent of QTL×environment interactions is often unknown because the QTL mapping studies have been limited to only a few years (replications) or locations. The existence of QTL×environment interactions must be carefully considered in order to develop an effective MAS scheme.
(h) High cost of marker-assisted selection
The cost of using MAS compared with conventional phenotypic selection may vary considerably, although only a relatively small number of studies have addressed this topic. Landmark papers by Dreher et al. (2003) and Morris et al. (2003) showed that the cost–benefit ratio of MAS will depend on several factors, such as the inheritance of the trait, the method of phenotypic evaluation, the cost of field and glasshouse trials and labour costs. It is also worth noting that large initial capital investments are required for the purchase of equipment, and regular expenses will be incurred for maintenance. Intellectual property rights, for example, licensing costs due to patents, may also affect the cost of MAS (Jorasch 2004; Brennan et al. 2005). One approach to this problem is to contract the marker work out to larger laboratories that can benefit from economies of scale and high-throughput equipment.
(i) ‘Application gap’ between research laboratories and plant breeding institutes
In many cases, QTL mapping research is undertaken at universities whereas breeding is generally undertaken at different locations such as research stations or private companies. Consequently, there may be difficulties in the transfer of markers and relevant information to breeders in situations where the two groups do not work closely together. More importantly, Van Sanford et al. (2001) also pointed out that transfer problems may be related to the culture of the scientific community. Given the emphasis on conducting innovative research, and on the publication of research results within academic institutions, scientists do not have much motivation to ensure that markers are developed into breeder-friendly ones and that they are actually applied in breeding programmes. This is even truer for activities in the private sector where publication of results is generally discouraged.
(j) ‘Knowledge gap’ among molecular biologists, plant breeders and other disciplines
DNA marker technology, QTL theory and statistical methodology for QTL analysis have undergone rapid developments in the past two decades. These concepts and the jargon used by molecular biologists may not be clearly understood by plant breeders and other plant scientists (Collard et al. 2005). In addition to this, many highly specialized pieces of equipment are based on sophisticated techniques used for molecular genotyping. Similarly, fundamental concepts in plant breeding may not be well understood by molecular biologists. This restricts the level of integration between conventional plant and molecular breeding and ultimately affects the development of new breeding lines.
5. Plant breeding in the future: the dawn of marker-assisted selection?
Despite the relatively small impact that MAS has had on variety development to date, there has been a ‘cautious optimism’ for the future (Young 1999). We predict that six main factors will give rise to a much greater level of adoption of MAS in plant breeding in the early part of the twenty-first century in many breeding programmes.
First, the extent to which DNA marker technology has already spread to plant breeding institutes coupled with the enormous amount of data from previous QTL mapping and MAS studies should lead to the greater adoption of MAS. Many such institutes now possess the essential equipment and expertise required for marker genotyping. Of course, the frequency of use will depend on available funding.
Second, since the landmark concept of ‘advanced BC QTL analysis’ directly integrated QTL mapping with plant breeding by combining QTL mapping with simultaneous variety development (Tanksley & Nelson 1996), there have been several encouraging examples of an efficient merging of plant and molecular breeding. Some of these excellent examples are Toojinda et al. (1998) and Castro et al. (2003) in which QTL mapping and MAS breeding were combined. There have also been encouraging reports of the combination of QTL validation and line development (Flint-Garcia et al. 2003b). The use of backcrossing and the development of near-isogenic lines (NILs) may be particularly advantageous in this context (Stuber et al. 1999; van Berloo et al. 2001). Ideally, QTL mapping and marker-assisted line development should now always be conceived together, in a holistic scheme.
Third, the increasing use of genetic transformation technology means that MAS can be used to directly select for progeny that possess transgenes via target gene selection. As discussed earlier, specific genotypes often with poor agronomic characteristics are routinely used for transformation. Therefore, MAS can be used to track the transgenes during elite line development.
Fourth, a rapid growth in genomics research has taken place within the last decade. Data generated from functional genomics studies have led to the identification of many candidate genes for numerous traits. SNPs within candidate genes could be extremely useful for ‘association mapping’ and ultimately MAS (Rafalski 2002; Flint-Garcia et al. 2003a; Gupta et al. 2005; Breseghello & Sorrells 2006). This approach also circumvents the requirement for constructing linkage maps and performing QTL analysis for new genotypes that have not been previously mapped, although genotyping and phenotyping of segregating populations (e.g. F2 or F3) is recommended for marker validation (Breseghello & Sorrells 2006). Furthermore, genome sequencing projects in rice and other crop species will provide considerable data that could be used for QTL mapping and marker development in other cereals (Gale & Devos 1998; Yuan et al. 2001; Varshney et al. 2005). However, the costs associated with genomics research may be considerable. This could be detrimental to breeding programmes if funding is diverted away from actual breeding efforts (Brummer 2004).
Fifth, many new high-throughput methods for DNA extraction and especially new high-throughput marker genotyping platforms have been developed (Syvanen 2001, 2005). A current trend in some crops is the adoption of high-throughput genotyping equipment for SSR and SNP markers, although the cost of these new platforms may be higher than for standard genotyping methods (Brennan et al. 2005). Some of these genotyping platforms use fluorescently labelled primers that permit high levels of multiplexing (Coburn et al. 2002). Some authors have predicted that SNP markers, due to their widespread abundance and potentially high levels of polymorphism, and the development of SNP genotyping platforms will have a great impact on MAS in the future (Rafalski 2002; Koebner & Summers 2003). Numerous SNP genotyping platforms have been recently developed, usually for medical applications; however, at present no superior platform has been universally adopted (Syvanen 2001). Array-based methods such as Diversity Array Technology (DArT; Jaccoud et al. 2001) and single feature polymorphism (SFP) detection (Hazen & Kay 2003; Rostoks et al. 2005) offer prospects for lower-cost marker technology that can be used for whole-genome scans.
Finally, the availability of large numbers of publicly available markers and the parallel development of user-friendly databases for the storage of marker and QTL data will undoubtedly encourage the more widespread use of MAS. In cereals, two of the most extensive and useful databases are ‘Gramene’ and ‘GrainGenes’ (Ware et al. 2002a,b; Matthews et al. 2003). The development and curation of these and other databases to keep pace with the continually growing amount of data generated will be critical for the efficient use of markers in the future (Lehmensiek et al. 2005).
Although we believe that these factors will lead to the greater adoption of MAS in many instances (especially for major cereals), there will clearly be situations in which the incorporation of MAS in plant breeding programmes will still be very slow or even non-existent, for example in orphan crop species and in developing countries (Naylor et al. 2004). In both of these situations, funding of research and breeding programmes is extremely limited. The improvement of orphan crop species, especially in developing countries—using any method—represents another great challenge for agricultural scientists.
Generally, the cost of MAS will continue to be a major obstacle for its application. Some cost estimates for consumables and labour associated with MAS are listed in table 3 in order to provide information for breeding programmes. It should be noted that MAS cost estimates may change depending on the number of samples and/or number of marker assays. The study by Dreher et al. (2003) indicated that costs may decrease as the number of samples and/or marker assays increases due to economies of scale and lack of divisibility for many components of MAS. One current trend is the establishment of marker genotyping companies, which will enable marker genotyping to be outsourced. Assuming that the costs for outsourcing genotyping are cheaper, and that logistical problems are not created or are minimal, this may provide breeding programmes with more opportunities for MAS. Furthermore, some new SNP high-throughput genotyping methods may also be comparable with or even cheaper than current methods, although a large initial investment is required for the purchase of equipment (Chen & Sullivan 2003).
Table 3.
Estimates of costs (consumables and labour) per data point for marker genotyping during MAS.
| institute | country | crop species | cost estimatea (US$) | reference |
|---|---|---|---|---|
| IRRIb | The Philippines | rice | 0.30c, 1.00 | this study |
| University of Guelph | Canada | bean | 2.74 | Yu et al. (2000) |
| CIMMYTd | Mexico | maize | 1.24–2.26 | Dreher et al. (2003) |
| University of Adelaide | Australia | wheat | 1.46 | Kuchel et al. (2005) |
| NSW Department of Agriculture | Australia | wheat | 4.16 | Brennan et al. (2005) |
| University of Kentucky, University of Minnesota, University of Oregon, Michigan State University, USDA-ARS | United States | wheat and barley | 0.50–5.00 | Van Sanford et al. (2001) |
Costs were converted to US dollars from other currencies based on exchange rates on August 26, 2005. Estimates did not include costs associated with the collection of plant samples or capital costs.
Conservative cost estimates at IRRI were performed using currently used routine marker genotyping methods for a single rice SSR marker using 96 samples. Cost estimates exclude gloves, paper towels, delivery charges, electricity and water and waste disposal.
$0.30—cost estimate when marker genotyping performed by a research technician. $1.00—cost estimate when marker genotyping performed by a postdoctoral research fellow.
$2.26—cost per data point estimated using a single SSR marker for 100 samples; $1.24—cost per data point estimated using over 200 markers for at least 250 samples.
6. Realizing the potential of marker-assisted selection for crop improvement
Considering the enormous potential of MAS in plant breeding, achieving a tangible impact on crop improvement represents the great challenge of molecular breeding in the early part of the twenty-first century. Solutions to the above-mentioned obstacles of MAS need to be developed in order to achieve a greater impact. In the short term, the most important factors that should enable the impact of MAS to be realized include:
a greater level of integration among conventional breeding, QTL mapping/validation and MAS,
careful planning and execution of QTL mapping studies (especially for complex quantitative traits) and an emphasis on validating results prior to MAS,
optimization of methods used in MAS such as DNA extraction and marker genotyping, especially in terms of cost reduction and efficiency, and
efficient systems for data storage (from in-house laboratory information management systems (LIMS) to publicly available databases).
For MAS to reach its full potential for crop improvement, the advantages of MAS over conventional breeding need to be fully exploited. This may depend on ex ante studies evaluating alternative schemes prior to experimentation. Computer simulations may indicate the most effective breeding schemes in order to maximize genetic gain and minimize costs (Kuchel et al. 2005). Based on the schemes of MAS reviewed in this paper, the most important areas to target include:
use of markers for the selection of parents in breeding programmes,
continued use of MAS for high-priority traits that are difficult, time consuming or expensive to measure,
using markers to minimize linkage drag via recombinant selection,
screening of multiple traits per line (i.e. per unit of DNA), especially populations derived from multiple F1s for pyramiding,
exploiting the ability to rapidly eliminate unsuitable lines after early generation selection or tandem selection in breeding programmes, thus allowing breeders to concentrate on the most promising materials, and
exploiting the time savings for line development (especially using background selection) for accelerated variety release.
For MAS in orphan crops and breeding programmes in developing countries, emphasis should be given to careful prioritization of traits for marker development as well as simplifying and optimizing methods to reduce marker genotyping costs. Currently at IRRI, we are investigating ways in which marker genotyping costs can be further reduced. Preliminary cost analysis indicates potential for cost reduction of standard genotyping methods, which was also reported to be the case at CIMMYT (Dreher et al. 2003). An effective strategy to increase the arsenal of DNA markers in orphan crops could be to conduct data mining of genomics databases. An excellent example of the use of publicly available DNA sequence data to develop new markers for an orphan crop was the development of single-strand conformational polymorphism (SSCP)–SNP markers in pearl millet (Bertin et al. 2005). Similarly, information on rice markers has been useful for genetics of American wild rice, Zizania palustris (Phillips et al. 2006).
Generally, innovation—big and small—may play an important role in obtaining tangible benefits from MAS. Dekkers & Hospital (2002) stated that there is considerable scope for innovative plant/molecular breeding schemes that are tailor-made for using DNA markers; such schemes could lead to a completely new plant breeding paradigm.
Advances in functional genomics will lead to the rapid identification of gene functions in the major cereal crops. This strategy usually relies on fine mapping using molecular markers, as well as other methods such as gene-expression studies (microarray), mutants and gene knockouts, RNAi and association genetics. The identification of gene function will allow the development of allele-specific markers that will be more efficient than using linked DNA markers. In addition, the identified genes can be used for transformation studies as well as mining of gene banks to find more useful alleles. Even though we can expect far-reaching advances in the area of gene function identification, the complex genetic interactions that produce different phenotypes may remain unexplained for the most part. However, even in these cases, we may identify chromosome fragments that are conducive to improved phenotype.
A breeding application resulting from the development of high-throughput genotyping equipment is the use of ‘whole-genome scans’ for determining allelic variation at many agronomically important loci in the genome (Langridge & Chalmers 2005; Langridge 2006). One recent approach called ‘breeding by design’ could enable breeders to exploit known allelic variation to design superior genotypes by combining multiple favourable alleles (Peleman & van der Voort 2003). This also means that plants with the desired combinations of genes can be pre-selected before extensive and expensive field testing. In many cases, the objective would be just to avoid advanced testing of a number of lines with similar genotypic constitutions. Current limitations to the application of breeding by design or similar approaches include the prohibitive cost, since thousands of marker loci need to be scored in breeding material and, perhaps more importantly, our current knowledge and understanding of the function of the majority of agronomically important genes and allelic interactions with respect to phenotype which remain unknown. Therefore, at least in the short term, such approaches will probably not have a great impact on crop improvement.
7. Conclusions
Plant breeding has made remarkable progress in crop improvement and it is critical that this continue. It seems clear that current breeding programmes continue to make progress through commonly used breeding approaches. MAS could greatly assist plant breeders in reaching this goal although, to date, the impact on variety development has been minimal. For the potential of MAS to be realized, it is imperative that there should be a greater integration with breeding programmes and that current barriers be well understood and appropriate solutions developed. The exploitation of the advantages of MAS relative to conventional breeding could have a great impact on crop improvement. The high cost of MAS will continue to be a major obstacle for its adoption for some crop species and plant breeding in developing countries in the near future. Specific MAS strategies may need to be tailored to specific crops, traits and available budgets. New marker technology can potentially reduce the cost of MAS considerably. If the effectiveness of the new methods is validated and the equipment can be easily obtained, this should allow MAS to become more widely applicable for crop breeding programmes.
Acknowledgments
We thank Ms Marichu Bernardo (IRRI), Dr Haydn Kuchel (University of Adelaide, Australia) and Dr Xiangning Chen (Virginia Commonwealth University, U.S.) for providing further information about cost estimates for MAS. We also thank Dr Bill Hardy (IRRI) for proofreading the manuscript and Dr J. R. Witcombe (University of Wales, UK) and an anonymous reviewer for helpful comments.
Footnotes
One contribution of 16 to a Theme Issue ‘Sustainable agriculture I’.
References
- Ahmadi N, Albar L, Pressoir G, Pinel A, Fargette D, Ghesquiere A. Genetic basis and mapping of the resistance to rice yellow mottle virus. III. Analysis of QTL efficiency in introgressed progenies confirmed the hypothesis of complementary epistasis between two resistance QTLs. Theor. Appl. Genet. 2001;103:1084–1092. doi:10.1007/s001220100642 [Google Scholar]
- Allard R.W. 2nd edn. Wiley; New York, NY: 1999. Principles of plant breeding. [Google Scholar]
- Beavis W. QTL analyses: power, precision and accuracy. In: Paterson A.H, editor. Molecular dissection of complex traits. CRC Press; Boca Raton, FL: 1998. [Google Scholar]
- Bertin I, Zhu J.H, Gale M.D. SSCP–SNP in pearl millet—a new marker system for comparative genetics. Theor. Appl. Genet. 2005;110:1467–1472. doi: 10.1007/s00122-005-1981-0. doi:10.1007/s00122-005-1981-0 [DOI] [PubMed] [Google Scholar]
- Bohn M, Groh S, Khairallah M.M, Hoisington D.A, Utz H.F, Melchinger A.E. Re-evaluation of the prospects of marker-assisted selection for improving insect resistance against Diatraea spp. in tropical maize by cross validation and independent validation. Theor. Appl. Genet. 2001;103:1059–1067. doi:10.1007/s001220100708 [Google Scholar]
- Bonnett D.G, Rebetzke G.J, Spielmeyer W. Strategies for efficient implementation of molecular markers in wheat breeding. Mol. Breed. 2005;15:75–85. doi:10.1007/s11032-004-2734-5 [Google Scholar]
- Borlaug, N. E. 1957 The development and use of composite varieties based upon the mechanical mixing of phenotypically similar lines developed through backcrossing. Report of the Third International Wheat Conference, pp. 12–18.
- Bouchez A, Hospital F, Causse M, Gallais A, Charcosset A. Marker-assisted introgression of favorable alleles at quantitative trait loci between maize elite lines. Genetics. 2002;162:1945–1959. doi: 10.1093/genetics/162.4.1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan, J. P., Rehman, A., Raman, H., Milgate, A. W., Pleming, D. & Martin, P. J. 2005 An economic assessment of the value of molecular markers in plant breeding programs. In 49th Annual Conf. of the Australian Agricultural and Resource Economics Society, Coffs Harbour, Australia, 9-11 February
- Breseghello F, Sorrells M.E. Association mapping of kernel size and milling quality in wheat (Triticum aestivum L.) cultivars. Genetics. 2006;172:1165–1177. doi: 10.1534/genetics.105.044586. doi:10.1534/genetics.105.044586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brummer E.C. Applying genomics to alfalfa breeding programs. Crop Sci. 2004;44:1904–1907. [Google Scholar]
- Castro A.J, et al. Mapping and pyramiding of qualitative and quantitative resistance to stripe rust in barley. Theor. Appl. Genet. 2003;107:922–930. doi: 10.1007/s00122-003-1329-6. doi:10.1007/s00122-003-1329-6 [DOI] [PubMed] [Google Scholar]
- Charcosset A, Moreau L. Use of molecular markers for the development of new cultivars and the evaluation of genetic diversity. Euphytica. 2004;137:81–94. doi:10.1023/B:EUPH.0000040505.65040.75 [Google Scholar]
- Chen X, Sullivan P.F. Single nucleotide polymorphism genotyping: biochemistry, protocol, cost and throughput. Pharmacogenom. J. 2003;3:77–96. doi: 10.1038/sj.tpj.6500167. doi:10.1038/sj.tpj.6500167 [DOI] [PubMed] [Google Scholar]
- Chen S, Lin X.H, Xu C.G, Zhang Q.F. Improvement of bacterial blight resistance of ‘Minghui 63’, an elite restorer line of hybrid rice, by molecular marker-assisted selection. Crop Sci. 2000;40:239–244. [Google Scholar]
- Chen S, Xu C.G, Lin X.H, Zhang Q. Improving bacterial blight resistance of ‘6078’, an elite restorer line of hybrid rice, by molecular marker-assisted selection. Plant Breed. 2001;120:133–137. doi:10.1046/j.1439-0523.2001.00559.x [Google Scholar]
- Coburn J.R, Temnykh S.V, Paul E.M, McCouch S.R. Design and application of microsatellite marker panels for semiautomated genotyping of rice (Oryza sativa L.) Crop Sci. 2002;42:2092–2099. [Google Scholar]
- Collard B.C.Y, Jahufer M.Z.Z, Brouwer J.B, Pang E.C.K. An introduction to markers, quantitative trait loci (QTL) mapping and marker-assisted selection for crop improvement: the basic concepts. Euphytica. 2005;142:169–196. doi:10.1007/s10681-005-1681-5 [Google Scholar]
- Datta K, Baisakh N, Thet K.M, Tu J, Datta S.K. Pyramiding transgenes for multiple resistance in rice against bacterial blight, yellow stem borer and sheath blight. Theor. Appl. Genet. 2002;106:1–8. doi: 10.1007/s00122-002-1014-1. [DOI] [PubMed] [Google Scholar]
- Dekkers J.C.M, Hospital F. The use of molecular genetics in the improvement of agricultural populations. Nat. Rev. Genet. 2002;3:22–32. doi: 10.1038/nrg701. doi:10.1038/nrg701 [DOI] [PubMed] [Google Scholar]
- Dreher K, Khairallah M, Ribaut J, Morris M. Money matters (I): costs of field and laboratory procedures associated with conventional and marker-assisted maize breeding at CIMMYT. Mol. Breed. 2003;11:221–234. doi:10.1023/A:1022820520673 [Google Scholar]
- Eagles H, Bariana H, Ogbonnaya F, Rebetzke G, Hollamby G, Henry R, Henschke P, Carter M. Implementation of markers in Australian wheat breeding. Aust. J. Agric. Res. 2001;52:1349–1356. doi:10.1071/AR01067 [Google Scholar]
- Eastwood R.F, Lagudah E.S, Appels R, Hannah M, Kollmorgen J.F. Triticum tauschii—a novel source of resistance to cereal cyst nematode (Heterodera avenae) Aust. J. Agric. Res. 1991;42:69–77. [Google Scholar]
- Evans L.T. Adapting and improving crops: the endless task. Phil. Trans. R. Soc. B. 1997;352:901–906. doi:10.1098/rstb.1997.0069 [Google Scholar]
- Flint-Garcia S.A, Thornsberry J.M, Buckler E.S. Structure of linkage disequilibrium in plants. Ann. Rev. Plant Biol. 2003a;54:357–374. doi: 10.1146/annurev.arplant.54.031902.134907. doi:10.1146/annurev.arplant.54.031902.134907 [DOI] [PubMed] [Google Scholar]
- Flint-Garcia S.A, Darrah L.L, McMullen M.D, Hibbard B.E. Phenotypic versus marker-assisted selection for stalk strength and second-generation European corn borer resistance in maize. Theor. Appl. Genet. 2003b;107:1331–1336. doi: 10.1007/s00122-003-1387-9. doi:10.1007/s00122-003-1387-9 [DOI] [PubMed] [Google Scholar]
- Francia E, Tacconi G, Crosatti C, Barabaschi D, Bulgarelli D, Dall'Aglio E, Valè G. Marker assisted selection in crop plants. Plant Cell Tissue Org. 2005;82:317–342. doi:10.1007/s11240-005-2387-z [Google Scholar]
- Frisch M, Bohn M, Melchinger A.E. Comparison of selection strategies for marker-assisted backcrossing of a gene. Crop Sci. 1999a;39:1295–1301. [Google Scholar]
- Frisch M, Bohn M, Melchinger A.E. Minimum sample size and optimal positioning of flanking markers in marker-assisted backcrossing for transfer of a target gene. Crop Sci. 1999b;39:967–975. [Google Scholar]
- Gale M.D, Devos K.M. Plant comparative genetics after 10 years. Science. 1998;282:656–659. doi: 10.1126/science.282.5389.656. doi:10.1126/science.282.5389.656 [DOI] [PubMed] [Google Scholar]
- Gupta P.K, Varshney R.K. The development and use of microsatellite markers for genetic analysis and plant breeding with emphasis on bread wheat. Euphytica. 2000;113:163–185. doi:10.1023/A:1003910819967 [Google Scholar]
- Gupta P.K, Varshney R.K, Sharma P.C, Ramesh B. Molecular markers and their applications in wheat breeding. Plant Breed. 1999;118:369–390. doi:10.1046/j.1439-0523.1999.00401.x [Google Scholar]
- Gupta P.K, Rustgi S, Kulwal P.L. Linkage disequilibrium and association studies in higher plants: present status and future prospects. Plant Mol. Biol. 2005;57:461–485. doi: 10.1007/s11103-005-0257-z. doi:10.1007/s11103-005-0257-z [DOI] [PubMed] [Google Scholar]
- Han F, Romagosa I, Ullrich S.E, Jones B.L, Hayes P.M, Wesenberg D.M. Molecular marker-assisted selection for malting quality traits in barley. Mol. Breed. 1997;3:427–437. doi:10.1023/A:1009608312385 [Google Scholar]
- Hayes P.M, et al. Quantitative trait locus effects and environmental interaction in a sample of North-American barley germplasm. Theor. Appl. Genet. 1993;87:392–401. doi: 10.1007/BF01184929. doi:10.1007/BF01184929 [DOI] [PubMed] [Google Scholar]
- Hazen S.P, Kay S.A. Gene arrays are not just for measuring gene expression. Trends Plant Sci. 2003;8:413–416. doi: 10.1016/S1360-1385(03)00186-9. doi:10.1016/S1360-1385(03)00186-9 [DOI] [PubMed] [Google Scholar]
- Hittalmani S, Parco A, Mew T.V, Zeigler R.S, Huang N. Fine mapping and DNA marker-assisted pyramiding of the three major genes for blast resistance in rice. Theor. Appl. Genet. 2000;100:1121–1128. doi:10.1007/s001220051395 [Google Scholar]
- Holland J. Epistasis and plant breeding. Plant Breed. Rev. 2001;21:27–92. [Google Scholar]
- Holland, J. B. 2004 Implementation of molecular markers for quantitative traits in breeding programs—challenges and opportunities. In Proc. 4th Int. Crop Sci. Congress., Brisbane, Australia, 26 September—1 October
- Hospital F. Selection in backcross programmes. Phil. Trans. R. Soc. B. 2005;360:1503–1511. doi: 10.1098/rstb.2005.1670. doi:10.1098/rstb.2005.1670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hospital F, Charcosset A. Marker-assisted introgression of quantitative trait loci. Genetics. 1997;147:1469–1485. doi: 10.1093/genetics/147.3.1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hospital F, Moreau L, Lacoudre F, Charcosset A, Gallais A. More on the efficiency of marker-assisted selection. Theor. Appl. Genet. 1997;95:1181–1189. doi:10.1007/s001220050679 [Google Scholar]
- Huang N, Angeles E.R, Domingo J, Magpantay G, Singh S, Zhang G, Kumaravadivel N, Bennett J, Khush G.S. Pyramiding of bacterial blight resistance genes in rice: marker-assisted selection using RFLP and PCR. Theor. Appl. Genet. 1997;95:313–320. doi:10.1007/s001220050565 [Google Scholar]
- Huang J.K, Pray C, Rozelle S. Enhancing the crops to feed the poor. Nature. 2002;418:678–684. doi: 10.1038/nature01015. doi:10.1038/nature01015 [DOI] [PubMed] [Google Scholar]
- Jaccoud D, Peng K, Feinstein D, Kilian A. Diversity arrays: a solid state technology for sequence information independent genotyping. Nucleic Acids Res. 2001;29:e25. doi: 10.1093/nar/29.4.e25. doi:10.1093/nar/29.4.e25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferies S.P, King B.J, Barr A.R, Warner P, Logue S.J, Langridge P. Marker-assisted backcross introgression of the Yd2 gene conferring resistance to barley yellow dwarf virus in barley. Plant Breed. 2003;122:52–56. doi:10.1046/j.1439-0523.2003.00752.x [Google Scholar]
- Jiang G.H, Xu C.G, Tu J.M, Li X.H, He Y.Q, Zhang Q.F. Pyramiding of insect- and disease-resistance genes into an elite indica, cytoplasm male sterile restorer line of rice, ‘Minghui 63’. Plant Breed. 2004;123:112–116. doi:10.1046/j.1439-0523.2003.00917.x [Google Scholar]
- Jones E.S, Liu C.J, Gale M.D, Hash C.T, Witcombe J.R. Mapping quantitative trait loci for downy mildew resistance in pearl millet. Theor. Appl. Genet. 1995;91:448–456. doi: 10.1007/BF00222972. doi:10.1007/BF00222972 [DOI] [PubMed] [Google Scholar]
- Jorasch P. Intellectual property rights in the field of molecular marker analysis. In: Lorz H, Wenzel G, editors. Biotechnology in agriculture and forestry, molecular marker system. vol. 55. Springer; Berlin, Germany: 2004. [Google Scholar]
- Jordan D.R, Tao Y, Godwin I.D, Henzell R.G, Cooper M, McIntyre C.L. Prediction of hybrid performance in grain sorghum using RFLP markers. Theor. Appl. Genet. 2003;106:559–567. doi: 10.1007/s00122-002-1144-5. [DOI] [PubMed] [Google Scholar]
- Jordan D.R, Tao Y, Godwin I.D, Henzell R.G, Cooper M, McIntyre C.L. Comparison of identity by descent and identity by state for detecting genetic regions under selection in a sorghum pedigree breeding program. Mol. Breed. 2004;14:441–454. doi:10.1007/s11032-005-0901-y [Google Scholar]
- Joseph M, Gopalakrishnan S, Sharma R.K, Singh V.P, Singh A.K, Singh N.K, Mohapatra T. Combining bacterial blight resistance and Basmati quality characteristics by phenotypic and molecular marker-assisted selection in rice. Mol. Breed. 2004;13:377–387. doi:10.1023/B:MOLB.0000034093.63593.4c [Google Scholar]
- Kearsey M.J. The principles of QTL analysis (a minimal mathematics approach) J. Exp. Bot. 1998;49:1619–1623. doi:10.1093/jexbot/49.327.1619 [Google Scholar]
- Kearsey M.J, Farquhar A.G.L. QTL analysis in plants; where are we now? Heredity. 1998;80:137–142. doi: 10.1046/j.1365-2540.1998.00500.x. doi:10.1046/j.1365-2540.1998.00500.x [DOI] [PubMed] [Google Scholar]
- Kloppers F.J, Pretorius Z.A. Effects of combinations amongst genes Lr13, Lr34 and Lr37 on components of resistance in wheat to leaf rust. Plant Pathol. 1997;46:737–750. doi:10.1046/j.1365-3059.1997.d01-58.x [Google Scholar]
- Koebner R.M.D, Summers R.W. 21st century wheat breeding: plot selection or plate detection? Trends Biotech. 2003;21:59–63. doi: 10.1016/S0167-7799(02)00036-7. doi:10.1016/S0167-7799(02)00036-7 [DOI] [PubMed] [Google Scholar]
- Kuchel H, Ye G.Y, Fox R, Jefferies S. Genetic and economic analysis of a targeted marker-assisted wheat breeding strategy. Mol. Breed. 2005;16:67–78. doi:10.1007/s11032-005-4785-7 [Google Scholar]
- Lande R, Thompson R. Efficiency of marker-assisted selection in the improvement of quantitative traits. Genetics. 1990;124:743–756. doi: 10.1093/genetics/124.3.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langridge, P. 2006 Lessons from applying genomics to wheat and barley improvement. In Fifth Int. Rice Genetics Symp., Manila, Philippines Los Baños, The Philippines: International Rice Research Institute.
- Langridge P, Chalmers K. The principle: identification and application of molecular markers. In: Lorz H, Wenzel G, editors. Biotechnology in agriculture and forestry. Molecular marker systems. vol. 55. Springer; Heidelberg, Germany: 2005. pp. 3–22. [Google Scholar]
- Langridge P, Lagudah E, Holton T, Appels R, Sharp P, Chalmers K. Trends in genetic and genome analyses in wheat: a review. Aust. J. Agric. Res. 2001;52:1043–1077. doi:10.1071/AR01082 [Google Scholar]
- Lee M, Godshalk E.B, Lamkey K.R, Woodman W.W. Association of restriction fragment length polymorphisms among maize inbreds with agronomic performance of their crosses. Crop Sci. 1989;29:1067–1071. [Google Scholar]
- Lehmensiek A, Eckermann P.J, Verbyla A.P, Appels R, Sutherland M.W, Daggard G.E. Curation of wheat maps to improve map accuracy and QTL detection. Aust. J. Agric. Res. 2005;56:1347–1354. [Google Scholar]
- Li, Z. K. 2000 QTL mapping in rice: a few critical considerations. In Proc. Fourth Int. Rice Genetics Symp. (eds G. S. Khush, D. S. Brar & B. Hardy), pp. 153–171. Los Baños, The Philippines: International Rice Research Institute.
- Li Z.K, et al. QTL × environment interactions in rice. I. Heading date and plant height. Theoret. Appl. Genet. 2003;108:141–153. doi: 10.1007/s00122-003-1401-2. doi:10.10071s00122-003-1401-2 [DOI] [PubMed] [Google Scholar]
- Liao C.Y, Wu P, Hu B, Yi K.K. Effects of genetic background and environment on QTLs and epistasis for rice (Oryza sativa L.) panicle number. Theor. Appl. Genet. 2001;103:104–111. doi:10.1007/s001220000528 [Google Scholar]
- Liu B. CRC Press; Boca Raton, FL: 1998. Statistical genomics: linkage, mapping and QTL analysis. [Google Scholar]
- Liu S.X, Anderson J.A. Marker assisted evaluation of Fusarium head blight resistant wheat germplasm. Crop Sci. 2003;43:760–766. [Google Scholar]
- Liu J, Liu D, Tao W, Li W, Wang S, Chen P, Cheng S, Gao D. Molecular marker-facilitated pyramiding of different genes for powdery mildew resistance in wheat. Plant Breed. 2000;119:21–24. doi:10.1046/j.1439-0523.2000.00431.x [Google Scholar]
- Liu S.P, Li X, Wang C.Y, Li X.H, He Y.Q. Improvement of resistance to rice blast in Zhenshan 97 by molecular marker-aided selection. Acta Bot. Sin. 2003;45:1346–1350. [Google Scholar]
- Mackill D.J. Breeding for resistance to abiotic stresses in rice: the value of quantitative trait loci. In: Lamkey K.R, Lee M, editors. Plant breeding: the Arnel R. Hallauer International Symposium. Blackwell Publication; Ames, IA: 2006. pp. 201–212. [Google Scholar]
- Mackill, D. J. & Ni, J. 2000 Molecular mapping and marker assisted selection for major-gene traits in rice. In Proc. Fourth Int. Rice Genetics Symp. (eds G. S. Khush, D. S. Brar & B. Hardy), pp. 137–151. Los Baños, The Philippines: International Rice Research Institute.
- Mackill D.J, Nguyen H.T, Zhang J. Use of molecular markers in plant improvement programs for rainfed lowland rice. Field Crops Res. 1999;64:177–185. doi:10.1016/S0378-4290(99)00058-1 [Google Scholar]
- Mackill, D. J., Collard, B. C. Y., Neeraja, C. N., Maghirang-Rodriquez, R., Heuer, S. & Ismail, A. M. 2006 QTLs in rice breeding: examples for abiotic stresses. In Fifth Int. Rice Genetics Symp., Manila, Philippines Los Baños, The Philippines: International Rice Research Institute.
- Matthews D.E, Carollo V.L, Lazo G.R, Anderson O.D. GrainGenes, the genome database for small-grain crops. Nucleic Acids Res. 2003;31:183–186. doi: 10.1093/nar/gkg058. doi:10.1093/nar/gkg058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCartney C.A, Somers D.J, Fedak G, Cao W. Haplotype diversity at fusarium head blight resistance QTLs in wheat. Theor. Appl. Genet. 2004;109:261–271. doi: 10.1007/s00122-004-1640-x. doi:10.1007/s00122-004-1640-x [DOI] [PubMed] [Google Scholar]
- Melchinger A.E, Utz H.F, Schon C.C. Quantitative trait locus (QTL) mapping using different testers and independent population samples in maize reveals low power of QTL detection and large bias in estimates of QTL effects. Genetics. 1998;149:383–403. doi: 10.1093/genetics/149.1.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohan M, Nair S, Bhagwat A, Krishna T.G, Yano M, Bhatia C.R, Sasaki T. Genome mapping, molecular markers and marker-assisted selection in crop plants. Mol. Breed. 1997;3:87–103. doi:10.1023/A:1009651919792 [Google Scholar]
- Mohler, V. & Singrun, C. 2004 General considerations: marker-assisted selection. In Biotechnology in agriculture and forestry, vol. 55: Molecular marker systems (eds H. Lorz & G. Wenzel), pp. 305–317. Berlin, Germany: Springer.
- Moreau L, Charcosset A, Gallais A. Experimental evaluation of several cycles of marker-assisted selection in maize. Euphytica. 2004;137:111–118. doi:10.1023/B:EUPH.0000040508.01402.21 [Google Scholar]
- Morris M, Dreher K, Ribaut J.M, Khairallah M. Money matters (II): costs of maize inbred line conversion schemes at CIMMYT using conventional and marker-assisted selection. Mol. Breed. 2003;11:235–247. doi:10.1023/A:1022872604743 [Google Scholar]
- Naylor R.L, Falcon W.P, Goodman R.M, Jahn M.M, Sengooba T, Tefera H, Nelson R.J. Biotechnology in the developing world: a case for increased investments in orphan crops. Food Policy. 2004;29:15–44. doi:10.1016/j.foodpol.2004.01.002 [Google Scholar]
- Okada Y, Kanatani R, Arai S, Ito K. Interaction between barley yellow mosaic disease-resistance genes rym1 and rym5, in the response to BaYMV strains. Breed. Sci. 2004;54:319–325. doi:10.1270/jsbbs.54.319 [Google Scholar]
- Ortiz R. Critical role of plant biotechnology for the genetic improvement of food crops: perspectives for the next millennium. Electron. J. Biotechnol. 1998;1(3), [cited 15 August] doi:10.2225/vol1-issue3-fulltext-7 [Google Scholar]
- Paterson A.H, Lander E.S, Hewitt J.D, Peterson S, Lincoln S.E, Tanksley S.D. Resolution of quantitative traits into Mendelian factors by using a complete linkage map of restriction fragment length polymorphisms. Nature. 1988;335:721–726. doi: 10.1038/335721a0. doi:10.1038/335721a0 [DOI] [PubMed] [Google Scholar]
- Peleman J.D, van der Voort J.R. Breeding by design. Trends Plant Sci. 2003;8:330–334. doi: 10.1016/S1360-1385(03)00134-1. doi:10.1016/S1360-1385(03)00134-1 [DOI] [PubMed] [Google Scholar]
- Phillips, R. L., Odland, W. E. & Kahler, A. L. 2006 Rice as a reference genome and more. In Proc. Fifth Int. Rice Genetics Symposium, Manila, Philippines, 19–23 November 2005
- Pingali, P. L. & Heisey, P. W. 1999 Cereal crop productivity in developing countries. CIMMYT Economics Paper 99-03. CIMMYT, Mexico, DF.
- Pinstrup-Andersen P, Pandya-Lorch R, Rosegrant M.W. International Food Policy Research Institute; Washington, DC: 1999. World food prospects: critical issues for the early twenty-first century. [Google Scholar]
- Price A.H. Believe it or not, QTLs are accurate! Trends Plant Sci. 2006;11:213–216. doi: 10.1016/j.tplants.2006.03.006. doi:10.1016/j.tplants.2006.03.006 [DOI] [PubMed] [Google Scholar]
- Rafalski A. Applications of single nucleotide polymorphisms in crop genetics. Curr. Opin. Plant Biol. 2002;5:94–100. doi: 10.1016/s1369-5266(02)00240-6. doi:10.1016/S1369-5266(02)00240-6 [DOI] [PubMed] [Google Scholar]
- Reif J.C, Melchinger A.E, Xia X.C, Warburton M.L, Hoisington D.A, Vasal S.K, Beck D, Bohn M, Frisch M. Use of SSRs for establishing heterotic groups in subtropical maize. Theor. Appl. Genet. 2003;107:947–957. doi: 10.1007/s00122-003-1333-x. doi:10.1007/s00122-003-1333-x [DOI] [PubMed] [Google Scholar]
- Reif J.C, Hamrit S, Heckenberger M, Schipprack W, Maurer H.P, Bohn M, Melchinger A.E. Trends in genetic diversity among European maize cultivars and their parental components during the past 50 years. Theor. Appl. Genet. 2005;111:838–845. doi: 10.1007/s00122-005-0004-5. doi:10.1007/s00122-005-0004-5 [DOI] [PubMed] [Google Scholar]
- Ribaut J.-M, Betran J. Single large-scale marker-assisted selection (SLS–MAS) Mol. Breed. 1999;5:531–541. doi:10.1023/A:1009631718036 [Google Scholar]
- Ribaut J.-M, Hoisington D. Marker-assisted selection: new tools and strategies. Trends Plant Sci. 1998;3:236–239. doi:10.1016/S1360-1385(98)01240-0 [Google Scholar]
- Ribaut J.-M, William H.M, Khairallah M, Worland A.J, Hoisington D. Genetic basis of physiological traits. In: Reynolds M.P, Ortiz-Monasterio J.I, McNab A, editors. Application of physiology in wheat breeding. CIMMYT; Mexico, DF: 2001. [Google Scholar]
- Ribaut J.-M, Jiang C, Hoisington D. Simulation experiments on efficiencies of gene introgression by backcrossing. Crop Sci. 2002;42:557–565. [Google Scholar]
- Romagosa I, Han F, Ullrich S.E, Hayes P.M, Wesenberg D.M. Verification of yield QTL through realised molecular marker-assisted selection responses in a barley cross. Mol. Breed. 1999;5:143–152. doi:10.1023/A:1009684108922 [Google Scholar]
- Rostoks N, Borevitz J.O, Hedley P.E, Russell J, Mudie S, Morris J, Cardel L, Marshall D.F, Waugh R. Single-feature polymorphism discovery in the barley transcriptome. Genome Biol. 2005;6:R54. doi: 10.1186/gb-2005-6-6-r54. doi:10.1186/gb-2005-6-6-r54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruttan V.W. The transition to agricultural sustainability. Proc. Natl Acad. Sci. USA. 1999;96:5960–5967. doi: 10.1073/pnas.96.11.5960. doi:10.1073/pnas.96.11.5960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salina E, Dobrovolskaya O, Efremova T, Leonova I, Roder M.S. Microsatellite monitoring of recombination around the Vrn-B1 locus of wheat during early backcross breeding. Plant Breed. 2003;122:116–119. doi:10.1046/j.1439-0523.2003.00817.x [Google Scholar]
- Sanchez A.C, Brar D.S, Huang N, Li Z, Khush G.S. Sequence tagged site marker-assisted selection for three bacterial blight resistance genes in rice. Crop Sci. 2000;40:792–797. [Google Scholar]
- Schmierer D.A, Kandemir N, Kudrna D.A, Jones B.L, Ullrich S.E, Kleinhofs A. Molecular marker-assisted selection for enhanced yield in malting barley. Mol. Breed. 2004;14:463–473. doi:10.1007/s11032-004-0903-1 [Google Scholar]
- Servin B, Martin O.C, Mezard M, Hospital F. Toward a theory of marker-assisted gene pyramiding. Genetics. 2004;168:513–523. doi: 10.1534/genetics.103.023358. doi:10.1534/genetics.103.023358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan X, Blake T.K, Talbert L.E. Conversion of AFLP markers to sequence-specific PCR markers in barley and wheat. Theor. Appl. Genet. 1999;98:1072–1078. doi:10.1007/s001220051169 [Google Scholar]
- Shanti M.L, George M.L.C, Cruz C.M.V, Bernardo M.A, Nelson R.J, Leung H, Reddy J.N, Sridhar R. Identification of resistance genes effective against rice bacterial blight pathogen in eastern India. Plant Dis. 2001;85:506–512. doi: 10.1094/PDIS.2001.85.5.506. doi:10.1094/PDIS.2001.85.5.506 [DOI] [PubMed] [Google Scholar]
- Sharma P.N, Torii A, Takumi S, Mori N, Nakamura C. Marker-assisted pyramiding of brown planthopper (Nilaparvata lugens Stål) resistance genes Bph1 and Bph2 on rice chromosome 12. Hereditas. 2004;140:61–69. doi: 10.1111/j.1601-5223.2004.01726.x. doi:10.1111/j.1601-5223.2004.01726.x [DOI] [PubMed] [Google Scholar]
- Sharp P.J, et al. Validation of molecular markers for wheat breeding. Aust. J. Agric. Res. 2001;52:1357–1366. doi:10.1071/AR01052 [Google Scholar]
- Shen L, Courtois B, McNally K.L, Robin S, Li Z. Evaluation of near-isogenic lines of rice introgressed with QTLs for root depth through marker-aided selection. Theor. Appl. Genet. 2001;103:75–83. doi:10.1007/s001220100538 [Google Scholar]
- Singh S, Sidhu J.S, Huang N, Vikal Y, Li Z, Brar D.S, Dhaliwal H.S, Khush G.S. Pyramiding three bacterial blight resistance genes (xa5, xa13 and Xa21) using marker-assisted selection into indica rice cultivar PR106. Theor. Appl. Genet. 2001;102:1011–1015. doi:10.1007/s001220000495 [Google Scholar]
- Slafer G.A, Araus J.L, Royo C, Del Moral L.F.G. Promising eco-physiological traits for genetic improvement of cereal yields in Mediterranean environments. Ann. Appl. Biol. 2005;146:61–70. doi:10.1111/j.1744-7348.2005.04048.x [Google Scholar]
- Spielmeyer W, Sharp P.J, Lagudah E.S. Identification and validation of markers linked to broad-spectrum stem rust resistance gene Sr2 in wheat (Triticum aestivum L.) Crop Sci. 2003;43:333–336. [Google Scholar]
- Steele K.A, Edwards G, Zhu J, Witcombe J.R. Marker-evaluated selection in rice: shifts in allele frequency among bulks selected in contrasting agricultural environments identify genomic regions of importance to rice adaptation and breeding. Theor. Appl. Genet. 2004;109:1247–1260. doi: 10.1007/s00122-004-1732-7. doi:10.1007/s00122-004-1732-7 [DOI] [PubMed] [Google Scholar]
- Steele K.A, Price A.H, Shashidhar H.E, Witcombe J.R. Marker-assisted selection to introgress rice QTLs controlling root traits into an Indian upland rice variety. Theor. Appl. Genet. 2006;112:208–221. doi: 10.1007/s00122-005-0110-4. doi:10.1007/s00122-005-0110-4 [DOI] [PubMed] [Google Scholar]
- Stoskopf N.C, Tomes D.T, Christie B.R. Westview Press Inc.; San Francisco, CA; Oxford: 1993. Plant breeding: theory and practice. [Google Scholar]
- Stuber C.W, Polacco M, Senior M.L. Synergy of empirical breeding, marker-assisted selection, and genomics to increase crop yield potential. Crop Sci. 1999;39:1571–1583. [Google Scholar]
- Syvanen A.C. Accessing genetic variation: genotyping single nucleotide polymorphisms. Nat. Rev. Genet. 2001;2:930–942. doi: 10.1038/35103535. doi:10.1038/35103535 [DOI] [PubMed] [Google Scholar]
- Syvanen A.C. Toward genome-wide SNP genotyping. Nat. Genet. 2005;37:S5–S10. doi: 10.1038/ng1558. doi:10.1038/ng1558 [DOI] [PubMed] [Google Scholar]
- Tanksley S.D. Molecular markers in plant breeding. Plant Mol. Biol. Rep. 1983;1:3–8. [Google Scholar]
- Tanksley S. Mapping polygenes. Ann. Rev. Genet. 1993;27:205–233. doi: 10.1146/annurev.ge.27.120193.001225. doi:10.1146/annurev.ge.27.120193.001225 [DOI] [PubMed] [Google Scholar]
- Tanksley S.D, Nelson J.C. Advanced backcross QTL analysis: a method for the simultaneous discovery and transfer of valuable QTLs from unadapted germplasm into elite breeding lines. Theor. Appl. Genet. 1996;92:191–203. doi: 10.1007/BF00223376. doi:10.1007/s001220050114 [DOI] [PubMed] [Google Scholar]
- Thomas W. Prospects for molecular breeding of barley. Ann. Appl. Biol. 2003;142:1–12. doi:10.1111/j.1744-7348.2003.tb00223.x [Google Scholar]
- Toojinda T, Baird E, Booth A, Broers L, Hayes P, Powell W, Thomas W, Vivar H, Young G. Introgression of quantitative trait loci (QTLs) determining stripe rust resistance in barley: an example of marker-assisted line development. Theor. Appl. Genet. 1998;96:123–131. doi:10.1007/s001220050718 [Google Scholar]
- Toojinda T, Tragoonrung S, Vanavichit A, Siangliw J.L, Pa-In N, Jantaboon J, Siangliw M, Fukai S. Molecular breeding for rainfed lowland rice in the Mekong region. Plant Prod. Sci. 2005;8:330–333. doi:10.1626/pps.8.330 [Google Scholar]
- Trethowan R.M, Reynolds M, Sayre K, Ortiz-Monasterio I. Adapting wheat cultivars to resource conserving farming practices and human nutritional needs. Ann. Appl. Biol. 2005;146:405–413. doi:10.1111/j.1744-7348.2005.040137.x [Google Scholar]
- van Berloo R, Aalbers H, Werkman A, Niks R.E. Resistance QTL confirmed through development of QTL–NILs for barley leaf rust resistance. Mol. Breed. 2001;8:187–195. doi:10.1023/A:1013722008561 [Google Scholar]
- Van Sanford D, Anderson J, Campbell K, Costa J, Cregan P, Griffey C, Hayes P, Ward R. Discovery and deployment of molecular markers linked to Fusarium head blight resistance: an integrated system for wheat and barley. Crop Sci. 2001;41:638–644. [Google Scholar]
- Varshney R.K, Graner A, Sorrells M.E. Genic microsatellite markers in plants: features and applications. Trends Biotech. 2005;23:48–55. doi: 10.1016/j.tibtech.2004.11.005. doi:10.1016/j.tibtech.2004.11.005 [DOI] [PubMed] [Google Scholar]
- Visscher P.M, Haley C.S, Thompson R. Marker-assisted introgression in backcross breeding programs. Genetics. 1996;144:1923–1932. doi: 10.1093/genetics/144.4.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ware D, et al. Gramene: a resource for comparative grass genomics. Nucleic Acids Res. 2002a;30:103–105. doi: 10.1093/nar/30.1.103. doi:10.1093/nar/30.1.103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ware D.H, et al. Gramene, a tool for grass genomics. Plant Physiol. 2002b;130:1606–1613. doi: 10.1104/pp.015248. doi:10.1104/pp.015248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner K, Friedt W, Ordon F. Strategies for pyramiding resistance genes against the barley yellow mosaic virus complex (BaMMV, BaYMV, BaYMV-2) Mol. Breed. 2005;16:45–55. doi:10.1007/s11032-005-3445-2 [Google Scholar]
- Willcox M.C, et al. Selection for resistance to southwestern corn borer using marker-assisted and conventional backcrossing. Crop Sci. 2002;42:1516–1528. [Google Scholar]
- Witcombe J.R, Hash C.T. Resistance gene deployment strategies in cereal hybrids using marker-assisted selection: gene pyramiding, three-way hybrids, and synthetic parent populations. Euphytica. 2000;112:175–186. doi:10.1023/A:1003836132603 [Google Scholar]
- Witcombe J.R, Virk D.S. Number of crosses and population size for participatory and classical plant breeding. Euphytica. 2001;122:451–462. doi:10.1023/A:1017524122821 [Google Scholar]
- Xu Y.B, Beachell H, McCouch S.R. A marker-based approach to broadening the genetic base of rice in the USA. Crop Sci. 2004;44:1947–1959. [Google Scholar]
- Yashitola J, Thirumurugan T, Sundaram R.M, Naseerullah M.K, Ramesha M.S, Sarma N.P, Sonti R.V. Assessment of purity of rice hybrids using microsatellite and STS markers. Crop Sci. 2002;42:1369–1373. [Google Scholar]
- Young N.D. A cautiously optimistic vision for marker-assisted breeding. Mol. Breed. 1999;5:505–510. doi:10.1023/A:1009684409326 [Google Scholar]
- Yu K, Park S, Poysa V. Marker-assisted selection of common beans for resistance to common bacterial blight: efficacy and economics. Plant Breed. 2000;119:411–415. doi:10.1046/j.1439-0523.2000.00514.x [Google Scholar]
- Yuan Q.P, Quackenbush J, Sultana R, Pertea M, Salzberg S.L, Buell C.R. Rice bioinformatics: analysis of rice sequence data and leveraging the data to other plant species. Plant Physiol. 2001;125:1166–1174. doi: 10.1104/pp.125.3.1166. doi:10.1104/pp.125.3.1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P.H, Tan Y.F, He Y.Q, Xu C.G, Zhang Q. Simultaneous improvement for four quality traits of Zhenshan 97, an elite parent of hybrid rice, by molecular marker-assisted selection. Theor. Appl. Genet. 2003a;106:326–331. doi: 10.1007/s00122-002-1023-0. [DOI] [PubMed] [Google Scholar]
- Zhou W.C, Kolb F.L, Bai G.H, Domier L.L, Boze L.K, Smith N.J. Validation of a major QTL for scab resistance with SSR markers and use of marker-assisted selection in wheat. Plant Breed. 2003b;122:40–46. doi:10.1046/j.1439-0523.2003.00802.x [Google Scholar]
- Zhou R.H, Zhu Z.D, Kong X.Y, Huo N.X, Tian Q.Z, Li P, Jin C.Y, Dong Y.C, Jia J.Z. Development of wheat near-isogenic lines for powdery mildew resistance. Theor. Appl. Genet. 2005;110:640–648. doi: 10.1007/s00122-004-1889-0. doi:10.1007/s00122-004-1889-0 [DOI] [PubMed] [Google Scholar]
- Zwart R.S, Thompson J.P, Godwin I.D. Genetic analysis of resistance to root-lesion nematode (Pratylenchus thornei) in wheat. Plant Breed. 2004;123:209–212. doi:10.1111/j.1439-0523.2004.00986.x [Google Scholar]