Abstract
Peroxisome proliferator-activated receptor gamma polymorphisms have been widely associated with type 2 diabetes, although their role in the pathogenesis of vascular complications is not yet demonstrated. In this study, a cohort of 211 type 2 diabetes, 205 obese, and 254 control individuals was genotyped for Pro12Ala, C1431T, C-2821T polymorphisms, and for a newly identified polymorphism (A-2819G). The above-mentioned polymorphisms were analyzed by gene-specific PCR and direct sequencing of all samples. A significant difference was found for -2819G frequency when patients with type 2 diabetes—particularly diabetic women with the proliferative retinopathy—were compared with healthy control individuals. In conclusion, we identified a novel polymorphism, A-2819G, in PPARG gene, and we found it to be associated with type 2 diabetes and proliferative retinopathy in diabetic females. In the analyzed population, this variant represents a genetic risk factor for developing the diabetic retinopathy, whereas Pro12Ala and C1431T do not.
1. Introduction
Peroxisome proliferator-activated receptor gamma (PPARG) gene is a member of nuclear hormone receptor superfamily, and it regulates the transcription of several genes involved in glucose metabolism, adipocyte differentiation, lipid oxidation, angiogenesis, and inflammation [1]. PPARγ displays different protein isoforms generated by different promoters and alternative splicing [2, 3]. The functional role of PPARγ is well documented, and its alterations have been widely associated with metabolic diseases, such as type 2 diabetes and obesity [4, 5].
Indeed, the most analyzed PPARG polymorphism, Pro12Ala in the exon B [6], exerts an unclear effect on receptor structure, and it seems to reduce the receptor binding affinity with the responsive elements, also inducing a decrease in transcriptional activity both with and without PPARγ agonists [7, 8]. The common Pro12Ala polymorphism has been associated with increased risk of developing type 2 diabetes, even though a recent hypothesis suggests Pro12Ala could be involved in the aetiology of metabolic disorders not by itself, but because of linkage disequilibrium with other single nucleotide polymorphisms (SNPs) in the PPARG promoter [9, 10].
Moreover, PPARG gene polymorphisms have also been associated with micro- and macrovascular complications of diabetes, although the few available studies do not unambiguously confirm the role of PPARG in the pathogenesis of these complications, especially diabetic retinopathy [11–13]. Indeed, recent studies have demonstrated that PPARγ ligands could both inhibit intraocular angiogenesis and upregulate VEGF gene expression in several cell types [14].
To date, most of the mutation analyses focused on the coding region of the PPARG gene [9], and only few studies have analyzed SNPs in the regulatory region of PPARG possibly associated with metabolic disorder susceptibility [15, 16]. A novel functional variant in the PPARγ2 promoter, C-2821T, that associates with predictors of type 2 diabetes and obesity, was recently reported in Pima Indians population [16].
In attempt to analyze this variant in Italian individuals affected by type 2 diabetes, we have performed an SNP analysis on 211 unrelated type 2 diabetic patients from Campania County, Southern Italy. Subsequently, we have extended this analysis to a cohort of 205 unrelated obese patients and 254 control individuals.
Furthermore, we have genotyped the above-mentioned cohort (diabetic, obese, and control individuals), for two extensively studied SNPs in high linkage disequilibrium, Pro12Ala and C1431T.
2. Materials and Methods
2.1. Patients and Clinical Characteristics
Genomic DNA was extracted from blood samples by standard procedures [17]. SNP analysis was performed on genomic DNA from 211 unrelated Italian type 2 diabetic patients (102 of which suffered from proliferative retinopathy), 205 unrelated obese patients, and 254 control individuals. All the subjects were recruited from the Department of Geriatrics and Metabolic Diseases, Second University of Naples, Italy, and from the Department of Neuroscience, “Federico II” University of Naples, Italy. All procedures were approved by the Ethics Board of the participating institutes and adhered to the tenets of the Declaration of Helsinki. All samples were acquired after informed consent was obtained from the patients.
The diabetes and obese status were determined according to the criteria of World Health Organization [18]. Patients with type 2 diabetes were eligible for the study if they had background or proliferative diabetic retinopathy, similar age (±1 year) and similar duration of diabetes (±1 year). No other inclusion criteria had to be met. The clinical characteristics (age, sex, duration of T2DM, 2h-plasma glucose, HbA1c and BMI) of the 102 diabetic patients with proliferative retinopathy were not different from those of the 109 diabetic patients with background retinopathy (see Table 1). Retinopathy condition was evaluated by ocular fundus photography.
Table 1.
Clinical characteristics of type 2 diabetic patients.
| Characteristic | Proliferative | Background | P value |
|---|---|---|---|
| DR group | DR group | ||
| n | 102 | 109 | |
| Age (years) | 63 ± 10 | 62 ± 9 | 0.44 |
| Male (%) | 45 (44,1) | 54 (49,5) | 0.51 |
| Female (%) | 57 (55,9) | 55 (50,5) | 0.72 |
| Duration of T2DM | 11,5 ± 2,9 | 12,0 ± 2.5 | 0.18 |
| 2h-plasma glucose (mg/dl) | 230 ± 90 | 172 ± 60 | 0.001 |
| HbA1c | 8,9 ± 1, 0 | 7,9 ± 0,9 | 0.001 |
| BMI (Kg/m2) | 31,2 ± 4,4 | 30,4 ± 4,5 | 0.19 |
2.2. SNP Analysis and Genotyping
Pro12Ala SNP analysis was performed with specific primers pairs (Pro12AlaF 5′-TTT TAA CGG ATT GAT CTT TTG C-3′ and Pro12AlaR 5′-ACC CTT ACA TAA ATG CCC CC-3′) amplifying a fragment of 214 bp. C1431T SNP in the exon 6 of PPARG gene was analyzed by PCR amplification of a 358 bp fragment using specific primers pairs (C1431TF 5′-CTG AAC CCC CTG TTG TGT TT-3′ and C1431TR 5′-GGT GTC AGA TTT TCC CTC AGA-3′). The screening of C-2821T and A-2819G SNPs was carried out on a 453 bp genomic region of PPARγ2-specific promoter, PCR amplified with specific oligonucleotides (C-2821TF 5′-TAC ACC AAT GGG TTG TCA-3′; C-2821TR 5′-GTA GCC AAA GAC AGG TTC TG-3′) by Muller et al. (2003) [16].
PCR reactions set-up with AmpliTaq Gold (Applied Biosystems, Calif, USA) were performed using an ABI 9700 automatic thermal cycler under the following conditions: 95°C for 10 minutes; then 35 cycles of 95°C for 1 minute, at annealing temperature (Ta) for 1 minute, of 72°C for 2 minutes; and a final step of 72°C for 10 minutes. PCR products were analyzed by denaturing high performance liquid chromatography (DHPLC), and those with a DHPLC pattern different from controls were sequenced using the ABI Prism Big Dye Terminator Cycle Sequencing V2.0 (Applied Biosystems).
2.3. Statistical Analysis
Statistical analysis was performed using standard chi-square, Fisher, and student's t-tests. For continuous variables, the general estimating equation procedure was used to adjust for covariates. The association of polymorphisms with type 2 diabetes, obesity, and retinopathy was analyzed by exact chi-square test, comparing SNPs frequencies.
Allele and genotype frequencies (Pearson χ2 statistics), O.R., and P values, as well as dominant and recessive genetic models, were analyzed using FINETTI (http://ihg2.helmholtz-muenchen.de/cgi-bin/hw/hwa1.pl). Hardy-Weinberg equilibrium of tested groups and Armitage's trend test (ATT) were also calculated using FINETTI. ATT takes into account genotypes rather than alleles, avoiding a possible bias due to doubling of sample size [19]. It assumes additive (or codominant) disease model where all disease allele are independent and have the same contribution to the disease risk.
Continuous clinical data, such as duration of type 2 diabetes (years), HbA1c and BMI (body mass index-Kg/m2), different between background and proliferative retinopathy populations, were compared by unpaired student's t-test (two-tailed test); a resulting P value less than 0.05 (C.I. 95%) was considered significant.
3. Results
To investigate whether SNPs in the PPARG gene could be a risk factor for type 2 diabetes and obesity, an extensive polymorphism analysis in a population of 670 individuals from Campania County, in Southern Italy, was performed. The diabetes and obese status were determined according to the criteria of World Health Organization.
We genotyped three SNPs, Pro12Ala, C1431T—in the coding region of PPARG gene—and C-2821T in the regulatory region [16], in 211 individuals with type 2 diabetes, 205 suffering from obesity, and 254 control individuals. Our analysis revealed that T-2821 allele was not found in 670 individuals, and consequently, C-2821 allele frequency was 100%.
Allelic frequencies of Pro12Ala and C1431T variants (listed in Table 2) are in agreement to the data previously reported by Doney et al., 2004 [20], although 1431T allele was shown to be less frequent in obese than in control individuals in the analyzed population.
Table 2.
Genotypes and allele frequencies in diabetic, obese, and control groups (statistical significant values are in bold).
(a).
| Control group | Diabetic group | Obese group | |
|---|---|---|---|
| n | 254 | 211 | 205 |
| Pro12Ala | |||
| Pro/Pro | 222 (87,5%) | 179 (84,6%) | 176 (86%) |
| Pro/Ala | 32 (12,5%) | 32 (15,4%) | 28 (13,6%) |
| Ala/Ala | 0 (0%) | 0 (0%) | 1 (0,4%) |
| C1431T | |||
| C/C | 199 (78,3%) | 171 (81%) | 176 (85,9%) |
| C/T | 44 (17,4%) | 38 (18%) | 27 (13,2%) |
| T/T | 11 (4,3%) | 2 (1%) | 2 (0,9%) |
| A-2819G | |||
| A/A | 208 (81,7%) | 141 (66,7%) | 165 (80,6%) |
| A/G | 39 (15,4%) | 64 (30,6%) | 36 (17,6%) |
| G/G | 7 (2,9%) | 6 (2,7%) | 4 (1,8%) |
(b).
| SNP | Risk allele | Frequency of risk allele | Risk allele odds ratio (95% C.I.) | P value; statistical power (%) | ||||
|---|---|---|---|---|---|---|---|---|
| Pro12Ala | Ala | Control group | Diabetic group | Obese group | Control/Diabetic | Control/Obese | Control/Diabetic | Control/Obese |
| 0.06 | 0.08 | 0.08 | 1.2 (0.73–2.03) | 1.1 (0.70–1.96) | 0.4; 23% | 0.5; 12.2% | ||
| C1431T | T | |||||||
| 0.13 | 0.10 | 0.07 | 0.74 (0.49–1.1) | 1.83 (1.17–2.86) | 0.1; 17.6% | 0.008; 68.3% | ||
| A-2819G | G | |||||||
| 0.10 | 0.18 | 0.10 | 2.25 (1.46–3.44) | 1.03 (0.67–1.57) | 1.9 × 10−4; 98% | 0.9; 9% | ||
Moreover, during the SNP analysis of the C-2821T SNP, we have characterized a novel SNP, A-2819G, located two base pair downstream, included within the same putative E2 box [16]. This novel SNP was genotyped on the entire cohort of 670 individuals using the same standard procedures. Genotypes and allelic frequencies of A-2819G are reported in Table 2.
The -2819G minor allele frequency, observed in 211 individuals with type 2 diabetes, was about 2-fold than the frequency of the risk allele in the control and obese groups (see Table 2). In logistic regression analyses, aimed to test the hypothesis of A-2819G as risk factor for type 2 diabetes under the assumption of a codominant, recessive and dominant model, the dominant model best fitted the data (P = 1.9 × 10−4, Table 3). According to this model, -2819G allele carriers (AG and GG genotype) showed an O.R. = 2.24 (95% C.I. 1.46–3.44) of developing type 2 diabetes compared to the noncarriers (Table 3). The A-2819G allele is in Hardy-Weinberg disequilibrium (DHW) in the control group (P = .01 see Table 3).
Table 3.
Hardy-Weinberg equilibrium and association tests for A-2819G SNP.
| Hardy-Weinberg equilibrium | Tests for association (C.I.: 95% confidence interval) | |||
|---|---|---|---|---|
| Controls | Diabetic group | Risk allele | Armitage's trend test | |
| n = 254 | n = 211 | [AA versus GG] | [AA versus AG+GG] | Common odd ratio |
| O.R. = 2.42 | O.R. = 2.24 | O.R. = 1.60 | ||
| C.I. = [0.41–3.84] | C.I. = [1.46–3.44] | |||
| P = .01 | P = .8 | P = .6 | P = 1.9 × 10−4 | P = .0013 |
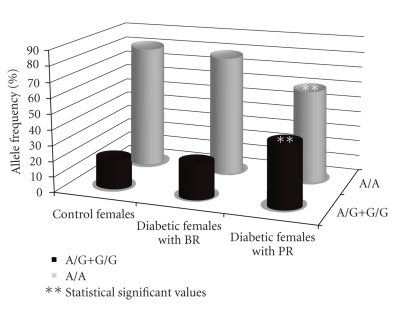
The newly identified risk allele -2819G was not found to be associated with obesity (Table 2) and it was estimated to be not in linkage disequilibrium with Pro12Ala and C1431T SNPs. The possible association of A-2819G SNP with some specific clinical features of both type 2 diabetes and obesity was also considered. Specifically, BMI in obese individuals, blood glucose, and HA1c levels and the presence of proliferative retinopathy in type 2 diabetic patients were evaluated. The analysis revealed that -2819G frequency in diabetic females with proliferative retinopathy was 2-fold increased with respect to background retinopathy female (P = .02; see Table 4 and Figure 1).
Table 4.
Genotypes and -2819G frequency in diabetic women with and without proliferative retinopathy (statistical significant values are in bold).
(a).
| Diabetic female with background retinopathy | Diabetic female with proliferative retinopathy | |
|---|---|---|
| n | 55 | 57 |
| A-2819G | ||
| A/A | 43 (78%) | 34 (60%) |
| A/G | 11 (22%) | 21 (36.7%) |
| G/G | 0 (0%) | 2 (3.3%) |
(b).
| SNP | Risk allele | Frequency of risk allele | Risk allele odds ratio (95% C.I.) | P value; statistical power (%) | |
|---|---|---|---|---|---|
| A-2819G | G | Diabetic female with background retinopathy | Diabetic female with proliferative retinopathy | Background/proliferative | Background/proliferative |
| 0.11 | 0.22 | 2.30 (1.09–4.83) | 0.02; 66.3% | ||
Figure 1.
Comparison of A-2819G frequencies in diabetic women with background or proliferative retinopathy and control women.
In logistic regression analyses, the dominant model best fitted the data (P = .03; see Table 5). According to this model, type 2 diabetes female with -2819G allele (AG and GG genotype) showed an O.R. = 2.42 (95% C.I. = 1.057–5.56) of developing proliferative retinopathy compared to the noncarrier diabetic female (Table 5). The A-2819G allele is in Hardy-Weinberg equilibrium in both background and proliferative groups (Table 5). Moreover, diabetic females with proliferative retinopathy had more than 3-fold risk compared to control females (data not shown).
Table 5.
Hardy-Weinberg equilibrium and association tests for A-2819G and proliferative retinopathy.
| Hardy-Weinberg equilibrium | Tests for association (C.I.: 95% confidence interval) | |||
|---|---|---|---|---|
| Background | Proliferative | Risk allele | Armitage's trend test | |
| n = 55 | n = 57 | [AA versus GG] | [AA versus AG+GG] | Common odd ratio |
| O.R. = 6.30 | O.R. = 2.42 | O.R. = 3.57 | ||
| C.I. = [0.29–135.68] | C.I. = [1.057–5.56] | |||
| P = .37 | P = .57 | P = .12 | P = .03 | P = .02 |
In conclusion, our results show that a novel nucleotide variant A-2819G, in the regulatory region of PPARG, is associated with type 2 diabetes and with proliferative retinopathy in females in an Italian cohort.
4. Discussion
Previous studies have shown that nucleotide variations, both point mutations and SNPs, in the PPARG coding region, are associated with an increased susceptibility of developing type 2 diabetes and obesity [9]. Particularly, the well-known Pro12Ala substitution has been widely associated with both type 2 diabetes and obesity, or predictors of these diseases, in different populations [10, 12]. The Pro12Ala polymorphism has been recently predicted to confer protection, through unknown mechanisms, from type 2 diabetes and nephropathy [12, 15, 21, 22].
Conversely, it has also been shown that Pro12Ala does not associate with diabetic retinopathy [11, 13]. The Pro12Ala SNP has been described in high linkage disequilibrium with C1431T polymorphism, located in exon 6 of PPARG gene. Several studies have shown T minor allele to be associated, in obese individuals, with low plasmatic leptin levels [14]. The same variation has been associated with high BMI values in obese female from Finland in coassociation with Ala12 allele [23].
In the present study, we demonstrated that Pro12Ala and C1431T variants are not associated with type 2 diabetes, obesity, BMI variation, and also microangiopatic complication of diabetes, in a cohort of 670 individuals from Campania County, in the Southern Italy. We also found that -2821T allele, described in the Pima Indians population from Arizona, was not present in the analyzed population.
In contrast, a novel nucleotide variation, A-2819G, two base pair downstream the C-2821T SNP described by Muller et al. [16], was identified. We demonstrated that the A-2819G nucleotide variant is associated with type 2 diabetes, but not with obesity or BMI variations (Tables 2 and 3). The A-2819G allele is in DHW in the control group. DHW may be due to several factors such as genotyping errors, population stratification, selection, inbreeding, the presence of a causative allele or simply by the chance. We can therefore exclude with high confidence that DHW in the control group could be due to genotyping errors. For A-2819G SNP, as well as for all the examined SNPs, all the subjects were analyzed by DHPLC and then sequenced. We cannot exclude that DHW is due to other confounding factors; however, at least for the autosome genes, the presence of DHW in controls is compatible with the segregation of a causative variant according to a dominant model [24].
The association tests for the newly identified A-2819G SNP, revealed statistical significant association with a common and detrimental microvascular complication of type 2 diabetes, namely, the diabetic proliferative retinopathy, in the diabetic females of the examined population (O.R. = 2.43; C.I. 1.09–4.83; P = 0.02). This microvascular complication represents a leading cause of blindness, characterized by an increased vascular permeability, haemostatic abnormalities, increased tissue ischemia, and also neoangiogenesis.
The newly identified A-2819G SNP and the C-2821T [16] are both located in a binding site for a transcriptional repressor δEF1. The polymorphisms described by Muller et al., 2003 [16], leading to higher levels of PPARG transcription, has been associated with an increased risk of development of type 2 diabetes and obesity in Pima Indians [16]. This finding is supported by the notion that supraphysiological activation of PPARγ by thiazolidinedione (TZD) agonists increases triglyceride (TG) content of white adipose tissue, thereby decreasing TG levels in the liver and muscles, resulting in increased insulin sensitivity and type 2 diabetes [25].
In this study, we demonstrated that a novel polymorphism, in the regulatory region of PPARG, associates to proliferative retinopathy in diabetic females, suggesting a possible involvement of the hormonal female component, such as the estrogen receptor signaling that interferes with the physiological PPARγ intracellular pathway [26]. We can reject sex differences due to genotype bias, since males and females were analyzed together, in blind fashion, and the gender was assigned after the analysis.
We postulate that PPARγ may be involved in the development and progression of diabetic retinopathy via a plethora of mechanisms, although its pathogenic effect on the development of the microvascular complications in type 2 diabetic patients is not yet known.
Indeed, Muller et al. (2003) concluded that -2821T allele, via its effect on PPARγ expression, may exert functional consequences on different PPARγ2-activated pathways and that its association with the well-known Pro12Ala may contribute to PPARγ2-related phenotypes [16].
PPARγ is a multivalent receptor able to bind different ligands. Full and partial TZD agonists can induce distinct receptor combinations, leading to selective gene expression. Ligand-receptor complexes, assuming different three-dimensional conformations, show unique and differential interactions with cofactors and other transcription factors [27].
Particularly, PPARγ2 is highly and heterogeneously expressed in the human eye, where its ligand-dependent activation evokes potent and robust inhibition of corneal angiogenesis [27]. Moreover, altered lipid oxidation in the retina supporting cells (retinal pigment epithelium, RPE) and in the eye vessels, and an increased glucose uptake from RPE cells, has pathological effects on eye vessels [28, 29]. However, it was already shown that lipid peroxidation is influenced by C1431T polymorphism, also involved in the susceptibility to premature myocardial infarction [30].
We hypothesize that the newly identified A-2819G nucleotide variation—possibly altering PPARγ2 transcript abundance—may affect the complex and highly regulated gene expression network in the eye, conferring in turn the susceptibility to diabetic retinopathy.
Further studies enrolling a greater number of subjects, both patients and controls, would be useful to better elucidate the involvement of this novel SNP in the susceptibility of developing diabetic retinopathy. However, these findings strengthen the hypothesis that PPARγ could be a novel pharmacological target of angiostatic agents in the treatment of diabetic retinopathy.
Acknowledgments
The authors thank Dr. Teresa Esposito (IGB-CNR) for helpful discussion and Mrs. Maria Terracciano for technical assistance. The authors are also grateful to Dr. Flora Beneduce (ASL NA5), Dr. Luciano Improta (ASL NA5), and Dr. Franco Saccomanno (Diabetic Clinic, Second University of Naples, Italy) for giving them the opportunity to study their patients. This work was supported by Legge 5, Regione Campania, to Amelia Casamassimi and by Ricerca di Ateneo, Second University of Naples, Italy, to D. G. Valerio Costa and Amelia Casamassimi contributed equally to this work.
References
- 1.Francis GA, Fayard E, Picard F, Auwerx J. Nuclear receptors and the control of metabolism. Annual Review of Physiology. 2003;65:261–311. doi: 10.1146/annurev.physiol.65.092101.142528. [DOI] [PubMed] [Google Scholar]
- 2.Mukherjee R, Jow L, Croston GE, Paterniti JR., Jr. Identification, characterization, and tissue distribution of human peroxisome proliferator-activated receptor (PPAR) isoforms PPARγ2 versus PPARγ1 and activation with retinoid X receptor agonists and antagonists. The Journal of Biological Chemistry. 1997;272(12):8071–8076. doi: 10.1074/jbc.272.12.8071. [DOI] [PubMed] [Google Scholar]
- 3.Sabatino L, Casamassimi A, Peluso G, et al. A novel peroxisome proliferator-activated receptor γ isoform with dominant negative activity generated by alternative splicing. The Journal of Biological Chemistry. 2005;280(28):26517–26525. doi: 10.1074/jbc.M502716200. [DOI] [PubMed] [Google Scholar]
- 4.Rosen ED, Spiegelman BM. PPARγ: a nuclear regulator of metabolism, differentiation, and cell growth. The Journal of Biological Chemistry. 2001;276(41):37731–37734. doi: 10.1074/jbc.R100034200. [DOI] [PubMed] [Google Scholar]
- 5.Hasstedt SJ, Ren Q-F, Teng K, Elbein SC. Effect of the peroxisome proliferator-activated receptor-γ2 Pro12Ala variant on obesity, glucose homeostasis, and blood pressure in members of familial type 2 diabetic kindreds. The Journal of Clinical Endocrinology & Metabolism. 2001;86(2):536–541. doi: 10.1210/jcem.86.2.7205. [DOI] [PubMed] [Google Scholar]
- 6.Yen C-J, Beamer BA, Negri C, et al. Molecular scanning of the human peroxisome proliferator activated receptor γ (hPPARγ) gene in diabetic Caucasians: identification of a Pro12Ala PPARγ2 missense mutation. Biochemical and Biophysical Research Communications. 1997;241(2):270–274. doi: 10.1006/bbrc.1997.7798. [DOI] [PubMed] [Google Scholar]
- 7.Semple RK, Chatterjee VKK, O'Rahilly S. PPARγ and human metabolic disease. The Journal of Clinical Investigation. 2006;116(3):581–589. doi: 10.1172/JCI28003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Masugi J, Tamori Y, Mori H, Koike T, Kasuga M. Inhibitory effect of a proline-to-alanine substitution at codon 12 of peroxisome proliferator-activated receptor-γ 2 on thiazolidinedione-induced adipogenesis. Biochemical and Biophysical Research Communications. 2000;268(1):178–182. doi: 10.1006/bbrc.2000.2096. [DOI] [PubMed] [Google Scholar]
- 9.Knouff C, Auwerx J. Peroxisome proliferator-activated receptor-γ calls for activation in moderation: lessons from genetics and pharmacology. Endocrine Reviews. 2004;25(6):899–918. doi: 10.1210/er.2003-0036. [DOI] [PubMed] [Google Scholar]
- 10.Andrulionytè L, Zacharova J, Chiasson J-L, Laakso M. Common polymorphisms of the PPAR-γ2 (Pro12Ala) and PGC-1α (Gly482Ser) genes are associated with the conversion from impaired glucose tolerance to type 2 diabetes in the STOP-NIDDM trial. Diabetologia. 2004;47(12):2176–2184. doi: 10.1007/s00125-004-1577-2. [DOI] [PubMed] [Google Scholar]
- 11.Zietz B, Barth N, Spiegel D, Schmitz G, Schölmerich J, Schäffler A. Pro12Ala polymorphism in the peroxisome proliferator-activated receptor-γ2 (PPARγ2) is associated with higher levels of total cholesterol and LDL-cholesterol in male caucasian type 2 diabetes patients. Experimental and Clinical Endocrinology & Diabetes. 2002;110(2):60–66. doi: 10.1055/s-2002-23487. [DOI] [PubMed] [Google Scholar]
- 12.Caramori ML, Canani LH, Costa LA, Gross JL. The human peroxisome proliferator-activated receptor γ2 (PPARγ2) Pro12Ala polymorphism is associated with decreased risk of diabetic nephropathy in patients with type 2 diabetes. Diabetes. 2003;52(12):3010–3013. doi: 10.2337/diabetes.52.12.3010. [DOI] [PubMed] [Google Scholar]
- 13.Petrovič MG, Kunej T, Peterlin B, Dovč P, Petrovič D. Gly482Ser polymorphism of the peroxisome proliferator-activated receptor-γ coactivator-1 gene might be a risk factor for diabetic retinopathy in Slovene population (Caucasians) with type 2 diabetes and the Pro12Ala polymorphism of the PPARγ gene is not. Diabetes/Metabolism Research and Reviews. 2005;21(5):470–474. doi: 10.1002/dmrr.546. [DOI] [PubMed] [Google Scholar]
- 14.Sassa Y, Hata Y, Aiello LP, Taniguchi Y, Kohno K, Ishibashi T. Bifunctional properties of peroxisome proliferator-activated receptor γ1 in KDR gene regulation mediated via interaction with both Sp1 and Sp3. Diabetes. 2004;53(5):1222–1229. doi: 10.2337/diabetes.53.5.1222. [DOI] [PubMed] [Google Scholar]
- 15.Meirhaeghe A, Amouyel P. Impact of genetic variation of PPARγ in humans. Molecular Genetics and Metabolism. 2004;83(1-2):93–102. doi: 10.1016/j.ymgme.2004.08.014. [DOI] [PubMed] [Google Scholar]
- 16.Muller YL, Bogardus C, Beamer BA, Shuldiner AR, Baier LJ. A functional variant in the peroxisome proliferator-activated receptor γ2 promoter is associated with predictors of obesity and type 2 diabetes in Pima Indians. Diabetes. 2003;52(7):1864–1871. doi: 10.2337/diabetes.52.7.1864. [DOI] [PubMed] [Google Scholar]
- 17.Costa V, Conte I, Ziviello C, et al. Identification and expression analysis of novel Jakmip1 transcripts. Gene. 2007;402(1-2):1–8. doi: 10.1016/j.gene.2007.07.001. [DOI] [PubMed] [Google Scholar]
- 18.World Health Organization. Diabetes mellitus: report of a WHO Study Group. Geneva, Switzerland: World Health Organization; 1985 Tech. Rep. 727. [PubMed]
- 19.Sasieni PD. From genotypes to genes: doubling the sample size. Biometrics. 1997;53(4):1253–1261. [PubMed] [Google Scholar]
- 20.Doney ASF, Fischer B, Cecil JE, et al. Association of the Pro12Ala and C1431T variants of PPARG and their haplotypes with susceptibility to type 2 diabetes. Diabetologia. 2004;47(3):555–558. doi: 10.1007/s00125-003-1323-1. [DOI] [PubMed] [Google Scholar]
- 21.Herrmann S-M, Ringel J, Wang J-G, Staessen JA, Brand E. Peroxisome proliferator-activated receptor-γ2 polymorphism Pro12Ala is associated with nephropathy in type 2 diabetes: The Berlin Diabetes Mellitus (BeDiaM) Study. Diabetes. 2002;51(8):2653–2657. doi: 10.2337/diabetes.51.8.2653. [DOI] [PubMed] [Google Scholar]
- 22.Altshuler D, Hirschhorn JN, Klannemark M, et al. The common PPARγ Pro12Ala polymorphism is associated with decreased risk of type 2 diabetes. Nature Genetics. 2000;26(1):76–80. doi: 10.1038/79216. [DOI] [PubMed] [Google Scholar]
- 23.Valve R, Sivenius K, Miettinen R, et al. Two polymorphisms in the peroxisome proliferator-activated receptor-γ gene are associated with severe overweight among obese women. The Journal of Clinical Endocrinology & Metabolism. 1999;84(10):3708–3712. doi: 10.1210/jcem.84.10.6061. [DOI] [PubMed] [Google Scholar]
- 24.Wittke-Thompson JK, Pluzhnikov A, Cox NJ. Rational inferences about departures from Hardy-Weinberg equilibrium. American Journal of Human Genetics. 2005;76(6):967–986. doi: 10.1086/430507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yamauchi T, Kamon J, Waki H, et al. The mechanisms by which both heterozygous peroxisome proliferator-activated receptor γ (PPARγ) deficiency and PPARγ agonist improve insulin resistance. The Journal of Biological Chemistry. 2001;276(44):41245–41254. doi: 10.1074/jbc.M103241200. [DOI] [PubMed] [Google Scholar]
- 26.Bonofiglio D, Gabriele S, Aquila S, et al. Estrogen receptor α binds to peroxisome proliferator-activated receptor response element and negatively interferes with peroxisome proliferator-activated receptor γ signaling in breast cancer cells. Clinical Cancer Research. 2005;11(17):6139–6147. doi: 10.1158/1078-0432.CCR-04-2453. [DOI] [PubMed] [Google Scholar]
- 27.Pershadsingh HA, Moore DM. PPARγ agonists: potential as therapeutics for neovascular retinopathies. PPAR Research. 2008;2008:13 pages. doi: 10.1155/2008/164273. Article ID 164273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Muranaka K, Yanagi Y, Tamaki Y, et al. Effects of peroxisome proliferator-activated receptor γ and its ligand on blood-retinal barrier in a streptozotocin-induced diabetic model. Investigative Ophthalmology and Visual Science. 2006;47(10):4547–4552. doi: 10.1167/iovs.05-1432. [DOI] [PubMed] [Google Scholar]
- 29.Kim J, Oh Y-S, Shinn S-H. Troglitazone reverses the inhibition of nitric oxide production by high glucose in cultured bovine retinal pericytes. Experimental Eye Research. 2005;81(1):65–70. doi: 10.1016/j.exer.2005.01.010. [DOI] [PubMed] [Google Scholar]
- 30.Chao T-H, Li Y-H, Chen J-H, et al. The 161TT genotype in the exon 6 of the peroxisome-proliferator-activated receptor γ gene is associated with premature acute myocardial infarction and increased lipid peroxidation in habitual heavy smokers. Clinical Science. 2004;107(5):461–466. doi: 10.1042/CS20040014. [DOI] [PubMed] [Google Scholar]