Abstract
Single molecule imaging and manipulation are powerful tools in describing the operations of molecular machines like molecular motors. The single molecule measurements allow a dynamic behaviour of individual biomolecules to be measured. In this paper, we describe how we have developed single molecule measurements to understand the mechanism of molecular motors. The step movement of molecular motors associated with a single cycle of ATP hydrolysis has been identified. The single molecule measurements that have sensitivity to monitor thermal fluctuation have revealed that thermal Brownian motion is involved in the step movement of molecular motors. Several mechanisms have been suggested in different motors to bias random thermal motion to directional movement.
Keywords: single molecule measurement, molecular motors, Brownian motion, mechanochemical coupling
1. Introduction
Progress in bioscience studies has been intimately related to the development of new technologies. These new technologies are in response to the need for acquiring new and better data in order to improve our understanding. New data with new technologies, in some cases, lead us to new fundamental concepts regarding the operation of life. Recently, single molecule detection techniques have been developed and applied to a wide range of biosciences. Single molecule manipulation techniques allow for the mechanical properties of biomolecule interaction to be measured. Single molecule imaging techniques can visualize the behaviour of individual biomolecules. In this paper, we describe the recently developed science and technologies in molecular motor studies, demonstrating how single molecule techniques have been needed to be developed in the studies of molecular motors and how the latest in single molecule techniques has led to understanding the mechanism of molecular motors.
Molecular motors are molecular machines that convert chemical energy released from the hydrolysis of ATP to mechanical work for unidirectional movement. In particular, the actomyosin motors have been extensively studied as contractile elements of muscle for more than a half century. However, until very recently, energy input and output have been studied separately in different experimental systems. The mechanical properties of muscle (output) have been studied using muscle fibres and myofibrils. Based on structural changes in these macroscopic apparatus, muscle contraction has been described by a sliding between myosin and actin filaments. Between these two filaments, cross-bridges or myosin heads (motor domain of myosin) that protrude from the myosin filaments towards the actin filament were observed as motors for the sliding movement. Two modes of cross-bridges were found in both the contracting and the relaxed muscle. These modes were interpreted as different orientations of the cross-bridge relative to the actin filaments. On the other hand, the biochemical studies have been performed using solubilized myosin and actin in solution. They showed that myosin and actin cycle between weak and strong interaction states depending on ATP hydrolysis. Considering these data, weak and strong interacting states are related to two cross-bridge states leading to a swinging cross-bridge model where the sliding movement is caused by swinging of the myosin head associated with the ATP hydrolysis (Huxley 1969). However, until recently, no assay system has been able to directly observe the sliding between actin and myosin. No direct evidence for the structural changes in myosin has been detected, either.
In the meantime, individual microtubules and the organelles moving along have been visualized using video-enhanced differential interference contrast microscopy (Allen et al. 1981). Kinesin was discovered as a microtubule-based motor responsible for the transport of organelles (Vale et al. 1985). These studies in the microtubule system have stimulated research in the field of actomyosin. Spudich and his colleagues demonstrated that HMM-coated beads slide along actin cables consisting of hundreds of filaments with identical polarity prepared from Nitella cells (Sheetz & Spudich 1983). Fujime also demonstrated the sliding movement between myosin filaments and actin bundles formed in the presence of 20–50 mM MgCl2 during and after superprecipitation (Higashi-Fujime 1985).
2. Visualization of actin filaments and thermal bending motion influenced by interaction with myosin heads
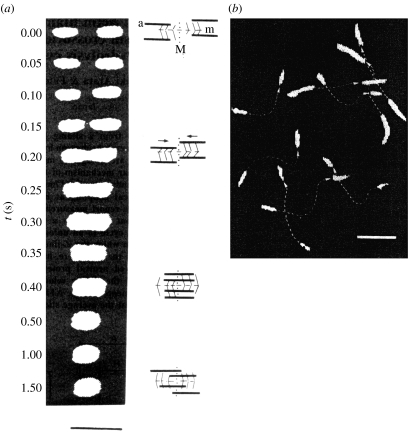
Visualization of individual actin filaments was a key to visualize the sliding movement of individual actin filaments. Actin filaments are too thin (5 nm in radius) to be observed under a light microscope but were visualized by attaching fluorescently labelled phalloidin (Yanagida et al. 1984; figure 1). Phalloidin specifically binds to the filamentous form of actin to stabilize against depolymerization, allowing the observation of the stable actin filament under a microscope (Wulf et al. 1979). The binding of phalloidin does not affect the physiological, biochemical or structural properties of the filaments. Fluorescently labelled actin filaments showed a thermal bending motion in solution (figure 1). The flexibility of the filaments deduced from the end-to-end distance distribution was consistent with that obtained from other techniques such as quasi-elastic light scattering based on ensemble measurements (Fujime & Ishiwata 1971). The bending motion of the filaments was enhanced and accelerated upon the interaction with HMM in the presence of Mg-ATP. However, the sliding movement of actin filaments was not observed in solution. The energy used for the directional movement of actomyosin appeared to dissipate as the thermal motion of the actin filaments increased in solution.
Figure 1.
Visualization of actin filament. Actin filament was labelled with phalloidin-rhodamine and imaged under fluorescence microscopy. Time lapse fluorescence micrographs of actin filament in the absence (upper images) and the presence (lower images) of interaction with HMM are shown. Actin filaments contained tropomyosin and troponin.
3. Visualization of sliding movement of actin and sliding distance during hydrolysis of single ATP molecules
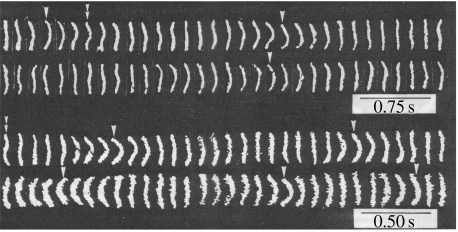
The visualization of actin filaments enabled the observation of their sliding movement (Yanagida et al. 1985; figure 2a). Single sarcomeres, a contractile machine that contains bipolar myosin filaments, and actin filaments sliding into myosin filaments from both sides were prepared by removing Z-lines from myofibrils of crab leg muscle. The actin filaments were specifically labelled with fluorescent phalloidin. The fluorescence micrographs showed that the actin filaments initially located at both ends of the sarcomere moved closer and overlapped at the centre when the sarcomere shortened upon the addition of ATP. The shortening velocity was estimated to be approximately 5 μm s−1 from the time-dependent fluorescence images, which is in agreement with the values estimated from light-scattering changes due to the overlap of myosin and actin filaments in the same suspension and reported for muscle contraction.
Figure 2.
In vitro motility of fluorescently labelled actin filament. (a) Sequence fluorescence micrographs of a sarcomere containing fluorescently labelled actin filament without Z-lines during shortening. Scale bar, 5 μm. (b) Sliding movement of fluorescently labelled actin filaments on a glass slide coated with single-headed myosin filaments. Two images taken 1.5 s apart were photographed on the same frame. Dotted lines represent traces of the movement of actin filaments with arrowheads indicating the direction of the movement. Scale bar, 5 μm.
Using fluorescently labelled actin filaments, Spudich and his colleagues observed the sliding movement of individual actin filaments on myosin filaments fixed on a glass slide (Kron & Spudich 1986). We also observed the sliding movement of individual actin filaments on single-headed myosin filaments instead of double-headed ones (Harada et al. 1987; figure 2b). The movement of actin filaments was smooth and the direction of the movement did not change in both cases. The sliding velocity of the one-headed myosin was the same as that of the two-headed myosin filament, indicating that two heads are not crucial for motility. This result was confirmed by using single-head myosin fragment (S1) and double-head fragment (HMM) fixed on a glass slide (Toyoshima et al. 1987).
To understand the mechanism of the sliding movement, the average sliding distance during hydrolysis of single ATP molecules was estimated by simultaneously measuring the velocity and the ATPase activity (Yanagida et al. 1985; Harada et al. 1990; Toyoshima et al. 1990; Uyeda et al. 1991). If the structural changes cause the movement, then the sliding distance should be of the order of the size of the structural changes. However, the results depended on the research groups, mainly because parameters such as working stroke time in a single ATPase cycle required an estimation that could not be determined directly from the measurements. Ideally, a direct observation of the unitary steps caused by single myosin motors during single ATP hydrolysis cycles would be made. To do this, one must be able to manipulate the actin filaments with nanometre- and millisecond-order sensitivity.
4. Manipulation of single actin filaments and force measurements using a microneedle
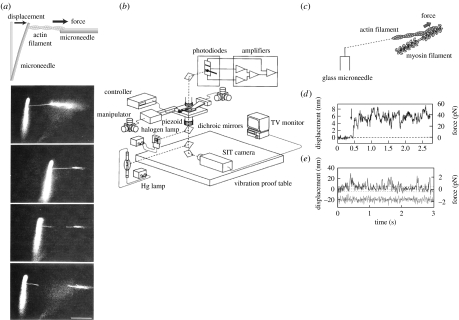
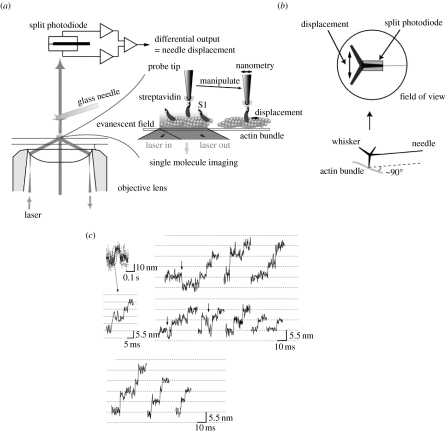
Microneedles have been used for the manipulation of single actin filaments (Kishino & Yanagida 1988; figure 3a). A similar method had originally been used for the manipulation and force measurements for microtubular sliding in flagella (Kamimura & Takahashi 1981). A thin glass thread (less than 1 μm radius) was prepared from a glass rod approximately 1 mm in diameter by heating and pulling it manually using a puller. Single actin filaments were attached to a glass microneedle while they were visualized under microscopy. For the measurement of force exerted by myosin, single actin filaments attached to the glass microneedle were brought to interact with a myosin filament fixed on the glass surface (figure 3c). When the actin filament moved, the flexible microneedle bent proportional to the force and the flexibility of the microneedle. Accurately measuring the displacement of the tip of the microneedle allowed the interaction with myosin molecules to be determined.
Figure 3.
Manipulation of single actin filaments and force measurement using a microneedle. (a) Measurement of the tensile strength of an actin filament. A single actin filament bound to a flexible glass microneedle at one end (at the left) was stretched through a stiff needle bound at the other end until the actin filament was broken. The flexible needle was bent as the filament was stretched. (b–d) Force measurement of the interaction between myosin molecules and actin filament. (b) Schematic of the measurement system with nanometre-, piconewton- and millisecond-order sensitivity. See Ishijima et al. (1996) for details. (c) A single actin filament was captured by a glass needle and interacted with small number of myosin heads in the cofilament of myosin and myosin rod. (d) The force fluctuation exerted by individual myosin molecules was measured as a displacement record of a tip of a microneedle. (e) Displacement record caused by single myosin heads. The lower trace indicates thermal vibrations of free needles. Broken lines show ±5 nm.
The accuracy of the displacement measurements of the glass microneedle was improved to nanometre and millisecond order using a detector composed of a pair of photodiodes (Ishijima et al. 1991; figure 3b). An image of the tip of the microneedle was projected onto the detector and the displacement was measured by recording the difference in intensity between the photodiodes. When single actin filaments interacted with a small number of myosin heads in artificially prepared cofilaments made from a mixture of myosins and myosin rods, force fluctuations generated by individual myosin molecules were observed in an isometric condition (Ishijima et al. 1996; figure 3d). The result suggested that individual myosin molecules underwent attachment, force generation and detachment in a cycle coupled to the hydrolysis of single ATP molecules randomly and independently. When actin filaments were sliding, the fluctuation of displacement observed diverged from the smoothed displacement curves and the amplitude of the variation greatly decreased with the increase in velocity or the decrease in load. In these cases, single mechanical events were not identified from each other.
In order to clearly distinguish the single mechanical events, the number of myosin molecules interacting with a single actin filament was decreased by decreasing the number of myosin heads in the myosin–myosin rod cofilament. Consequently, distinct attachment, force generation and detachment in the unitary step movement were directly observed (figure 3e). The mean displacement of single mechanical events was 17 nm at low needle stiffness (0.09 pN nm−1).
5. Manipulation of actin filaments using laser trap
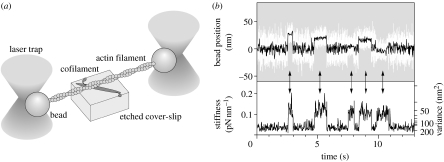
A single actin filament was also manipulated using laser trap (Saito et al. 1994). Laser trap is a method to capture dielectric particles on a focused laser beam (Ashkin & Dziedzic 1985). Dielectric particles such as polystyrene beads are attached to biomolecules because the biomolecules themselves are too small to be trapped. Two dielectric beads of submicron order were trapped by a laser and attached to both the ends of a single actin filament, while being visualized under a fluorescence microscope. Using this method, a single actin filament was suspended taut in solution and the movement of fluorescently labelled myosin filaments could be monitored along it.
The laser trap method had been used successfully for the detection of the step movement of processive motor kinesin (Svoboda et al. 1993). Beads attached to single kinesin molecules could trace the successive step movement of kinesin. In the case of muscle myosin, in contrast to kinesin, myosin motors dissociate after hydrolysis of each ATP molecule. Here, a single actin filament is caught by two beads trapped by a laser at each of the ends of the actin (Finer et al. 1994; figure 4a). The actin filament held taut was brought to interact with a single myosin head attached to the glass surface. The interaction with myosin was detected by an increase in stiffness calculated from the variance of the bead's position (figure 4b). However, the step size of individual steps could not be determined because the starting position of the step was not known due to a fast thermal fluctuation (Molloy et al. 1995). The average step size determined was 5–10 nm for myosin attached directly onto the glass surface (Finer et al. 1994; Molloy et al. 1995; Guilford et al. 1997). To avoid direct interaction between myosin and the glass surface, we measured mixed single-headed myosin in the cofilament with a large excess amount of myosin rod (Tanaka et al. 1998). In this measurement system, we could also measure the effects of the angles between actin and myosin. The average step size was 10–15 nm when myosin and actin filaments were oriented in approximately the same direction and no displacement spikes were observed when they were crossed nearly perpendicularly. Therefore, under the condition where myosin heads were adsorbed randomly in multiple directions, the step size was an average that included less than optimum values. Owing to these differences in the experimental systems and difficulties in the measurements of muscle myosin, the values of the step size varied from 3.5 to 15 nm with the research groups.
Figure 4.
Manipulation and force measurement between single actin filaments and single myosin using laser trap. (a) Schematic of the measurement. (b) Upper image: the time course of displacements by a single myosin molecule. White line represents raw data and black line represents the same data passed through a low pass filter of 20 Hz bandwidth. Lower image: changes in stiffness calculated from the variance of the displacement data as a monitor of the interaction of myosin with actin.
The nucleotide-dependent atomic structures of the motor domain of muscle myosin have been reported (Rayment et al. 1993; Whittaker et al. 1995), providing a basis for a model describing structural changes in the myosin head as the cause for sliding movement. Small changes in the conformation at a nucleotide-binding region caused by a change in the chemical state of the nucleotide are amplified into a rotation at the neck domain relative to the motor domain. Based on this result, the swinging cross-bridge model has been extended to a more elaborate model called the lever-arm model in which the neck domain acts as a lever arm to cause myosin steps. The model predicts a step size of approximately 5 nm. The data from some groups are consistent with the lever-arm model, whereas others are not.
6. Visualization of single molecules and single chemical reactions
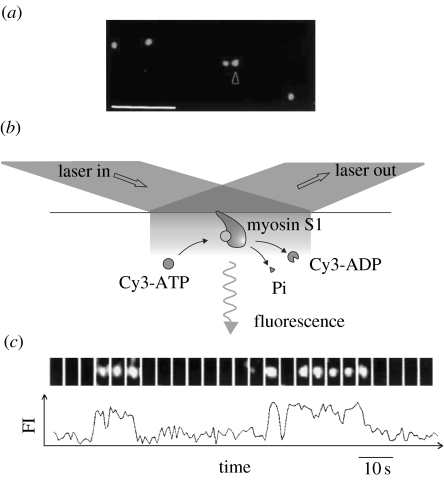
The observed mechanical events are caused by ATP hydrolysis catalysed by myosin. Therefore, it is important to know how these mechanical events are coupled to the chemical reaction of ATP. However, in the mechanical measurements described above, it was not known when ATP molecules were bound to myosin, hydrolysed or when the products were released. In order to detect the ATP turnover coupled to the mechanical events caused by single myosin molecules, single ATP molecules were visualized using fluorescently labelled ATP. In 1995, the visualization of single fluorophores in aqueous solution was accomplished using total internal reflection fluorescence microscopy (TIRFM; Funatsu et al. 1995; figure 5a). In this technique, only those molecules located in the vicinity of the glass slide (approx. 100 nm) are excited by an evanescent field created when the incident laser light is totally reflected at the surface. Local illumination such as the evanescent illumination resulted in a large reduction in background noise, increasing the signal-to-noise ratio. The visualization of single fluorescence molecules allows the observation of dissociation and association events in single molecules. The binding and dissociation of fluorescently labelled ATP associated with the ATP hydrolysis by single myosin molecules was observed as the appearance and disappearance of fluorescence spots at the position where myosin molecules were fixed to the glass surface (figure 5b,c). The time duration from the binding of ATP to the dissociation of ADP varied for individual events and was distributed as an exponential function in the histogram. The decay time statistically determined was consistent with the ATPase rate of myosin in solution in the ensemble measurement.
Figure 5.
Imaging of single fluorescent molecules using TIRFM. (a) Fluorescent micrograph of single Cy5-labelled S1 molecules bound to the surface of a glass slide. Scale bar, 5 μm. (b) Schematic of the visualization of the turnovers of a fluorescent analogue of ATP, Cy3-ATP, catalysed by single S1 molecules. (c) ATP turnovers by a single S1 molecule. Upper image: fluorescent image from Cy3-nucleotides bound to S1. Lower image: time course of the corresponding fluorescence intensity.
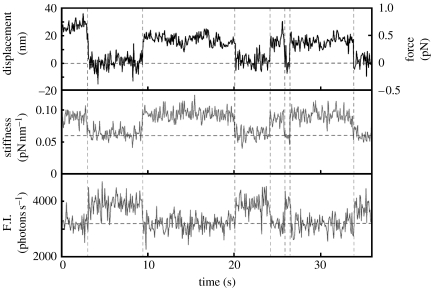
7. Direct determination of coupling between chemical and mechanical events
Based on biochemical studies, the interaction of myosin with actin is described depending on the nucleotide state of myosin. The myosin head interacts with the actin filament weakly in the ATP bound and hydrolysed products (ADPPi) bound state. The displacement of the actin filament is triggered by the release of inorganic phosphate (Pi) and ADP. The actin filament then remains strongly bound to the myosin head until a new ATP binds, which causes detachment of myosin from the actin filament. It should be very important to experimentally test this consensus scheme summarized from the ensemble measurements. Simultaneous measurements using a laser trap and imaging of the turnover of single ATP molecules makes it possible to directly determine the coupling between mechanical events and the hydrolysis of ATP (Ishijima et al. 1998; figure 6). The results were consistent with the biochemical studies. However, a more precise comparison of the timing between the chemical and mechanical events raised a question. The release of ADP and the reattachment of myosin to actin filaments did not always couple whereas the binding of ATP to myosin and the dissociation of myosin from actin was tightly coupled. In half of the cases the displacement occurred at the same time as the release of ADP, while in the other half the displacement was delayed after the dissociation of ADP. It is possible that the energy released from the hydrolysis of ATP is stored at some place in myosin and/or actin for later use.
Figure 6.
The simultaneous observation of individual ATPase and mechanical events. The upper and lower traces show the time course of displacements and changes in fluorescence intensity from the Cy3-nucleotide bound to the myosin head, respectively. The middle trace shows the changes in the stiffness.
8. Manipulation of a single myosin molecule and substeps
The mechanical events usually occurred too quickly to be resolved. The resolution of the measurement system could be improved by increasing the stiffness. The probes, either microneedles or trapped beads, undergo thermal motion due to collisions with water molecules in the surroundings. The variance of the probe is dependent on the stiffness of the probe system. In the laser trap system, the trapped beads are allowed to rotate freely and are attached to the side of actin filaments, which contribute to the compliance in the measurement system. To overcome this problem, we captured single myosin heads to a cantilever attached to the tip of a microneedle, preventing the probe from rotating (Kitamura et al. 1999; figure 7). To achieve this, scanning probes must be prepared and myosin molecules must be captured one by one. The confirmation of single molecules also must be made one by one using single molecule fluorescence imaging. This is in contrast to the laser trap method in which beads and myosin molecules are mixed in solution and the single molecules are confirmed statistically. The result was an improvement in the stiffness reaching 1 pN nm−1 compared with less than 0.2 pN nm−1 for the manipulation of actin filaments by the laser trap system, resulting in a decrease in the variance to 2.0 nm from 4.5 nm. This improvement in space resolution allowed us to monitor the substeps in the rising phase of the displacement during hydrolysis of single ATP molecules. The step generated during a single cycle of ATP hydrolysis was found to contain several substeps of 5.5 nm corresponding to the size of the actin monomer, giving rise to a mean total displacement of approximately 13 nm (figure 7c). These substeps cannot be explained by a conformational change of myosin tightly coupled with the hydrolysis of ATP. The substeps occur stochastically and some of them (less than 10% of the total) occur in the backward direction. The movement is described as a particle diffusing on an energy landscape in which the energy barrier difference for the forward and backward steps is 2–3 kBT. The energy landscape may reflect the interaction between myosin head and actin filament. Several models have been proposed for the mechanism of the substeps (Terada et al. 2002; Esaki et al. 2003).
Figure 7.
Manipulation of single myosin molecules using a scanning probe and measurement of substeps of myosin. (a) Schematic of the measurement. See reference Kitamura et al. (2005) for details. (b) The schematic of the tip of the scanning probe. A single myosin head was captured at the tip of a whisker attached at the glass microneedle. Motion of myosin head is restricted geometrically. (c) Stepwise movements in the rising phase of the displacements. Some backward steps were observed as indicated by arrows.
In the microneedle method, myosin heads are attached to large scanning probes. The movement of the head is restricted by the microneedle (figure 7b). The myosin head is allowed to move along a protofilament. Given that the head cannot go through the bottom of the filament for geometrical reasons, it does not move beyond the pitch of the actin helix unless it changes the path to another protofilament. In fact, the number of substeps was less than seven, in agreement with the number of steps possible along a single protofilament, based on the number of actin monomers in the pitch of the protofilament helix. It should be noted that this situation is similar to myosin heads on the filament in muscle. That is, the myosin head is attached to a large myosin filament and is therefore restricted by the movement of the filament. Thus, the myosin heads attached to a scanning probe behave like myosin molecules in muscle and the mechanical properties of muscle can be understood based on the conclusions obtained in this method.
In muscle, many myosin motors are present, which may modulate the properties of the isolated myosin. Previous reports have suggested that the step size of myosin measured in muscle is longer than that obtained for isolated single myosin molecules (Yanagida et al. 1985; Higuchi & Goldman 1991). A collaboration between myosin heads in the muscle is very probable. A model describing this collaboration was proposed based on the results of the scanning probe measurements. When many myosin motors interact with a single actin filament, the motors may switch a track to a different actin protofilament, resulting in a longer step than the single myosin step. This change is facilitated by rotating the actin filament (Kitamura et al. 2005). Given that the properties of an isolated single motor are well described, the next step is to understand the behaviour of motors in systems where many motors interact.
9. Roles of actin filament in the motility of myosin
These manipulation techniques have been used to study the physical properties of actin filaments. Actin filaments serve as a track for myosin motility. The physical properties of actin filaments are fundamental parameters to understand the mechanism of myosin motility. The basic premises of these techniques have also been applied to studies of the structure and interaction of single biomolecules.
Taking advantage of the laser trap method in which the rotational motion of a trapped bead is allowed, the torsional motion of actin filaments is measured while the actin is manipulated. The actin filaments are held taut vertically by trapping a bead attached at the end by a laser while the other end is fixed on the glass surface (Tsuda et al. 1996). The bead trapped by the laser rotates around the axis of the actin filaments due to thermal torsional motion, while the rotational motion is stopped at the other end. The rotational motion of the bead is measured by monitoring the motion of small fluorescent beads attached to it. The torsional rigidity obtained from the variance of the angle (8.0×10−26 Nm2) was similar to that deduced from longitudinal rigidity (5.8×10−26 Nm2). This method was used for the detection of the rotation of single DNA molecules associated with the step movement of RNA polymerase moving along them (Harada et al. 2001).
The force that breaks the monomer–monomer bond of the actin filament (tensile strength) was determined using both microneedle and laser trap methods (see figure 3a). The tensile strength was determined by pulling the microneedle or trapped bead until it broke by measuring the displacement of the pull (Kishino & Yanagida 1988; Tsuda et al. 1996). The maximum value of tensile strength was 600 pN at the untwisted condition. The tensile strength of the actin filament significantly decreased under a torsion of approximately 100 pN, which was not much greater than the force applied on the filament in a contracting muscle. When a sinusoidal displacement is applied at one end of the actin filament, the displacement at the other end is damped and delayed owing to the extensibility of the filament, allowing the stiffness of the actin filament to be measured. These elastic properties determined by single molecule techniques will be fundamental in considering the mechanical role of actin in muscle contraction, cell motility and formation of cell shape.
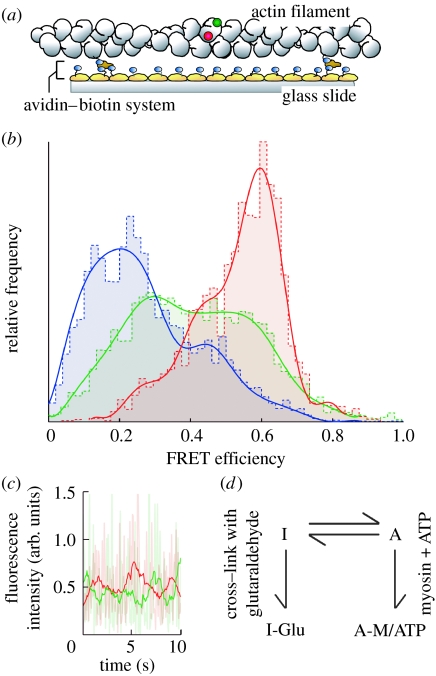
Actin has been suggested to play an active role in myosin motility. The motility of myosin is inhibited while the ATPase activity of myosin is not affected when actin is chemically cross-linked. Recent single molecule studies on actin conformation have revealed that the change in the conformational state of actin is responsible for the activation of myosin motility. The conformational changes of actin monomers in the filaments were observed using the single molecule fluorescence resonance energy transfer (FRET) technique (Kozuka et al. 2006; figure 8). The actin monomers in the filaments spontaneously changed conformation between two states, one in which the motility of myosin is inhibited due to the cross-linking of actin and the other in which it is activated upon the binding of myosin. The binding of the myosin heads shifted the population in the state towards a state that activates the motility of myosin. The conformational changes may involve the relative rotation of two subdomains of the monomer. These conformational changes may result in an increase in the flexibility of actin filaments as demonstrated in the imaging of actin filaments in solution.
Figure 8.
Single molecule FRET measurement from single actin molecules in the filament. (a) Schematic of the experiment. Actin monomer in the filament was specifically labelled with tetramethyl-rhodamine (TMR) attached as a donor and IC5 attached as an acceptor. (b) Histograms of FRET efficiency for actin molecules. Blue bars and line, actin molecules cross-linked with glutaraldehyde. Red bars and line, actin molecules in the presence of myosin V. Green bars and line, intact actin filaments. (c) Time course of the donor (green line) and the acceptor (red line) fluorescence from a double-labelled actin. (d) The conformational dynamics of actin molecules in the actin filament.
10. Processive movement of unconventional myosins
Owing to the development in molecular and cell biology, a variety of myosin motors have recently been available for experimental use. Unlike muscle myosin, unconventional myosins, such as myosin V and VI, move in a processive manner and the step size is much larger (36 nm) than that of muscle myosin. These properties make it easier to measure the mechanical properties of single molecule motors and to interpret the data. The stepwise movement has been detected using high spatial resolution fluorescence imaging as well as laser trap (Yildiz et al. 2003). These data have shown that myosin moves in a hand-over-hand manner where one head steps while the other head stays bound to the actin filament. The large step size of myosin V (36 nm) has been attributed to its long neck region, which is in agreement with the lever-arm model. To further test the lever-arm model, we measured step movements of a mutant of myosin V, in which the neck domain was shortened to one-sixth its natural length (Tanaka et al. 2002). Even then the step size was 36 nm, the same as that of native myosin V. However, the linear relationship between the length of the lever arm and the step size has been reported in many cases. Flexible regions created in the mutated neck domain may contribute to a diffusive motion in unbound heads. This diffusive motion may be involved in the stepping process in addition to structural changes in the myosin head. Recently, the diffusive motion of unbound myosin V heads has been directly observed during the stepping process (Dunn & Spudich 2007; Shiroguchi & Kinosita 2007). The random twisting motion of the neck domain around its axis was observed associated with the step movement (Komori et al. 2007).
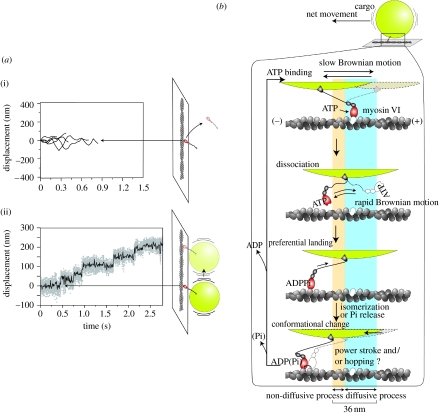
The involvement of diffusive motion was more evident for the processive step movement of the two-headed myosin VI, which has a large step size in spite of its naturally short neck domain (Rock et al. 2001; Nishikawa et al. 2002). A flexible region adjacent to the neck domain contributes to the diffusion of the unbound head and extends to bind to actin. The wide distribution of the step size is consistent with the diffusion mechanism. It has been reported that wild-type myosin VI has only one head when it transports vesicles in cells. Generally, single-headed motors must readily dissociate and diffuse away after a step. In the case of single-headed kinesin, additional interactions with microtubules prevent kinesin from dissociating (Inoue et al. 1997). In the case of myosin VI, single-headed myosin VI did not move processively but the attachment of 200 nm polystyrene beads as a cargo changed the movement of single-headed myosin VI to a processive one with a large step of 40 nm (Iwaki et al. 2006; figure 9a).
Figure 9.
Movements of single-headed myosin VI. (a) Time course of displacement of single-headed myosin VI (i) without bead and (ii) with bead. The traces in (i) were measured by the fluorescence position of GFP fused with myosin head and the trace in (ii) was measured by the bead position. (b) Stepping model for cargo-bound single-headed myosin VI. Huge cargo keeps the myosin head near the actin filament and the head repeatedly attaches and detaches to several actin monomers during diffusion. Finally, the head preferentially lands on the forward binding site.
The cargo-assisted stepwise movement is explained by a slow diffusion of cargoes relative to the diffusion of the head, because the diffusion is dependent on the size of the particles. When a myosin VI head dissociates from actin, the head diffuses rapidly to the next site on an actin filament, while the bead diffuses slowly preventing the head–bead complex from diffusing away from the actin filament. The preferential binding to a site in one direction biases the diffusive motion to that direction (figure 9b). In the case of myosin VI, among many possible explanations, we believe that the myosin head senses the strain applied to it, causing an acceleration in ATP binding and the release of ATP hydrolysis products.
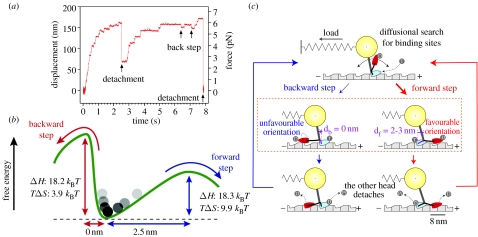
11. Biased step movement of microtubule-based motor kinesin
Kinesin has been extensively studied, partly because it has several advantages over myosin for various experiments. Kinesin is a processive motor, which was expressed before muscle myosin. The movement of individual kinesin molecules along microtubules was visualized immediately after single fluorophores were visualized in aqueous solution (Vale et al. 1996) and the stepwise movement was detected using a laser trap (Svoboda et al. 1993). Kinesin moves in a hand-over-hand manner with regular 8 nm intervals along microtubules (Svoboda et al. 1993; Kojima et al. 1997; figure 10a). Although kinesin is smaller than myosin and a microtubule-based motor, the fundamental structure is similar to that of myosin, leading to the expectation that basic mechanisms are shared with myosin. From structural studies, it has been suggested that the conformational docking at a short region called the neck linker, which is located between the head and coiled-coil tail domain, is responsible for the step movement of kinesin.
Figure 10.
Forward and backward step movements of kinesin. (a) Time course of the displacement of single kinesin molecule measured by optical trapping nanometry. (b) Free-energy landscape for kinesin forward and backward steps. (c) Biased stepping model for kinesin. Random steps are biased by preferential binding of the detached head to the forward binding site on microtubule, leading directional movement of kinesin.
The step movement of kinesin occurs overwhelmingly in one direction at no load. At high load, backward stepping has been observed. The number of the backward steps increases relative to the forward steps as the external force increases (Nishiyama et al. 2002). Finally, successive 8 nm backward steps occur in the presence of excessive load (Carter & Cross 2005). The stochastic processes of kinesin step movement were statistically analysed and the data were interpreted based on the thermal diffusion of the head in a free energy landscape (figure 10b). In the energy landscape of forward and backward step movement, the directional movement has a lower activation free-energy barrier for the forward step as compared with the backward step. The difference in the free-energy barrier between the forward and backward directions was 6kBT, suggesting that the energy reported for docking (1–2kBT) was not sufficient for the bias to the directional movement. The temperature dependence of the step movement of kinesin showed that the difference in the activation free energy between the forward and backward movements is mainly entropic (Taniguchi et al. 2005). One possible explanation is the sterically restricted geometry between the kinesin head and the binding sites on microtubules (figure 10c). The kinesin head orients itself in the opposite direction relative to the orientation of the microtubule when it is located in the forward or backward position. The orientation of the kinesin head in the forward position coincides with the orientation of the binding site on the microtubule biasing forward movement.
12. Perspectives
These single molecule detection techniques allowed us to directly measure the behaviour of individual molecules. For molecular motors, unitary steps during the hydrolysis of single ATP molecules have been described experimentally. Because the single molecule measurements were improved to have the sensitivity to detect thermal fluctuations, it was found that a diffusive motion was involved in the step movement of molecular motors. The random thermal motion of the molecules is biased by preferential landing in one direction resulting in an overall directional movement. The mechanism for the bias is important to explore. These processes will be studied experimentally and theoretically.
In the case of processive motors, especially myosin V and kinesin, that move cargo as only a few molecules, the unit steps are clearly characterized and the mechanism for the bias seems robust against thermal fluctuation, as indicated by a constant step size and almost unidirectional at no load. This property is advantageous for these molecules' biological functions which must be achieved unequivocally in single molecules. In contrast to these processive motors, muscle myosin works when large numbers of myosin motors participate. The step movement of muscle myosin seems rather stochastic when it is isolated, for example, the size of the step has a distribution attributed to stochastic properties of the substeps. This stochastic property may be advantageous in the viewpoint of adaptability, flexibility and perhaps efficiency when it assembles individual myosin molecules into a large machine like the muscle. Understanding the mechanism of the formation of assembly from individual molecules will be the theme that we will attack experimentally and theoretically in the next step.
Footnotes
One contribution of 17 to a Theme Issue ‘Japan: its tradition and hot topics in biological sciences’.
References
- Allen R.D, Allen N.S, Travis J.L. Video-enhanced contrast, differential interference contrast (AVEC-DIC) microscopy: a new method capable of analyzing microtubule-related motility in the reticulopodial network of Allogromia laticollaris. Cell Motil. 1981;1:291–302. doi: 10.1002/cm.970010303. doi:10.1002/cm.970010303 [DOI] [PubMed] [Google Scholar]
- Ashkin A, Dziedzic J.M. Observation of radiation-pressure trapping of particles by alternating light beams. Phys. Rev. Lett. 1985;54:1245–1248. doi: 10.1103/PhysRevLett.54.1245. doi:10.1103/PhysRevLett.54.1245 [DOI] [PubMed] [Google Scholar]
- Carter N.J, Cross R.A. Mechanics of the kinesin step. Nature. 2005;435:308–312. doi: 10.1038/nature03528. doi:10.1038/nature03528 [DOI] [PubMed] [Google Scholar]
- Dunn A.R, Spudich J.A. Dynamics of the unbound head during myosin V processive translocation. Nat. Struct. Mol. Biol. 2007;14:246–248. doi: 10.1038/nsmb1206. doi:10.1038/nsmb1206 [DOI] [PubMed] [Google Scholar]
- Esaki S, Ishii Y, Yanagida T. Model describing the biased Brownian movement of myosin. Proc. Jpn Acad. 2003;79:9–14. doi:10.2183/pjab.79B.9 [Google Scholar]
- Finer J.T, Simmons R.M, Spudich J.A. Single myosin molecule mechanics: piconewton forces and nanometre steps. Nature. 1994;368:113–119. doi: 10.1038/368113a0. doi:10.1038/368113a0 [DOI] [PubMed] [Google Scholar]
- Fujime S, Ishiwata S. Dynamic study of F-actin by quasielastic scattering of laser light. J. Mol. Biol. 1971;62:251–265. doi: 10.1016/0022-2836(71)90144-6. doi:10.1016/0022-2836(71)90144-6 [DOI] [PubMed] [Google Scholar]
- Funatsu T, Harada Y, Tokunaga M, Saito K, Yanagida T. Imaging of single fluorescent molecules and individual ATP turnovers by single myosin molecules in aqueous solution. Nature. 1995;374:555–559. doi: 10.1038/374555a0. doi:10.1038/374555a0 [DOI] [PubMed] [Google Scholar]
- Guilford W.H, Dupuis D.E, Kennedy G, Wu J, Patlak J.B, Warshaw D.M. Smooth muscle and skeletal muscle myosins produce similar unitary forces and displacements in the laser trap. Biophys. J. 1997;72:1006–1021. doi: 10.1016/S0006-3495(97)78753-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada Y, Noguchi A, Kishino A, Yanagida T. Sliding movement of single actin filaments on one-headed myosin filaments. Nature. 1987;326:805–808. doi: 10.1038/326805a0. doi:10.1038/326805a0 [DOI] [PubMed] [Google Scholar]
- Harada Y, Sakurada K, Aoki T, Thomas D.D, Yanagida T. Mechanochemical coupling in actomyosin energy transduction studied by in vitro movement assay. J. Mol. Biol. 1990;216:49–68. doi: 10.1016/S0022-2836(05)80060-9. doi:10.1016/S0022-2836(05)80060-9 [DOI] [PubMed] [Google Scholar]
- Harada Y, Ohara O, Takatsuki A, Itoh H, Shimamoto N, Kinosita K., Jr Direct observation of DNA rotation during transcription by Escherichia coli RNA polymerase. Nature. 2001;409:113–115. doi: 10.1038/35051126. doi:10.1038/35051126 [DOI] [PubMed] [Google Scholar]
- Higashi-Fujime S. Unidirectional sliding of myosin filaments along the bundle of F-actin filaments spontaneously formed during superprecipitation. J. Cell Biol. 1985;101:2335–2344. doi: 10.1083/jcb.101.6.2335. doi:10.1083/jcb.101.6.2335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi H, Goldman Y.E. Sliding distance between actin and myosin filaments per ATP molecule hydrolysed in skinned muscle fibres. Nature. 1991;352:352–354. doi: 10.1038/352352a0. doi:10.1038/352352a0 [DOI] [PubMed] [Google Scholar]
- Huxley H.E. The mechanism of muscular contraction. Science. 1969;164:1356–1365. doi: 10.1126/science.164.3886.1356. doi:10.1126/science.164.3886.1356 [DOI] [PubMed] [Google Scholar]
- Inoue Y, Toyoshima Y.Y, Iwane A.H, Morimoto S, Higuchi H, Yanagida T. Movements of truncated kinesin fragments with a short or an artificial flexible neck. Proc. Natl Acad. Sci. USA. 1997;94:7275–7280. doi: 10.1073/pnas.94.14.7275. doi:10.1073/pnas.94.14.7275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishijima A, Doi T, Sakurada K, Yanagida T. Sub-piconewton force fluctuations of actomyosin in vitro. Nature. 1991;352:301–306. doi: 10.1038/352301a0. doi:10.1038/352301a0 [DOI] [PubMed] [Google Scholar]
- Ishijima A, Kojima H, Higuchi H, Harada Y, Funatsu T, Yanagida T. Multiple- and single-molecule analysis of the actomyosin motor by nanometer–piconewton manipulation with a microneedle: unitary steps and forces. Biophys. J. 1996;70:383–400. doi: 10.1016/S0006-3495(96)79582-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishijima A, Kojima H, Funatsu T, Tokunaga M, Higuchi H, Tanaka H, Yanagida T. Simultaneous observation of individual ATPase and mechanical events by a single myosin molecule during interaction with actin. Cell. 1998;92:161–171. doi: 10.1016/s0092-8674(00)80911-3. doi:10.1016/S0092-8674(00)80911-3 [DOI] [PubMed] [Google Scholar]
- Iwaki M, Tanaka H, Iwane A.H, Katayama E, Ikebe M, Yanagida T. Cargo-binding makes a wild-type single-headed myosin-VI move processively. Biophys. J. 2006;90:3643–3652. doi: 10.1529/biophysj.105.075721. doi:10.1529/biophysj.105.075721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamimura S, Takahashi K. Direct measurement of the force of microtubule sliding in flagella. Nature. 1981;293:566–568. doi: 10.1038/293566a0. doi:10.1038/293566a0 [DOI] [PubMed] [Google Scholar]
- Kishino A, Yanagida T. Force measurements by micromanipulation of a single actin filament by glass needles. Nature. 1988;334:74–76. doi: 10.1038/334074a0. doi:10.1038/334074a0 [DOI] [PubMed] [Google Scholar]
- Kitamura K, Tokunaga M, Iwane A.H, Yanagida T. A single myosin head moves along an actin filament with regular steps of 5.3 nanometres. Nature. 1999;397:129–134. doi: 10.1038/16403. doi:10.1038/16403 [DOI] [PubMed] [Google Scholar]
- Kitamura K, Tokunaga M, Esaki S, Iwane A.H, Yanagida T. Mechanism of muscle contraction based on stochastic properties of single actomyosin motors observed in vitro. Biophysics. 2005;1:1–19. doi: 10.2142/biophysics.1.1. doi:10.2142/biophysics.1.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima H, Muto E, Higuchi H, Yanagida T. Mechanics of single kinesin molecules measured by optical trapping nanometry. Biophys. J. 1997;73:2012–2022. doi: 10.1016/S0006-3495(97)78231-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komori Y, Iwane A.H, Yanagida T. Myosin-V makes two Brownian 90° rotations per 36-nm step. Nat. Struct. Mol. Biol. 2007;14:968–973. doi: 10.1038/nsmb1298. doi:10.1038/nsmb1298 [DOI] [PubMed] [Google Scholar]
- Kozuka J, Yokota H, Arai Y, Ishii Y, Yanagida T. Dynamic polymorphism of single actin molecules in the actin filament. Nat. Chem. Biol. 2006;2:83–86. doi: 10.1038/nchembio763. doi:10.1038/nchembio763 [DOI] [PubMed] [Google Scholar]
- Kron S.J, Spudich J.A. Fluorescent actin filaments move on myosin fixed to a glass surface. Proc. Natl Acad. Sci. USA. 1986;83:6272–6276. doi: 10.1073/pnas.83.17.6272. doi:10.1073/pnas.83.17.6272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molloy J.E, Burns J.E, Kendrick-Jones J, Tregear R.T, White D.C. Movement and force produced by a single myosin head. Nature. 1995;378:209–212. doi: 10.1038/378209a0. doi:10.1038/378209a0 [DOI] [PubMed] [Google Scholar]
- Nishikawa S, et al. Class VI myosin moves processively along actin filaments backward with large steps. Biochem. Biophys. Res. Commun. 2002;290:311–317. doi: 10.1006/bbrc.2001.6142. doi:10.1006/bbrc.2001.6142 [DOI] [PubMed] [Google Scholar]
- Nishiyama M, Higuchi H, Yanagida T. Chemomechanical coupling of the forward and backward steps of single kinesin molecules. Nat. Cell Biol. 2002;4:790–797. doi: 10.1038/ncb857. doi:10.1038/ncb857 [DOI] [PubMed] [Google Scholar]
- Rayment I, Holden H.M, Whittaker M, Yohn C.B, Lorenz M, Holmes K.C, Milligan R.A. Structure of the actin–myosin complex and its implications for muscle contraction. Science. 1993;261:58–65. doi: 10.1126/science.8316858. doi:10.1126/science.8316858 [DOI] [PubMed] [Google Scholar]
- Rock R.S, Rice S.E, Wells A.L, Purcell T.J, Spudich J.A, Sweeney H.L. Myosin VI is a processive motor with a large step size. Proc. Natl Acad. Sci. USA. 2001;98:13 655–13 659. doi: 10.1073/pnas.191512398. doi:10.1073/pnas.191512398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Aoki T, Aoki T, Yanagida T. Movement of single myosin filaments and myosin step size on an actin filament suspended in solution by a laser trap. Biophys. J. 1994;66:769–777. doi: 10.1016/s0006-3495(94)80853-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheetz M.P, Spudich J.A. Movement of myosin-coated fluorescent beads on actin cables in vitro. Nature. 1983;303:31–35. doi: 10.1038/303031a0. doi:10.1038/303031a0 [DOI] [PubMed] [Google Scholar]
- Shiroguchi K, Kinosita K., Jr Myosin v walks by lever action and Brownian motion. Science. 2007;316:1208–1212. doi: 10.1126/science.1140468. doi:10.1126/science.1140468 [DOI] [PubMed] [Google Scholar]
- Svoboda K, Schmidt C.F, Schnapp B.J, Block S.M. Direct observation of kinesin stepping by optical trapping interferometry. Nature. 1993;365:721–727. doi: 10.1038/365721a0. doi:10.1038/365721a0 [DOI] [PubMed] [Google Scholar]
- Tanaka H, Ishijima A, Honda M, Saito K, Yanagida T. Orientation dependence of displacements by a single one-headed myosin relative to the actin filament. Biophys. J. 1998;75:1886–1894. doi: 10.1016/S0006-3495(98)77629-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka H, Homma K, Iwane A.H, Katayama E, Ikebe R, Saito J, Yanagida T, Ikebe M. The motor domain determines the large step of myosin-V. Nature. 2002;415:192–195. doi: 10.1038/415192a. doi:10.1038/415192a [DOI] [PubMed] [Google Scholar]
- Taniguchi Y, Nishiyama M, Ishii Y, Yanagida T. Entropy rectifies the Brownian steps of kinesin. Nat. Chem. Biol. 2005;1:342–347. doi: 10.1038/nchembio741. doi:10.1038/nchembio741 [DOI] [PubMed] [Google Scholar]
- Terada T.P, Sasai M, Yomo T. Conformational change of the actomyosin complex drives the multiple stepping movement. Proc. Natl Acad. Sci. USA. 2002;99:9202–9206. doi: 10.1073/pnas.132711799. doi:10.1073/pnas.132711799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyoshima Y.Y, Kron S.J, McNally E.M, Niebling K.R, Toyoshima C, Spudich J.A. Myosin subfragment-1 is sufficient to move actin filaments in vitro. Nature. 1987;328:536–539. doi: 10.1038/328536a0. doi:10.1038/328536a0 [DOI] [PubMed] [Google Scholar]
- Toyoshima Y.Y, Kron S.J, Spudich J.A. The myosin step size: measurement of the unit displacement per ATP hydrolyzed in an in vitro assay. Proc. Natl Acad. Sci. USA. 1990;87:7130–7134. doi: 10.1073/pnas.87.18.7130. doi:10.1073/pnas.87.18.7130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda Y, Yasutake H, Ishijima A, Yanagida T. Torsional rigidity of single actin filaments and actin–actin bond breaking force under torsion measured directly by in vitro micromanipulation. Proc. Natl Acad. Sci. USA. 1996;93:12 937–12 942. doi: 10.1073/pnas.93.23.12937. doi:10.1073/pnas.93.23.12937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uyeda T.Q, Warrick H.M, Kron S.J, Spudich J.A. Quantized velocities at low myosin densities in an in vitro motility assay. Nature. 1991;352:307–311. doi: 10.1038/352307a0. doi:10.1038/352307a0 [DOI] [PubMed] [Google Scholar]
- Vale R.D, Reese T.S, Sheetz M.P. Identification of a novel force-generating protein, kinesin, involved in microtubule-based motility. Cell. 1985;42:39–50. doi: 10.1016/s0092-8674(85)80099-4. doi:10.1016/S0092-8674(85)80099-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vale R.D, Funatsu T, Pierce D.W, Romberg L, Harada Y, Yanagida T. Direct observation of single kinesin molecules moving along microtubules. Nature. 1996;380:451–453. doi: 10.1038/380451a0. doi:10.1038/380451a0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittaker M, Wilson-Kubalek E.M, Smith J.E, Faust L, Milligan R.A, Sweeney H.L. A 35-A movement of smooth muscle myosin on ADP release. Nature. 1995;378:748–751. doi: 10.1038/378748a0. doi:10.1038/378748a0 [DOI] [PubMed] [Google Scholar]
- Wulf E, Deboben A, Bautz F.A, Faulstich H, Wieland T. Fluorescent phallotoxin, a tool for the visualization of cellular actin. Proc. Natl Acad. Sci. USA. 1979;76:4498–4502. doi: 10.1073/pnas.76.9.4498. doi:10.1073/pnas.76.9.4498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagida T, Nakase M, Nishiyama K, Oosawa F. Direct observation of motion of single F-actin filaments in the presence of myosin. Nature. 1984;307:58–60. doi: 10.1038/307058a0. doi:10.1038/307058a0 [DOI] [PubMed] [Google Scholar]
- Yanagida T, Arata T, Oosawa F. Sliding distance of actin filament induced by a myosin crossbridge during one ATP hydrolysis cycle. Nature. 1985;316:366–369. doi: 10.1038/316366a0. doi:10.1038/316366a0 [DOI] [PubMed] [Google Scholar]
- Yildiz A, Forkey J.N, McKinney S.A, Ha T, Goldman Y.E, Selvin P.R. Myosin V walks hand-over-hand: single fluorophore imaging with 1.5-nm localization. Science. 2003;300:2061–2065. doi: 10.1126/science.1084398. doi:10.1126/science.1084398 [DOI] [PubMed] [Google Scholar]