Abstract
The use of predicted binding sites (binding sites calculated from the protein structure alone) is evaluated here as a tool to focus the docking of small molecule ligands into protein structures, simulating cases where the real binding sites are unknown. The resulting approach consists of a few independent docking runs carried out on small boxes, centered on the predicted binding sites, as opposed to one larger blind docking run that covers the complete protein structure. The focused and blind approaches were compared using a set of 77 known protein-ligand complexes and 19 ligand-free structures. The focused approach is shown to: (1) identify the correct binding site more frequently than blind docking; (2) produce more accurate docking poses for the ligand; (3) require less computational time. Additionally, the results show that very few real binding sites are missed in spite of focusing on only 3 predicted binding sites per target protein. Overall the results indicate that, by improving the sampling in regions that are likely to correspond to binding sites, the focused docking approach increases accuracy and efficiency of protein ligand docking for those cases where the ligand-binding site is unknown. This is especially relevant in applications such as reverse virtual screening and structure-based functional annotation of proteins.
Keywords: docking protocol, binding site discovery, reverse virtual screening, AutoDock, Astex Diverse Set
Introduction
The goal of protein-ligand docking is to predict the position and orientation of a ligand (usually a small molecule) when it is bound to a receptor protein. When the binding site to be targeted by the small-molecule is known, selecting a reasonably small docking box around this site facilitates docking by focusing sampling of the translational, rotational and torsional degrees of freedom of the ligand. This is the usual situation in lead optimization, where predicting the binding mode or pose of the ligand is needed for rational design of improved potency and selectivity, and in hit identification through virtual screening where the goal is the discovery of ligands, out of a large library, that are likely to bind a protein target. The reverse question is more difficult to address. Given a ligand, is it possible to discover its most likely target? In this “reverse virtual screening” case, since the binding site is not known it becomes necessary to explore the entire protein surface by docking, a procedure that has been named “blind docking”1,2. Since the space where blind docking takes place must accommodate the entire protein and is therefore much larger than a regular docking box, the number of energy evaluations carried out by the docking program is usually set up to a proportionally higher value1,2, with a corresponding increase in the running time. This shortcoming has been partially overcome by using known protein binding sites as targets for reverse-virtual screening3. While this approach enables faster reverse virtual screening, it limits the universe of candidate targets to those proteins that have clearly identified binding sites and only to those sites within the protein. Ideally, a reverse virtual screening approach would require only the knowledge of the three-dimensional structure of the candidate target proteins and would allow for the discovery of unexpected interactions that may occur at previously unidentified binding sites. One such approach has been described by Brown and Vander Jagt4 in which a macromolecule encapsulating surface (MES) was used to geometrically define the boundaries of predicted binding sites and guide the docking search. On a set of 14 protein-ligand complexes the MES approach was shown to improve the efficiency of the genetic algorithm-based optimizer in the AutoDock5 docking software.
In this paper the use of binding sites calculated directly from the docking grid (i.e. interaction energy-based calculation) is evaluated as a tool to focus the docking searches of the AutoDock5,6 software. This results in an approach consisting of multiple independent docking runs carried out on smaller boxes, centered on a few predicted binding sites, as opposed to one larger blind docking run that covers the complete protein structure. By comparing the focused docking approach with reference blind docking runs over a set of 77 ligand-protein complexes and 19 ligand-free proteins we address the following questions: Is focused docking more accurate than blind docking? Is there a real gain in computational efficiency when using focused docking? Is there a penalty paid (e.g. missed binding sites) when using focused docking?
Materials and Methods
Selection of Complexes
Both focused and blind docking experiments were carried out on the same set of complexes obtained from the Astex Diverse Set7, a published collection of 85 protein-ligand crystal structures extracted from the Protein Data Bank (PDB)8 and specifically selected to evaluate the performance of docking algorithms. All water molecules and heteroatoms (including the ligands) were removed and for the cases that contained identical sets of chains, only one set was retained.
Preparation of the proteins and ligands for docking
Gasteiger charges were added to both ligands and proteins, using the programs included in the AutoDockTools suite (version 1.4.5). At that stage, 8 cases that issued warnings and would have required manual intervention were removed resulting in a final set of 77 complexes. The PDB codes of the selected chains are: 1gkcA, 1gm8, 1hnnA, 1hp0A, 1hq2, 1hvyD, 1hwiA+B, 1hww, 1ia1B, 1ig3, 1j3jA, 1jd0B, 1jjeA, 1jlaA, 1k3u, 1ke5, 1kzk, 1l2sB, 1l7f, 1lpz, 1lrhD, 1m2zA, 1meh, 1mzc, 1n1mA, 1n2jA, 1n2v, 1n46A, 1nav, 1of1B, 1opk, 1oq5, 1owe, 1oyt, 1p2y, 1p62, 1pmn, 1q1gF, 1q41A, 1q4gB, 1r1h, 1r55, 1r58, 1r9o, 1s19, 1s3v, 1sg0B, 1sj0, 1sq5A, 1sqnB, 1t40, 1t46, 1tow, 1tt1A, 1tz8B, 1u1cF, 1uml, 1unlA+D, 1uou, 1v0pA, 1v48, 1v4s, 1vcj, 1w1pB, 1w2gB, 1x8x, 1xm6A, 1xoqB, 1xoz, 1ygc, 1yqy, 1yvf, 1ywr, 1z95, 2bm2B, 2br1, 2bsm.
Unbound proteins dataset
For each single-chain binding site entry in the Astex Diverse Set a BLAST9 search was performed against the PDB database selecting all the entries that had a sequence identity > 95% and a coverage > 0.95. Subsequently, the cases that had mutated residues in the binding site were eliminated from the dataset. Finally, from the remaining cases only the entries that did not have any ligand in the binding site were selected. This procedure led to 19 unbound proteins corresponding to a subset of the 77 complexes described above. The PDB codes of the bound_unbound pairs are: 1hq2_1hka, 1t46_1t45, 1ke5_1hcl, 1v0pA_1ob3A, 1l2sB_2blsA, 1v48_1pbn, 1l7f_1nmaN, 1w1pB_1e15A, 1n1mA_1r9mA, 1yvf_2girA, 1n2v_1pud, 1ywr_2okrA, 1oq5_2cbe, 2br1_1ia8, 1oyt_1vr1H, 2bsm_1uyl, 1q41A_1i09A, 1s3v_1pdb, 1t40_1xgd. To facilitate comparison of docking results the binding site residues in the unbound proteins was superimposed on the corresponding residues of the bound proteins using the backbone atoms of the residues that had at least one atom within 6.0 Å of the ligand heavy atoms in the complex. A site is considered to have been detected if the fraction of overlapping heavy atoms between the lowest energy pose and the ligand in the complex is >= 0.15.
Binding site detection
The algorithm to predict the location of potential binding sites for drug-like molecules is based on principles similar to those that underlie the QSiteFinder algorithm10. Both algorithms identify regions characterized by favorable van der Waals interactions, which have been shown to play an important role in the binding of drug-like molecules to proteins11,12.
The first step requires the computation of a low resolution (1.0Å) Carbon affinity map with AutoGrid (part of the AutoDock suite v. 4), using a box large enough to accommodate the entire protein. In the next step, a predefined energy cut-off (−0.3 kcal/mol for all cases) is applied to filter out all the affinity map points corresponding to unfavorable interaction energies. Subsequently, the remaining points are clustered according to spatial proximity with an agglomerative hierarchical clustering algorithm using average linkage, as implemented in the C Cluster Library13. This step yields a hierarchical dendrogram, which is finally cut into non-overlapping clusters by applying a distance cut-off (7.8Å for all cases). This last step is made possible by the fact that average linkage clustering produces monotonic hierarchies. In other words, the distance between clusters at each merging step never decreases. Therefore, the number of clusters needs not be determined a priori, but only the value for the distance cut-off must be chosen. Finally, these so-obtained clusters are ranked by Total Interaction Energy (TIE, the sum of the energy values of all the points that belong to the same cluster) and the first three are selected for focused docking (see below). The spatial localization of the clusters is characterized by their Center of Energy (COE, the average of their coordinates weighted by energy).
The algorithm described above was implemented in a Python/C program called SiteHound (available upon request). The only two parameters that the program requires are the energy cutoff to filter the grid points and the distance cut-off for the clustering step. A range of values for these two parameters was tested, and a combination (−0.3 kcal/mol and 7.8Å, respectively) was chosen that yielded the most accurate binding site prediction as defined by the accuracy measure introduced by Laurie10.
In terms of computational overhead for the binding site prediction step, it is noteworthy to mention that the time required to run the SiteHound program is negligible with respect to the time required for a full docking experiment. The median time calculated on the dataset was < 1 minute per protein on a Pentium IV machine.
Blind docking set-up
The proteins and the ligands were prepared for docking as described above. The docking parameters recommended by Hetenyi and van der Spoel2 were used, with the most relevant for this analysis being the docking box size and the number of energy evaluations. The dimensions of the boxes were calculated in such a way to allow a clearance of 5 Å from each side of the box, and the resolution was set to 0.55 Å. The average number of points per box for this dataset amounted to ~ 1.6×106. The number of energy evaluations was set to 107 and for comparison with the faster focused docking (see below) an additional set of blind docking experiments was carried out with 106 energy evaluations. We refer to these two groups of docking experiments as “slow” and “fast” blind docking respectively.
Focused docking set-up
In the focused docking experiments the search space was restricted to the vicinity of the top 3 binding sites predicted by the SiteHound program (Figure 1). Thus, each focused docking experiment consisted of three independent runs, with the docking box centered on the COE of the predicted first, second and third binding site respectively (ranked by TIE). The size of the box for the focused docking experiments (23 Å × 23 Å × 23 Å) was chosen on the base of the results shown in Figure 2, where it is shown that in 95% of the cases the center of the ligand falls within 10.0 Å of the COE of one of the first three predicted sites. The candidate solution was defined as the one that had the lowest docking energy among the three putative sites explored. Two alternative ranking methods were explored, one based on the selection of the largest cluster, and the one proposed by Ruvinsky14 that corrects for cluster occupancy. In both cases the ranking was less accurate than using the lowest docking energy. To mimic the two blind docking runs (slow and fast) the number of energy evaluations was also varied for the focused runs. Additionally, the smaller size of the 3 focused docking boxes enabled the use of a second set with a higher resolution box. Table I describes the four sets of focused docking experiments that result from varying the number of energy evaluations and the docking box resolution. Since the number of jobs per docking was set to 33 (instead of 100 as in the case of blind docking), set 1 and set 2 are comparable in running times to slow blind docking, whereas set 3 and set 4 are comparable to fast blind docking.
Figure 1. Blind docking and focused docking.
The blind protocol consists of a single docking experiment, carried out on the whole protein surface, whereas the focused protocol breaks up the problem into multiple smaller docking experiments, focusing on predicted binding sites.
Figure 2. SiteHound binding site detection performance.
Distribution of distances between the center of the ligand in the crystal structure of the complex and the Center of Energy of the best site (i.e. closest to the ligand) out of the first three ranking sites predicted by SiteHound for 77 protein-ligand complexes.
Table I.
Parameters for the different sets of focused docking experiments
| Set | Description | Box resolution | Box Dimensions | Number of energy evaluations |
|---|---|---|---|---|
| 1 | slow, low-resolution | 0.55 Å | 40×40×40 points | 107 |
| 2 | slow, high-resolution | 0.375 Å | 60×60×60 points | 107 |
| 3 | fast, low-resolution | 0.55 Å | 40×40×40 points | 106 |
| 4 | fast, high-resolution | 0.375 Å | 60×60×60 points | 106 |
Focused docking with masked grids
Another approach for biasing the docking towards the predicted binding sites was explored as an alternative to running independent docking experiments with smaller grids centered on the predicted site. The approach consists in masking all the Carbon grid points that are outside a sphere of 11.0Å radius centered at the predicted sites by assigning to them extremely high energy values (105 kcal/mol), so that the regions outside the binding sites become forbidden. The docking is then carried out as described for blind docking.
Comparison of blind vs. focused docking
As a first step to compare blind and focused docking, it was determined whether the docking results identified the correct binding pocket, as defined by the crystal structure protein/ligand complex. This was done by measuring the overlap between the candidate solution for blind and focused docking (lowest docking energy pose of the ligand) and the ligand in the experimental structure. The overlap was defined as the fraction of ligand heavy atoms that fell within 2.0A of a ligand heavy atom in the crystal structure. A docking solution was said to have identified the correct binding site if the overlap was >= 0.15. For those cases where both blind and focused docking identified the correct binding site the results were further characterized by comparing the Root Mean Squared Deviation of ligand heavy atoms (RMSD) of the candidate solutions for blind and focused docking with respect to the experimental structure, using the values reported in the output produced by AutoDock. The RMSD comparisons were restricted only to those complexes where both protocols correctly identified the binding sites because comparison of RMSDs for solutions in incorrect binding sites would not be meaningful. The statistical significance of the RMSD difference between blind and focused docking was assessed with a paired Student t-test.
Results and Discussion
As illustrated in Figure 1, the main idea behind the focused docking protocol is to break up the exploration of the protein surface into a few smaller independent docking jobs. The benefits that result from using a smaller sampling space focused on candidate binding sites include a better chance to identify the native binding mode of the ligand and the possibility to perform docking in a much faster way, as will be shown below. The assumption behind the use of a few predicted binding sites is that only a handful of possible small-molecule binding sites exist on protein structures, and that these sites can be reliably identified, thus it is not necessary to explore a very large number of sites and a gain in speed is possible without a significant loss in coverage. These assumptions are tested in the results shown below.
Binding site identification
The identification of candidate binding sites by the SiteHound algorithm (see methods) is the first step in the focused docking protocol. Since the predicted binding site was used to center the docking box, it is important to assess whether the centers of energy (COE) of the clusters representing the predicted binding sites are close to the real center of the ligand. Figure 2 shows the performance of the SiteHound binding site identification procedure on the Astex Diverse Set, expressed as a histogram of distances between the center of the ligand in the crystal structure of the complex and the COE of the predicted binding sites. In 95% of the cases the center of the ligand falls within 10.0 Å of the COE of one of the first three predicted sites (the first site alone yields 77% of the cases). For this reason the focused docking experiments, shown below, used the first three predicted sites.
Comparison of blind and focused docking protocols
Two sets of docking experiments, one with 107 and the other with 106 energy evaluations for both focused and blind docking protocols were carried out. These sets are referred to as “slow” and “fast” docking respectively, with fast mode docking being ten times faster than slow mode docking. As described in the Methods section, the number of runs per job was reduced for focused docking in such a way that the three individual runs (one for each predicted binding site) that make up one focused docking experiment taken together require the same amount of time as one blind docking experiment.
As a first step to assess the performance of the two protocols in predicting the native binding mode of the ligands as defined in the crystal structures, we selected the poses with the lowest docking energy and calculated the fraction of the ligand heavy atoms that overlapped with atoms of the ligand in the crystal structure. This was used as a measure of the ability of the docking protocol to identify the correct ligand binding site in the complete protein structure (blind docking) or among the top three predicted binding sites (focused docking). In the case of good overlap between the docking pose and the ligand in the crystal structure the fraction will be close or equal to 1, whereas in cases where the docking protocol misses the binding site the overlap will be close or equal to 0. As shown in Figure 3, the focused docking protocol outperformed the blind docking protocol in terms of ligand binding site identification in both fast and slow mode irrespective of the overlap cutoff used to measure accuracy. Furthermore, the data shows that there is a penalty to be paid when using blind docking in fast mode, since more cases are missed in the faster mode. In contrast, there is no significant difference between fast and slow mode for focused docking, or between high and low resolution focused docking. Thus, focused docking is able to achieve a higher accuracy of binding site detection than the best blind docking protocol (slow blind docking) even while requiring only one tenth of the computing time (set 3 and set 4, fast focused docking).
Figure 3. Binding site detection accuracy of focused and blind docking protocols for 77 protein-ligand complexes.
The number of cases that have a fraction of overlapping atoms equal to or greater than a threshold is represented. The fraction of overlapping atoms is calculated as the fraction of ligand heavy atoms in the lowest energy pose that are within 2.0 Å of a ligand heavy atom in the crystal structure – red (solid): slow blind docking; red (dashed): fast blind docking; purple(dashed): fast focused docking (masked sites); blue (solid): slow focused docking (set 1); green (solid): slow focused docking (set 2); blue (dashed): fast focused docking (set 3); green (dashed): fast focused docking (set 4). See Table 1 for description of docking sets.
Masked grids
The focused docking approach using masked grids (see methods) was tested on the Astex Diverse Set using the 106 energy evaluations protocol. All but the first three predicted sites were masked. Even though the results were better than the blind docking protocol (Figure 3), the overall accuracy is still much lower than with any of the other focused docking protocols. To evaluate whether the lower accuracy of the masked approach is a consequence of the competition of the three sites present simultaneously during docking, or the masking itself, the same experiment was repeated by masking one site at a time. In this case the masked approach yielded results that were indistinguishable from the ones produced by the other focused docking protocols. This suggests that the simultaneous presence of the hot-spots regions is suboptimal for achieving a thorough exploration of the correct binding site, and that there is an advantage in individually exploring the predicted sites one at a time either by reducing the size of the docking box or by masking the sites individually.
As mentioned before, for the Astex diverse set in 95% of the cases the ligand center falls within 10.0 Å of at least one of the first three predicted sites. Even though the first site alone accounts for 77% of the cases, the other two sites cannot be neglected if one wants to achieve high accuracy of binding site prediction. Using the overlap measure described above to assess whether the real binding site has been identified in docking, the accuracy ranges from 80% to 84% for focused docking. The same measure applied to the blind docking protocol yields a binding site detection accuracy of 71% and 66%, for slow and fast blind docking, respectively (Table II). These results suggest that focused docking can provide a small improvement over the initial SiteHound binding site detection step by identifying some of the correct binding sites that ranked in the second or third position. Blind docking is unable to do so probably because the large search space prevents the exhaustive exploration of the three candidate sites, resulting in poor discrimination. It is interesting to note that in most cases the incorrect sites identified in blind docking corresponds to one of the three sites predicted by SiteHound, thus the incorrect solution is a consequence of incomplete sampling rather than scoring. We also observed two cases (PDB chains 1l2sB and 1hww) where blind docking identified the correct site and focused docking did not. In both cases the correct binding site was not among the top three SiteHound sites (the correct sites ranked 4th and 8th, respectively).
Table II.
Accuracy of binding site detection
| Docking Experiment | Correct cases | Incorrect cases | Fraction Correct |
|---|---|---|---|
| Blind Slow | 55 | 22 | 71% |
| Blind Fast | 51 | 26 | 66% |
| Focused Slow (set 1) | 64 | 13 | 83% |
| Focused Slow (set 2) | 65 | 12 | 84% |
| Focused Fast (set 3) | 63 | 14 | 82% |
| Focused Fast (set 4) | 62 | 15 | 80% |
In those cases where the binding site was missed by the blind docking protocol, but correctly identified by the focused docking protocol, a tendency towards a higher number of rotatable bonds in the ligand was observed. On the other hand, in those cases where the docking performance was poor for both protocols no clear correlation with the number of rotatable bonds in the ligand was observed. This observation is consistent with the benefits provided by focused docking being simply a smaller sampling space, where the number of energy evaluations can be spent more efficiently exploring the torsional degrees of freedom of the ligand.
To further compare the performance of the two docking protocols, for fast and slow modes, the cases where both protocols correctly identified the binding site were selected (arbitrarily defined as the cases where the overlap was >= 0.15) and the distributions of ligand heavy atoms RMSD from the crystal structure were compared (Figure 4). For both fast and slow mode, focused docking outperformed blind docking (p-value < 0.05 and < 0.01 for slow and fast mode respectively). Thus, even in those cases where both methods identify the correct binding site focused docking is able to produce ligand poses that are more accurate than those produced by blind docking. In a few examples blind docking produced a slightly lower RMSD than focused docking, with the largest RMSD difference being 0.39Å. For comparison the largest RMSD improvement due to focused docking was 4.61Å. As regarding as the comparison among the different focused docking set-up, no statistically significant difference was observed in terms of binding site detection and RMSD of the poses from the crystal structure. Thus, focused docking is able to achieve a higher ligand docking accuracy than the best blind docking protocol (slow blind docking) even while requiring only one tenth of the computing time (set 3 and set 4, fast focused docking). This observation can be explained by considering that, on average, the smaller box used in the focused experiments yields convergence of the docking algorithm with a lower number of energy evaluations, thanks to the reduced sampling space. Therefore, for focused docking no penalty has to be paid when using the fast mode, whereas this does not hold true for the blind docking protocol. It is to be expected that the performance of blind docking will further increase with an even higher number of energy evaluations, however with the corresponding increase in computational cost.
Figure 4. Accuracy of blind and focused docking.
Distribution of RMSD of the lowest energy poses with respect to the crystal structures for the focused and blind docking protocols. Only “low-resolution” focused docking results are shown (see Table 1). The comparison includes only cases where both blind and focused docking identified the correct binding site. For slow docking 53 out of 77 cases are included. For fast docking 49 out of 77 cases are included.
Comparison of blind vs. focused docking in the unbound proteins dataset
Further testing of the docking protocols was carried out on a subset of the Astex dataset for which unbound forms of the proteins are available. The performance of blind docking (slow mode protocol) and focused docking (fast mode, low resolution) was compared on the unbound dataset of 19 proteins. As expected the overall docking accuracy on this set is lower than on the set of complexed proteins. However, the focused docking protocol produced a marked increase in accuracy with respect to the blind protocol. While the blind protocol identified 6 out of 19 binding sites, the focused protocol correctly identified 11, while using one tenth of the computational time. This corresponds to an increase in accuracy from 32% to 58%. In those few cases where blind docking identified the correct site, focused docking outperformed it in terms of the accuracy (RMSD) of the lowest energy pose (Table III).
Table III.
Accuracy of blind and focused docking in unbound proteins
| Target Protein | Focused RMSD (Å) | Blind RMSD (Å) |
|---|---|---|
| 1hq2 | 4.75 | n/a |
| 1ke5 | 6.00 | n/a |
| 1n2v | 3.95 | n/a |
| 1oq5 | 3.10 | 3.10 |
| 1oyt | 0.56 | 0.42 |
| 1q41A | 1.62 | n/a |
| 1s3v | 0.70 | 3.10 |
| 1v0pA | 3.40 | 3.29 |
| 1ywr | 4.64 | n/a |
| 2br1 | 3.03 | 2.96 |
| 2bsm | 1.72 | 1.72 |
In summary, the results on the Astex Diverse Set indicate that the focused docking protocol outperforms the blind docking approach both in terms of binding site identification and RMSD from the crystal structure in the cases where the binding site was successfully detected by both protocols (see Figure 5 for examples). Furthermore, for focused docking no significant advantage was observed for slow mode docking, probably due to the more thorough sampling achieved by focusing on a smaller region. This results in higher accuracy using only one tenth of the computing time necessary for blind docking.
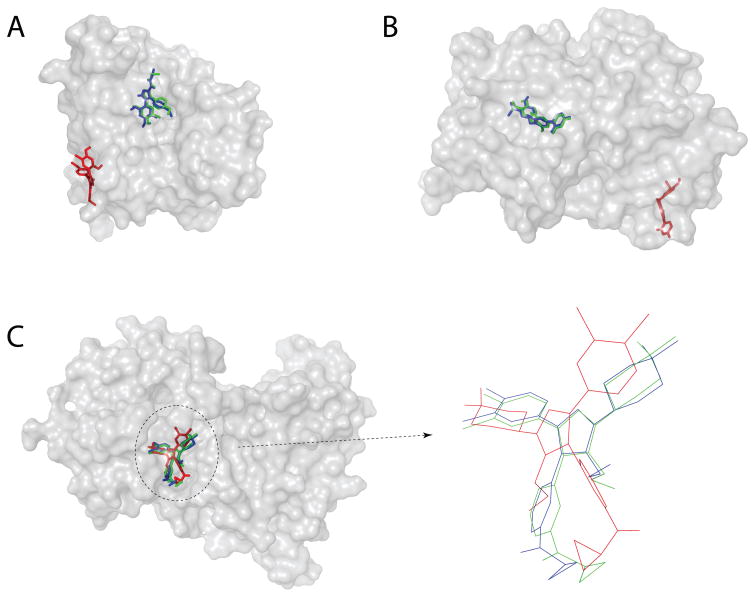
Figure 5. Examples of improved results with focused docking.
Red: blind docking (slow); blue: focused docking (set 3, fast, low-resolution) – green: crystal structure. (A) and (B), the ligand is placed in the correct binding site by focused docking, but missed by blind docking (PDB codes: 2bsm and 1n46, respectively). (C) The ligand is placed in correct site by focused and blind docking, but the focused docking results is more accurate (PDB code: 1pmn).
Conclusions
A protocol to carry out protein-ligand docking suitable for cases where the binding sites are not known a priori was developed. Using first a simple and fast algorithm to predict binding sites, the approach then performs independent docking jobs around each predicted site. The results show that docking focused on a small number of predicted binding sites not only reduces the computational time required to compute the solution, but the docking results are also more accurate both in terms of binding site identification and of RMSD of the lowest energy docked pose with respect to the experimental solution. Focused docking is able to improve the binding site detection of the SiteHound algorithm since it is able to identify the correct ligand binding site even in some cases where the binding site did not rank first in the SiteHound results. Overall the results suggest that the benefits of focused docking are a consequence of improved sampling in relevant regions (predicted binding sites) and not due to removing unwanted decoy sites that would interfere with scoring. The fact that very few binding sites were missed by the focused docking approach confirms that, at least in this set, it is sufficient to explore only a few of the putative binding sites per protein. The results, taken together, suggest that since focused docking achieves higher accuracy at a fraction of the computational cost of blind docking it is well suited as an effective and fast protocol to enable reverse virtual screening on a large number of proteins. It is also possible to envision the application of this approach to aid the process of characterization of newly determined structures, especially in the context of structural genomics initiatives. Many protein structures produced by structural genomics projects do not have functional annotations, and computational methods are often employed to provide clues for further experimental investigations15. Characterizing these protein structures from the perspective of potential ligands could be very valuable for functional annotation, and could also suggest novel therapeutic targets.
Acknowledgments
The authors thank Dr. Mihaly Mezei and the members of the Sanchez lab for useful suggestions and discussions. This work is supported by NIH grant GM081713 and NSF grant MCB 0517352. RS is an Irma T. Hirschl Career Award recipient.
References
- 1.Hetenyi C, van der Spoel D, Qj Efficient docking of peptides to proteins without prior knowledge of the binding site. Protein Sci. 2002;11(7):1729–1737. doi: 10.1110/ps.0202302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hetenyi C, van der Spoel D, Cg Blind docking of drug-sized compounds to proteins with up to a thousand residues. FEBS Lett. 2006;580(5):1447–1450. doi: 10.1016/j.febslet.2006.01.074. [DOI] [PubMed] [Google Scholar]
- 3.Paul N, Kellenberger E, Bret G, Muller P, Rognan D. Recovering the true targets of specific ligands by virtual screening of the protein data bank. Proteins-Structure Function and Bioinformatics. 2004;54(4):671–680. doi: 10.1002/prot.10625. [DOI] [PubMed] [Google Scholar]
- 4.Brown WM, Vander Jagt DL. Creating Artificial Binding Pocket Boundaries to Improve the Efficiency of Flexible Ligand Binding. J Chem Inform Comp Sci. 2004;44:1412–1422. doi: 10.1021/ci049853r. [DOI] [PubMed] [Google Scholar]
- 5.Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, Olson AJ, Nc Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J Comput Chem. 1998;19(14):1639–1662. [Google Scholar]
- 6.Huey R, Morris GM, Olson AJ, Goodsell DS, Uz A semiempirical free energy force field with charge-based desolvation. J Comput Chem. 2007;28(6):1145–1152. doi: 10.1002/jcc.20634. [DOI] [PubMed] [Google Scholar]
- 7.Hartshorn MJ, Verdonk ML, Chessari G, Brewerton SC, Mooij WTM, Mortenson PN, Murray CW, Lr Diverse, high-quality test set for the validation of protein-ligand docking performance. J Med Chem. 2007;50(4):726–741. doi: 10.1021/jm061277y. [DOI] [PubMed] [Google Scholar]
- 8.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28(1):235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Laurie ATR, Jackson RM, Rs Q-SiteFinder: an energy-based method for the prediction of protein-ligand binding sites. Bioinformatics. 2005;21(9):1908–1916. doi: 10.1093/bioinformatics/bti315. [DOI] [PubMed] [Google Scholar]
- 11.Jain AN. Scoring noncovalent protein-ligand interactions: a continuous differentiable function tuned to compute binding affinities. J Comput Aided Mol Des. 1996;10(5):427–440. doi: 10.1007/BF00124474. [DOI] [PubMed] [Google Scholar]
- 12.Ringe D. What makes a binding site a binding site? Curr Opin Struct Biol. 1995;5(6):825–829. doi: 10.1016/0959-440x(95)80017-4. [DOI] [PubMed] [Google Scholar]
- 13.de Hoon MJ, Imoto S, Nolan J, Miyano S. Open source clustering software. Bioinformatics. 2004;20(9):1453–1454. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- 14.Ruvinsky AM. Role of binding entropy in the refinement of protein-ligand docking predictions: analysis based on the use of 11 scoring functions. J Comput Chem. 2007;28(8):1364–1372. doi: 10.1002/jcc.20580. [DOI] [PubMed] [Google Scholar]
- 15.Lee D, Redfern O, Orengo C. Predicting protein function from sequence and structure. Nat Rev Mol Cell Biol. 2007;8(12):995–1005. doi: 10.1038/nrm2281. [DOI] [PubMed] [Google Scholar]