Abstract
Brain tumors can arise following deregulation of signaling pathways normally activated during brain development and may derive from neural stem cells. Given the requirement for Hedgehog in non-neoplastic stem cells, we investigated whether Hedgehog blockade could target the stem-like population in glioblastoma multiforme (GBM). We found that Gli1, a key Hedgehog pathway target, was highly expressed in 5 of 19 primary GBM and in 4 of 7 GBM cell lines. Shh ligand was expressed in some primary tumors, and in GBM-derived neurospheres, suggesting a potential mechanism for pathway activation. Hedgehog pathway blockade by cyclopamine caused a 40%–60% reduction in growth of adherent glioma lines highly expressing Gli1 but not in those lacking evidence of pathway activity. When GBM-derived neurospheres were treated with cyclopamine and then dissociated and seeded in media lacking the inhibitor, no new neurospheres formed, suggesting that the clonogenic cancer stem cells had been depleted. Consistent with this hypothesis, the stem-like fraction in gliomas marked by both aldehyde dehydrogenase activity and Hoechst dye excretion (side population) was significantly reduced or eliminated by cyclopamine. In contrast, we found that radiation treatment of our GBM neurospheres increased the percentage of these stem-like cells, suggesting that this standard therapy preferentially targets better-differentiated neoplastic cells. Most importantly, viable GBM cells injected intracranially following Hedgehog blockade were no longer able to form tumors in athymic mice, indicating that a cancer stem cell population critical for ongoing growth had been removed.
Keywords: . Hedgehog, Glioma, Stem cell
Introduction
Glioblastoma multiforme (GBM) are incurable brain cancers that exhibit various degrees of astrocytic differentiation [1, 2]. It has recently been proposed that GBM derive from neural stem or progenitor cells and that signaling pathways that play a key role in such primitive neural cells may also be required in glial tumors [3–5]. We investigated the role of the Hedgehog (Hh) pathway in malignant gliomas, as it plays an essential role in cerebellar precursor cells [6–8], in growth of the cerebral cortex [9], and in neural stem cells in the adult brain [10]. Although Hh signaling is clearly dysregulated in a subset of human medulloblastomas, its role in malignant gliomas is less clear. However, Gli1 was originally identified as a gene amplified in a human malignant glioma, strongly suggesting such a link [11]. In addition, Hh pathway members are expressed in primary astrocytoma samples [12], and Hh blockade has been shown to inhibit the growth of a small number of glioma cell lines [9].
Hh ligands are secreted glycoproteins that bind the cell surface receptor Patched (Ptch). Ligand binding to Ptch relieves its inhibition of Smoothened (Smo) and allows signaling to proceed. Smo activates the canonical Hh pathway through Gli-dependent transcription of multiple targets, including N-myc, cyclin D, Ptch, Gli1, and Gli2 [13, 14]. Gli proteins are large, multifunctional transcription factors, and their activities are tightly regulated (reviewed in [15, 16]). Hedgehog pathway blockade has emerged as a promising therapy for multiple types of cancer, including brain tumors and carcinomas of the prostate, digestive tract, and lung [17–21]. In these latter tumors, the Hh pathway is activated predominantly by ligands, rather than by the Ptch mutations seen in medulloblastoma.
Because Hh signaling plays a critical role in non-neoplastic stem cells, it has been suggested that stem-like neoplastic cells may also be susceptible to Hh pathway blockade. The nature of such cancer stem cells, and how one might target them therapeutically, has been the subject of several recent reviews [22–26]. In brief, the cancer stem cell hypothesis suggests that only the stem cell compartment in tumors is capable of unlimited self-renewal and that elimination of these cells will ultimately halt neoplastic expansion, as better-differentiated cells have limited mitogenic capacity and will not contribute to long-term tumor growth. The first well-documented example of cancer stem cells was in leukemia, where only a small fraction of the tumors identified by expression of stem-cell markers possessed the capacity for tumor propagation [27, 28]. Similar cells have subsequently been isolated from brain tumors using markers such as CD133 and side population, which were originally developed to study non-neoplastic stem cells [29–33]. The study of stem cells in gliomas also benefits from the capacity of these tumors to grow as neurospheres, allowing the clonogenic potential and differentiation capacity to be measured [33, 34]. In this study, we demonstrate that markers of Hh pathway activity are detected in malignant gliomas samples and that the pathway appears to regulate the stem cell fraction in GBM cell lines.
Materials and Methods
Clinical Specimens and Immunohistochemistry
Snap-frozen primary glioma tissues were obtained from the Department of Pathology, Johns Hopkins University School of Medicine, with Institutional Review Board approval. RNA was extracted from gliomas using Trizol reagent (Invitrogen, Carlsbad, CA, http://www.invitrogen.com) and further purified using RNeasy columns (Qiagen, Valencia, CA, http://www1.qiagen.com) according to the manufacturers’ instructions. The construction of the tissue microarray containing high-grade astrocytomas has been previously described [38]. Immunohistochemistry was performed on freshly cut, deparaffinized tumor or tissue microarray sections essentially as previously described [52]. The following antibodies were used for immunocytochemistry: Nestin, clone MAB5326 (1:2,500; Chemicon, Temecula, CA, http://www.chemicon.com); glial fibrillary acidic protein (GFAP), Z0334 (1:2,500; DAKO, Carpinteria, CA, http://www.dako.com). Statistical analyses were performed using GraphPad Prism4 (GraphPad Software, Inc., San Diego, http://www.graphpad.com).
Cell Culture
The U87-MG, A172, U251, and SW1088 cell lines were obtained from American Type Culture Collection (ATCC) (Manassas, VA, http://www.atcc.org) and maintained in ATCC’s recommended growth medium supplemented with 10% fetal bovine serum (FBS). The CJ-MG and BK-MG cell lines were a kind gift from Dr. Carol A. Kruse [53]. JHH-GBM2 was derived as follows: fresh tumor was chopped to small pieces using sterile razor blade and then mechanically triturated through a 21-gauge needle and filtered through a 40 μm cell strainer (BD Falcon; BD Biosciences, Bedford, MA, http://www.bdbiosciences.com). Cells were concentrated by centrifugation and then seeded in Dulbecco’s modified Eagle’s medium (DMEM)/Ham’s F-12 medium supplemented with 10% FBS. The glioblastoma-derived neurosphere line HSR-GBM1 was derived and propagated as previously described [54]. 293T, 293T/SHH-N′, and Light2 cells were the kind gift of Dr. Phil Beachy.
The Gli reporter assay was performed in either U87-MG by transient transfection of the Gli firefly luciferase reporter construct or in NIH3T3 cells stably transfected with Gli firefly luciferase and Renilla luciferase reporter constructs (Light2), as previously described with minor modifications [55]. Briefly, Light2 cells were plated in a 24 well plates at 7 × 104 cells per well in DMEM supplemented with 10% FBS. When confluent (normally the next day), the monolayers were washed once each with phosphate-buffered saline (PBS) and DMEM supplemented with 0.5% FBS. Cells were incubated overnight in a humidified incubator kept at 37°C, 5% CO2, and then growth medium was removed and monolayers were overlaid with conditioned medium from 293T, 293T/Shh-N′, or HSR-GBM1 and incubated for an additional 48 hours. Gli reporter activity was quantified using the dual-luciferase reporter assay system (Promega, Madison, WI, http://www.promega.com) and normalized to Renilla luciferase activity. In U87-MG, Gli reporter activity was quantified using the luciferase reporter assay system (Promega) and normalized to total protein.
For mRNA quantification and 3-(4,5-dimethylthiazol-2-yl)-5(3-carboxymethonyphenol)-2-(4-sulfophenyl)-2H-tetrazolium (MTS) assays, cells were plated in six-well plates (RNA) or 96-well plated (MTS) and incubated overnight, and the monolayers were washed once with PBS, followed by overlay with low-serum (0.5% fetal bovine serum) media. The following day (approximately 16 hours after serum withdrawal), medium was replaced with medium supplemented with 0.5% FBS and either ethanol or cyclopamine (5 or 10 μM). RNA was extracted at various time points after cyclopamine addition using Qiagen RNeasy kits. MTS assays were performed at various time points using the CellTiter96 assay (Promega) according to the manufacturer’s instructions.
To assay neurosphere differentiation, HSR-GBM1 cells were triturated and plated at 2 × 104 cells per cm2 in 10-cm2 cell culture dishes precoated with Matrigel (BD Biosciences) diluted 1:100 in Neurocult medium (Stem Cell Technologies, Vancouver, BC, Canada, http://www.stemcell.com) without epidermal growth factor (EGF). After attachment of cells to the plate surface, medium was replaced with Neurocult medium lacking mitogens and heparin but supplemented with 1% bovine calf serum (HyClone, Logan, UT, http://www.hyclone.com). Cells were incubated for 5 days as before and then washed, and RNA was extracted as described above.
shRNA targeting Gli1 was purchased from Sigma-Aldrich (St. Louis, http://www.sigmaaldrich.com). pLKO.1-puro was a kind gift from Dr. Bob A. Weinberg and Dr. Carl D. Novina [56]. U87-MG cells were plated at 2.5 × 105 cells per well or 7.5 × 105 cells per dish in six-well plates or 10-cm culture dishes, respectively. The next day, medium was replaced with 2 ml of transduction medium. Dishes were incubated overnight in humidified incubator at 37°C. RNA was extracted from cells grown in six-well plates 48 hours after infection. For cells grown in 10-cm dishes, medium was replaced 72 hours postinfection with fresh medium containing 10 μg/ml puromycin (Sigma-Aldrich). Cells were stained with 4,6-diamidino-2-phenylindole, and pictures of 10 random high-power fields were taken for each condition.
For Gli1 and Gli2 short interfering RNA (siRNA) experiments, 2 × 105 HSR-GBM1 cells were plated in duplicates in six-well plate format for RNA analysis and at 1.5 × 104 cells per well in a 48-well format for growth assays (MTS). Cells were incubated overnight and then transfected every other day according to the manufacturer’s directions using RNAiFect (Qiagen) with 8 nM/well of a nonspecific siRNA (Dharmacon, Lafayette, CO, http://www.dharmacon.com) as negative control, Gli1 pooled siRNA, or a single Gli2 siRNA duplex (Dharmacon) with the following oligo sequences: Gli1 forward, 5′-GGAAAUGACUGGCAAUGCAUU-3′, and Gli1 reverse, 5′-PUGCAUUGCCAGUCAUUUCCUU-3′ for duplex 1; Gli1 forward, 5′-GCACUGGUCUGUCCACUCUUU-3′, and Gli1 reverse, 5′-PAGAGUGGACAGACCAGUGCUU-3′, for duplex 2; Gli1 forward, 5′-GUCCUCACUUGAACAUUAUU-3′, and Gli1 reverse, 5′-PUAAUGUUAAGUCGAGGACUU-3′, for duplex 3; Gli1 forward, 5′-AGGCUCAGCUUGUGUGUAAUU-3′, and Gli1 reverse, 5′-PUUACACACAAGCUGAGCCUUU-3′, for duplex 4; Gli2 forward, 5′-CGUCAACCCUGUCGCCAUUUU-3′, and Gli2 reverse, 5′-PAAUGGCGACAGGGUUGACGUU-3′.
Clonogenic Assays
Clonogenic assays were performed as previously described [57]. Briefly, U87-MG cells plated at 3 × 103 cells per well. Cells were treated with either ethanol (vehicle [V]) or cyclopamine (5 and 10 μM). Each condition was tested in triplicate, and each assay was repeated twice with similar results. Colony number and size were scored with the ChemiDoc-XRS imager, using the QuantityOne software package (Bio-Rad, Hercules, CA, http://www.bio-rad.com).
HSR-GBM1 cells were seeded in T75 cm2 culture dishes at 1 × 105 cells per dish. Cells were treated with either ethanol or cyclopamine (10 μM) in a final volume of 20 ml of Neurocult medium (Stem Cell Technologies) supplemented with 0.002% heparin, 10 ng/ml human EGF, and 10 ng/ml human fibroblast growth factor-b (Peprotech, Rocky Hill, NJ, http://www.peprotech.com). Dishes were incubated upright for 7 days in a humidified incubator with 5% CO2 at 37°C. Cells were retreated with 2 ml of medium containing the indicated concentration of drug on days 2, 4, and 6. Seven days after treatment, spheres were triturated and replated in T25 cm2 at 2 × 103 cells per dish. Subsequent sphere formation was monitored and scored by light microscopy after 8 days. Ten random fields were photographed for both vehicle and cyclopamine-treated conditions, and the number of spheres over 50 μm in size was scored.
RNA Analyses
RNA levels were analyzed by real-time PCR analysis performed in triplicate with SYBR Green reagents (Applied Biosystems, Foster City, CA, http://www.appliedbiosystems.com), according to the manufacturer’s instructions on an I-Cycler IQ real-time detection system (Bio-Rad) as previously described [58]. To minimize contaminating genomic DNA, a 15-minute on-column DNase step (RNase-free DNase kit; Qiagen) was included during RNA extraction. In addition, all primer sets amplify a single product of the appropriate size and were designed to cross intron-exon boundaries. Oligo sequences were previously described [52].
Xenograft and Side Population Assays
For side population analyses, U87-MG and C6 cells were treated for 7 days in low-serum (0.5% FBS) media containing either ethanol (V) or cyclopamine (5 or 10 μM), with medium changes every 2 days. HSR-GBM1 cells were treated in 10 ml of media, with 2 ml of additional medium containing drug or vehicle supplemented every 2 days. Side population analyses on treated cells were performed as previously described [57]. For xenograft studies, HSR-GBM1 cells were treated as described above, and an aliquot collected from each flask following treatment was scored for viability by Guava-PCA flow cytometry system and ViaCount reagent, according to the manufacturer’s instructions (Guava Technologies, Hayward, CA, http://www.guavatechnologies.com). Next, viable cells were diluted with fresh medium and injected over 10 minutes into the right striatum of athymic (nu/nu) mice (Harlan, Indianapolis, IN, http://www.harlan.com). Mice were monitored daily and sacrificed at the first indication of tumor development (ataxia, seizure, lethargy, or cachexia). Brains were surgically removed and fixed immediately in formalin before submission for histological analysis, as previously described [58].
Radiation Treatment
HSR-GBM1 cells were plated at 5 × 103 cells per well (in triplicate) in 48-well culture plates and treated with either ethanol (0) or cyclopamine (5 or 10 μM). After 3 days of growth, the cultures were irradiated at either mock (0 Gy) or 10 Gy using a Gammacell 40 irradiator equipped with a 132cesium source (MDS Nordion, Ottawa, ON, Canada, http://www.mds.nordion.com). Cell mass was subsequently measured on day 7 using the MTS assay (Promega), and growth rate calculated from the first day of cyclopamine treatment.
For cotreatment with cyclopamine and radiation, HSR-GBM1 cells were plated at 1 × 106 cells per T75 culture flask and treated with either ethanol (0) or cyclopamine (5 or 10 μM) at plating and every other day thereafter. Cells were irradiated 4 days after plating, and percentages of side and aldehyde dehydrogenase (ALDH)-positive populations were determined on day 7.
Results
The Hh Pathway Is Active in a Subset of Primary GBM and Established GBM Cell Lines
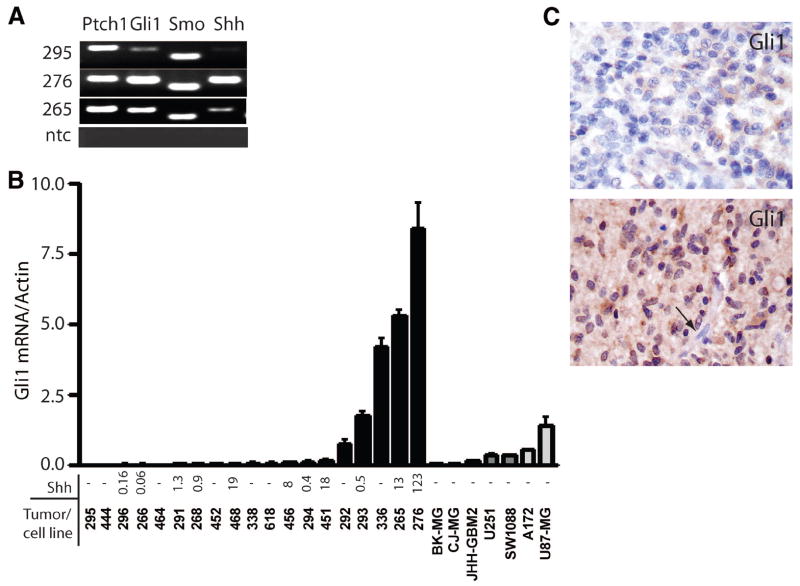
To evaluate the possibility that Hh signaling is active in malignant gliomas, we examined expression of pathway receptors, ligands, and targets in primary tumor samples and in GBM cell lines. The membrane-associated receptors Smo and Ptch were detected in all tested samples (eight primary GBM and six established GBM cell lines) using standard reverse transcription-polymerase chain reaction (RT-PCR). Bands representing Gli1 mRNA were detected in seven of eight primary GBM and five of six established GBM cell lines. Representative data from the analysis of three primary tumors is shown in Figure 1A. Gli1 expression is consistently correlated with Hh pathway activation [35–37], suggesting that Hh signaling is active to some degree in these tumors.
Figure 1.
Hedgehog pathway members are expressed in gliomas. (A): Ptch, Gli1, Smo, and Shh mRNAs were detected in primary glioblastoma multiforme (GBM) samples. (B): Measurement of Gli1 and Shh mRNA using quantitative reverse transcription-polymerase chain reaction in 19 primary GBM samples and seven GBM cell lines disclosed high Gli1 expression in five tumors. (C): Gli1 immunostaining was present in all malignant glioma samples examined, with some showing weak cytoplasmic staining (upper panel) and others showing more abundant cytoplasmic and nuclear protein (lower panel). Endothelial cells (arrow, lower panel) were negative for Gli1 and served as internal controls. Abbreviations: ntc, no template control; Ptch, Patched; Smo, Smoothened.
To more accurately quantitate the level of pathway activity, we examined Gli1 mRNA expression by real time RT-PCR (Fig. 1B). Gli1 expression was detected in all primary tumor samples (n = 19), and Gli1 was highly expressed (Gli1/actin ratio >0.5) in 5 of 19 (26%) of the GBM. Among the established GBM cell lines, Gli1 was most highly expressed in U87-MG cells. In the A172, SW1088, and U251 lines, Gli1 expression was modest, with Gli1/actin ratios of 0.3–0.5. The Gli1/actin ratio in JHH-GBM2, CJ-MG, and BK-MG was quite low, <0.2. We measured Shh expression to determine whether Hh activity might be ligand-driven in some or all cases (Fig. 1B). Relatively high levels of Shh mRNA were detected in several of the primary GBM, including five of nine with above-median Gli1 levels. Interestingly, the case with the highest level of Shh also had the most prominent Gli1 expression. However, other primary tumors containing Gli1 lacked Shh, suggesting that the pathway can be activated by a ligand other than Shh or by ligand-independent means. We did not identify Shh activity in any of the seven adherent GBM cell lines grown in high levels of serum, raising the possibility that these growth conditions have a negative effect on ligand expression. Alternatively, it is possible that the Shh mRNA we detected in primary glioma samples was being generated by non-neoplastic cells and that pure tumor cultures are therefore negative.
The presence of nuclear Gli1 protein expression in formalin-fixed, paraffin-embedded tumor samples would also suggest that Hh signaling is active in gliomas. We therefore examined whether Gli1 is expressed in a nonrelated set of astrocytic tumors using a previously described tissue microarray [38]. All high-grade astrocytic tumors on our tissue microarray had at least trace levels of cytoplasmic Gli1 protein (Fig. 1C, top panel). In approximately half of the tumors, cytoplasmic staining was stronger, and Gli1 was also detected in the nucleus, suggesting that the pathway was active (Fig. 1C, bottom panel). Nuclear Gli1 was detected in cytologically atypical tumor cells but not in non-neoplastic cells, such as blood vessels. We did not observe a significant difference between the percentage of cases with nuclear Gli1 among the grade III anaplastic astrocytoma (10 of 20 positive) and grade IV glioblastoma (8 of 18 positive).
Cyclopamine Decreases Gli1 Expression and Inhibits Growth and Clonogenicity of Adherent GBM Cell Lines
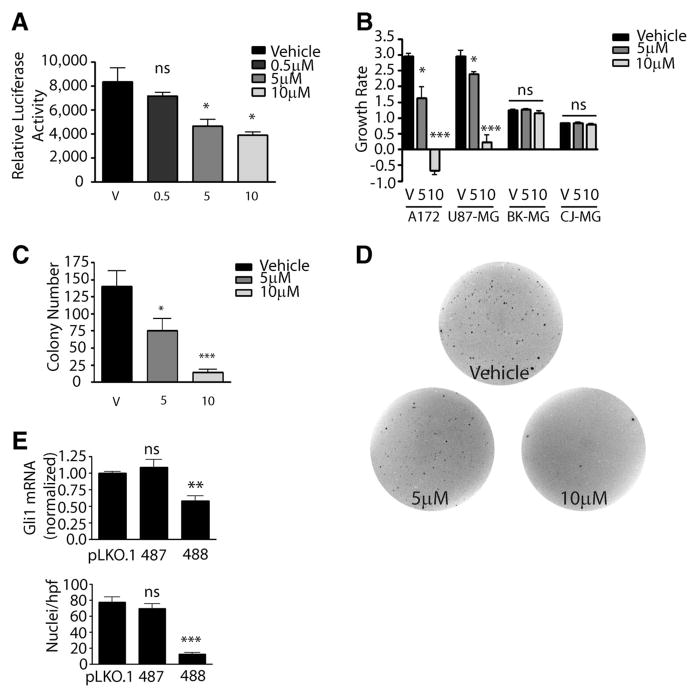
We used the Hh pathway inhibitor cyclopamine to test the ongoing requirement for Hh signaling in glioma cells. All experiments using this inhibitor were performed in low (0.5%) serum. In U87-MG cells transfected with an Hh reporter construct, cyclopamine reduced pathway activity in a dose-dependent fashion, with 5 μM or higher levels required to effect significant pathway inhibition (Fig. 2A). Cyclopamine administration also inhibited endogenous Gli1 mRNA expression in U87-MG cells by 21% at 5 μM and 51% at 10 μM levels (data not shown). Gli1 mRNA expression was also inhibited by 30% or more in the A172 and SW1088 glioma lines, but not in a dose-dependent fashion (data not shown). Significantly, Hh pathway inhibition using cyclopamine resulted in reduced growth in lines with high baseline levels of Gli1 expression but not in cells lacking significant Hh activity. In the high-Gli U87-MG and A172 lines, growth was essentially abolished by 10 μM cyclopamine, whereas the increase in cell mass in the low-Gli CJ-MG and BK-MG lines was unaffected (Fig. 2B). The fact that growth inhibition was observed only in tumor lines expressing moderate or high levels of Gli1 mRNA suggests that the effects of cyclopamine treatment are Hh pathway-specific, rather than generally cytotoxic.
Figure 2.
Hedgehog signaling is required in established glioma cultures. (A): Effect of 48-hour cyclopamine treatment on the activity of a Gli-dependent luciferase reporter in U87-MG. (B): The growth rate of glioma cultures was significantly inhibited by cyclopamine. (C): U87-MG cells treated with cyclopamine formed significantly fewer colonies in agar. (D): Representative wells with U87-MG colonies in soft agar. (E): U87-MG: Gli1 mRNA level (top panel) and total viable cells per hpf (bottom panel) were reduced by specific shRNA targeting lentivirus 488 compared with either empty vector pLKO.1-puro or the nonfunctional Gli1 shRNA 487. Two-sided t test was used throughout. *, p < .05; **, p < .01; ***, p < .001. Abbreviations: hpf, high-power field; ns, not significant; V, vehicle (ethanol) control.
We next examined the ability of cyclopamine to block colony formation of GBM cells. U87-MG cells seeded in soft agar and allowed to establish initial growth were then treated with vehicle, 5 μM drug, or 10 μM drug for an additional 14 days, and the resulting colonies were stained and analyzed. Colony formation was inhibited in a dose-dependent manner, with a statistically significant 46% reduction in colonies grown in 5 μM cyclopamine (p = .03) and an approximately 90% reduction in colonies grown in 10 μM cyclopamine (p < .0001; Fig. 2C, 2D).
To test whether the effect of cyclopamine is mediated by reducing levels of Gli1, rather than noncanonical effects on Hh signaling or nonspecific toxicity, we infected U87-MG cells with lentivirus carrying short hairpin against Gli1. Gli1 mRNA levels were reduced by 40%–50% 48 hours after virus transduction with shRNA 488, whereas a second construct (487) proved ineffective in reducing Gli1 levels (Fig. 2E, upper panel). To test the requirement for Gli1 only in cells containing the construct, we took advantage of the puromycin resistance gene in the pLKO.1-puro backbone. Consistent with an ongoing requirement for Gli1 in the glioma cells, introduction of Gli1 shRNA 488 and short-term puromycin selection significantly reduced the number of cells per high-power field, whereas virus carrying an empty vector or a nonfunctional Gli1 shRNA had no significant effect on viability (Fig. 2E, lower panel).
GBM-Derived Neurospheres Secrete and Respond to Shh Ligand
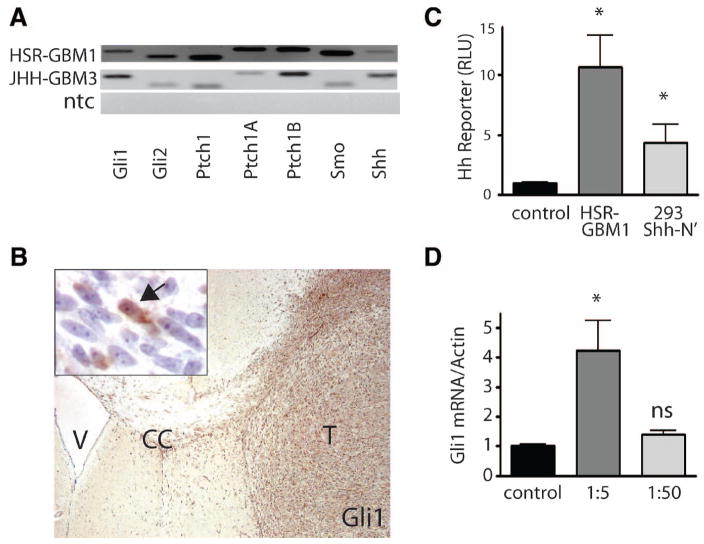
We used GBM-derived neurospheres to further examine the role of Hh in gliomas, including low-passage cells from tumors resected at Johns Hopkins Hospital and grown as neurospheres (JHH-GBM3) and a neurosphere line established at the Hospital San Raphael (HSR-GBM1), as previously described [34]. Such GBM-derived neurospheres demonstrate an infiltrative growth pattern similar to that of human gliomas in vivo and have improved karyotypic stability compared with adherent glioma lines [34, 39]. We detected expression of the transcription factors Gli1 and Gli2, as well as the receptor Smo and all three Ptch-receptor isoforms, in the GBM-derived neurospheres (Fig. 3A). Gli1 protein is also highly expressed in the infiltrating gliomas that arise when these cells are injected into athymic mice (Fig. 3B). Most of the immunopositive cells in this panel are found in or near the tumor mass and have the morphology of neoplastic astrocytes. However, some periventricular cells and scattered cells within the brain parenchyma are also positive, suggesting that some background staining of murine cells is also occurring. Interestingly, unlike the seven adherent glioma lines shown in Figure 1, the two glioma neurosphere cultures both expressed Shh mRNA, suggesting that cultured glioma cells are capable of generating this ligand when grown in this fashion (Fig. 3A).
Figure 3.
Glioblastoma multiforme (GBM)-derived neurospheres express Hedgehog (Hh) ligands, receptors, and targets. (A): mRNAs for Gli1, Gli2, Smo, and Ptch1 (including the inducible isoform Ptch1b) were all expressed in both long-term (HSR-GBM1) and low-passage (JHH-GBM3) neurosphere cultures. (B): Immunohistochemistry. Gli1 protein was highly expressed in HSR-GBM1 intracranial xenografts. Note the highly infiltrating cells invading the CC. Arrow in inset points to a nucleus positive for Gli1; surrounding cells were largely negative in this region. (C): HSR-GBM1 secretes biologically active Shh-ligand, as evidenced by the induction of Hh reporter activity in Light2 cells when conditioned medium from the GBM neurosphere cultures was applied. Medium isolated from 293T cells engineered to secrete Shh-N was a positive control. (D): Following application of concentrated (1:5 dilution) conditioned media from Shh-N secreting 293T cultures, Gli1 mRNA levels were elevated fourfold in low-density HSR-GBM1 cells. Two-sided t test was used throughout. *, p < .05. Abbreviations: CC, corpus callosum; ns, not significant; ntc, no template control; Ptch, Patched; Smo, Smooth-ened; T, tumor; V, lateral ventricle.
To verify that Shh expressed by GBM cells is biologically active, we treated a Gli-luciferase reporter cell line (NIH 3T3-Light2) that is dependent on exogenous Shh ligand with conditioned medium isolated from the HSR-GBM1 neurosphere culture. As a positive control, we used conditioned medium isolated from Shh-N-producing HEK 293 cells [40]. Conditioned medium from the GBM neurospheres activated Light2 cells approximately 10-fold, whereas Shh-N had an approximately 5-fold inductive effect (Fig. 3C), indicating that highly active Shh protein was being secreted by the GBM cells. We next examined the ability of HSR-GBM1 cells to respond to exogenous Shh ligand produced by Shh-N cells. HSR-GBM1 cells were dissociated and plated at low density in 1:5 or 1:50 dilutions of conditioned medium from densely confluent Shh-N cultures. This conditioned medium induced Gli1 mRNA upregulation in a dose-dependent manner, suggesting that HSR-GBM1 is capable of responding to exogenous ligand (Fig. 3D). Thus, glioma cells growing as neurospheres appear to be capable of secreting and responding to biologically active Shh ligand.
Hedgehog Pathway Blockade Inhibits Glioma Neurosphere Formation and Growth and Targets Radioresistant Cells
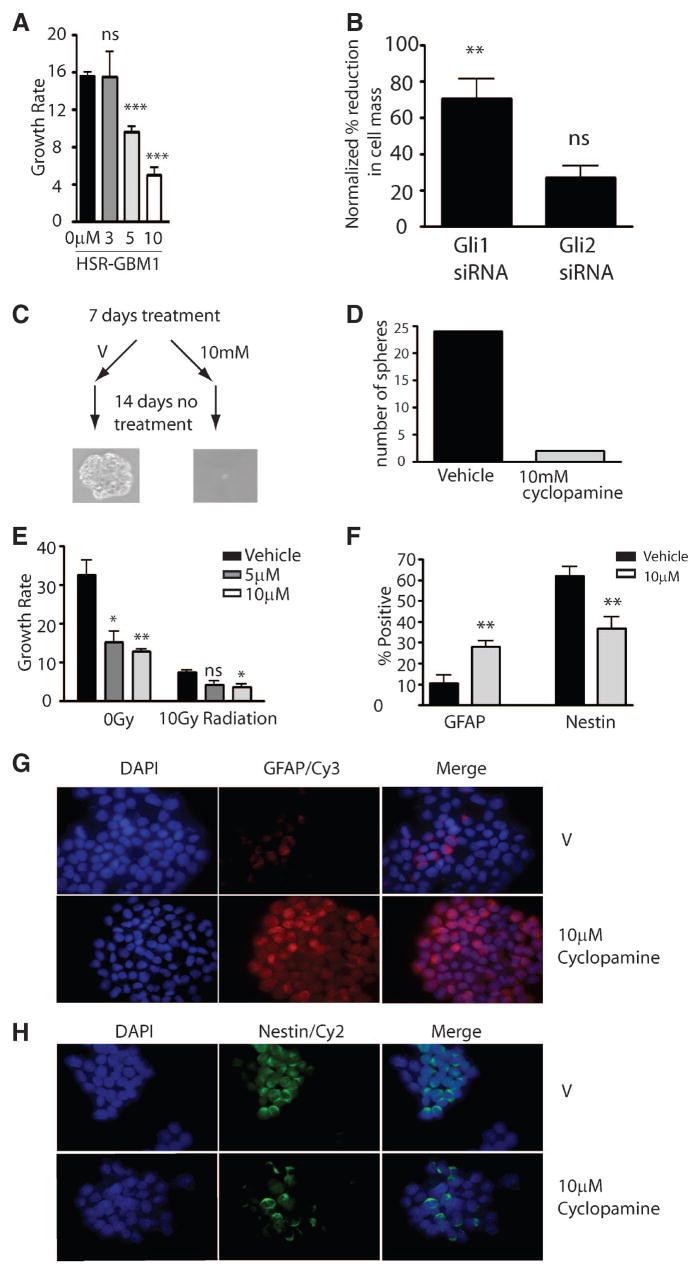
To examine the effects of Hh inhibition on the formation and growth of glioma-derived neurospheres, we treated mechanically separated spheres for 4 days with cyclopamine. Treatment using 3 μM or higher concentrations inhibited Gli1 mRNA expression by 50%–75% compared with vehicle. Cyclopamine-mediated Hh pathway blockade inhibited the overall growth rate of the culture by 30%–75% in a dose-dependent fashion (Fig. 4A). This experiment was repeated three times with similar results. Microscopic examination of these cultures showed that few well-developed neurospheres had formed during the period of Hh blockade, suggesting that the cells required to initiate or sustain neurosphere growth had been inhibited or killed. Transient transfection of siRNA directed against Gli1 but not Gli2 into the neurospheres also significantly reduced cell mass after 7 days compared with control siRNA (Fig. 4B).
Figure 4.
Hedgehog (Hh) pathway inhibition reduces glioma neurosphere initiation and growth. (A): The formation and growth of HSR-glioblastoma multiforme 1 (GBM1) neurospheres was inhibited by cyclopamine in a dose-dependent fashion. (B): Gli1 but not Gli2 siRNAs significantly inhibited growth of HSR-GBM1 neurospheres relative to non-Hh targeting siRNA control (Gli2 siRNA, p = .061). (C, D): Hh pathway blockade affected the ability of cells to initiate sphere formation, as HSR-GBM1 cultures treated for 7 days with cyclopamine (but not vehicle) had a drastically reduced ability to generate new spheres upon transfer into fresh medium lacking cyclopamine. (E): Cyclopamine modestly augmented the effect of radiation treatment on the growth rate of HSR-GBM1 cells. (F, G, H): Cyclopamine treatment for 7 days significantly increased the fraction of GFAP-expressing cells (F, G) and reduced the fraction of nestin-expressing cells (F, H). All images were taken at a magnification of ×60. Two-sided t test was used throughout. *, p < .05; **, p < .01; ***, p < .001. Abbreviations: DAPI, 4,6-diamidino-2-phenylindole; GFAP, glial fibrillary acidic protein; siRNA, short interfering RNA; V, vehicle.
To more stringently test the effect of Hh blockade on the ability of GBM cells to generate new spheres, actively growing HSR-GBM1 cultures were treated with cyclopamine for 7 days and then dissociated, and equal numbers of viable cells were replated in fresh, drug-free medium. Cyclopamine pretreatment almost completely blocked large sphere formation (Fig. 4C, 4D). Because no cyclopamine was present in the media after replating, the lack of spheres cannot be due to ongoing inhibition of sphere growth. Rather, these data strongly suggest that a 7-day period of Hh blockade can inactivate or remove the clonogenic stem-like cells required to initiate sphere formation.
Bao et al. have previously shown that stem-like CD133-positive tumor cells in GBM are resistant to radiation therapy [41]. However, radiation is clearly effective in eliminating at least a subset of cells in GBM clinically, and any patients receiving a novel chemotherapy will also likely have their tumor irradiated. We therefore examined the effects of Hh blockade given in combination with radiation. We treated HSR-GBM1 cultures with cyclopamine alone and in addition to 10 Gy of radiation. As expected, both radiation and cyclopamine significantly reduced the GBM neurosphere growth rate as monotherapies (Fig. 4E). However, we observed a significantly lower growth rate when 10 μM cyclopamine was given in combination with 10 Gy of radiation, as compared with radiation alone. Although 5 μM cyclopamine did not significantly reduce the growth rate, a trend in this direction was seen (p = .057, two-sided t-test). These data suggest that Hh blockade may target GBM cells not affected by radiation.
Cyclopamine Targets Stem-Like Glioma Cells and Blocks Tumor Engraftment
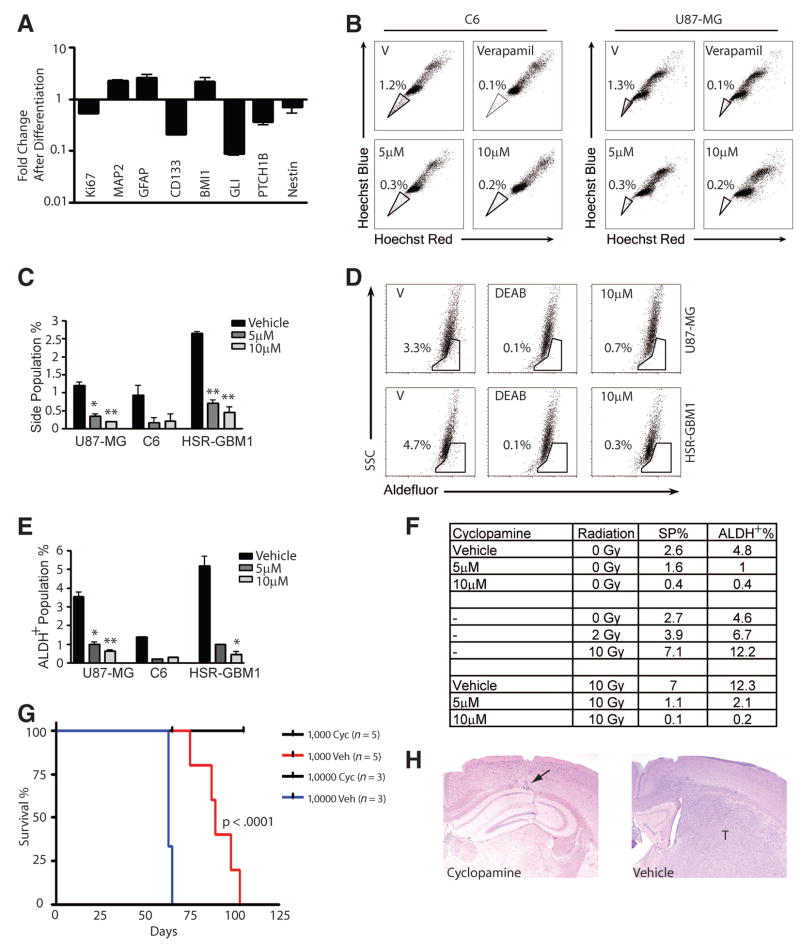
To quantitate the effects of Hh blockade at a single-cell level, we treated GBM neurospheres with 10 μM cyclopamine for 7 days and then used immunofluorescence to examine markers of stem/progenitor cells (nestin) and glial cells (GFAP). This revealed a significant decrease in the percentage of Nestin-positive cells and an increase of more than twofold in glial differentiation (Fig. 4F–4H). We also predicted that if Hh plays an important role in maintaining stem-like cells in glioma neuro-sphere cultures, markers of Hh activity would be reduced when such cultures are forced to differentiate. As previously described, when EGF, fibroblast growth factor, and heparin were removed and 1% serum added, tumor neurospheres adhered to the plastic culture dish and differentiated [34]. A reduction in the expression of the proliferation marker Ki67 was observed as cells differentiated and exited the cell cycle (Fig. 5A). In addition, we observed a 2.6-fold increase in the glial differentiation marker GFAP and a 2.2-fold increase in the neuronal marker microtubule-associated protein 2. Expression of the marker CD133 was reduced more than eightfold (Fig. 5A), indicating a loss of stem cells. As we predicted, the Hh pathway targets Ptch1b and Gli1 were downregulated sixfold and ninefold, respectively, in conjunction with tumor differentiation. Surprisingly, the expression of Bmi1, a gene that is involved in stem cell self-renewal, was increased almost twofold. One possible explanation for this observation is that Bmi1 expression has been reported to be maintained at high levels even in differentiation-inducing conditions [42].
Figure 5.
Cyclopamine depletes stem-like cells from glioblastoma multiforme (GBM) neurospheres and blocks tumor engraftment. (A): When GBM neurospheres were forced to differentiate, mRNA levels of the proliferation marker Ki67 and the neural stem-cell markers CD133 and Nestin decreased, along with the Hedgehog pathway targets Gli1 and Ptch1b. In contrast, markers of glial differentiation (GFAP) and neuronal differentiation (microtubule-associated protein 2) increased. (B, C): The stem-like side population was significantly decreased by cyclopamine (two-sided t test; *, p < .05; **, p < .01). (D, E): The aldehyde dehydrogenase (ALDH)-positive subpopulation of stem/progenitor cells was also depleted by cyclopamine. (F): The side- and ALDH-positive stem/progenitor cell subpopulations were depleted by cyclopamine and increased by radiation when each was given alone. Cotreatment with cyclopamine and radiation depleted side- and ALDH-positive subpopulations. (G, H): HSR-GBM1 cultures pretreated with vehicle always engrafted when either 1,000 or 10,000 viable cells counted at the end of treatment were injected into the athymic mice. In contrast, when the same number of viable cells was injected after 7 days of treatment with 10 μM cyclopamine, no detectable tumors formed (the arrow points to degenerative changes along the injection needle tract). Log-rank analysis of Kaplan-Meier survival curves indicates that the prolongation of survival associated with cyclopamine pretreatment is significant (p < .0001). Abbreviations: ALDH, aldehyde dehydrogenase; Cyc, cyclopamine; DEAB, 4-(diethylamino)benzaldehyde; GFAP, glial fibrillary acidic protein; SP, side population; SSC, side scatter; T, t test; V, vehicle; Veh, vehicle.
A more direct assessment of the effect of Hh on glioma stem cells was performed using flow cytometric analysis of side population, which represents a small fraction of cells within tumors expressing ATP-binding cassette (ABC)-type transporters and able to efflux both Hoechst dye and chemotherapeutic agents [43–45]. Side population has been shown to define a small population of stem-like cells in several established glioma cells lines, including U87-MG [43–45] and C6 [31]. In C6 glioma cultures, it was shown that only prospectively sorted side population cells are able to form tumor xenografts [31]. To confirm that cyclopamine is targeting glioma stem cells, we analyzed its effect on side population. In our experiments, side population represented approximately 1% of the overall cellularity in both the U87-MG and C6 lines (Fig. 5B, 5C). Verapamil treatment totally ablated these cells, confirming that we were measuring a bona fide side population. Cyclopamine treatment dramatically diminished the side population percentage, from 1.2% to 0.3% (5 μM) and 0.2% (10 μM) in C6 cells and from 1.3% to 0.3% (5 μM) and 0.2% (10 μM) in U87-MG cells. The HSR-GBM1 neurosphere line also had a small side population (2.6%) that was reduced to 0.6% and 0.3% by 5 and 10 μM cyclopamine, respectively. All experiments were repeated at least twice, on different days, with means reported above. Interestingly, cyclopamine also depleted the population of tumor cells expressing ALDH, which has been shown to mark both hematopoietic and neural stem cells [46, 47]. This expression is assessed using the fluorescent marker Aldefluor. Cyclopamine treatment dramatically diminished the Aldefluor-positive population of U87-MG cells from 3.3% to 0.9% (5 μM) and 0.7% (10 μM), in C6 cells from 1.4% to 0.2% (5 μM) and 0.3% (10 μM), and in HSR-GBM1 cells from 5.7% to 1.0% (5 μM) and 0.6% (10 μM) (Fig. 5D, 5E).
It has recently been reported by Bao et al. that radiation treatment does not effectively target stem/progenitor cells, as indicated by increased CD133-positive cell populations following radiation exposure [41]. Radiation alone also dramatically increased the percentages of side and ALDH-positive populations in our GBM neurospheres, supporting the concept that this standard therapy is not able to kill cancer stem cells (Fig. 5F). To test the ability of Hh blockade to augment radiation therapy, we treated HSR-GBM1 cells with cyclopamine for a total of 7 days. Four days after the beginning of cyclopamine treatment, we irradiated the cells with various doses. Side population and ALDH-positive population percentages were determined on the seventh day. When given alone, cyclopamine treatment reduced the side and ALDH-positive populations from 2.6% and 4.8% to 1.6% and 1.0% at 5 μM cyclopamine and to 0.4% and 0.4% at 10 μM cyclopamine (Fig. 5F). Strikingly, in cultures treated with 10 Gy radiation and cyclopamine, side and ALDH-positive populations were dramatically reduced from 7.0% and 12.3% to 1.1% and 2.1% at 5 μM cyclopamine and to 0.1% and 0.2% at 10 μM cyclopamine.
Collectively, these data suggest that cyclopamine targets glioma stem cells and might therefore prevent tumor engraftment and long-term growth. To test this, HSR-GBM1 cells were treated with vehicle or cyclopamine for 7 days in culture before intracerebral implantation of either 1 × 103 (n = 5 animals per group) or 1 × 104 (n = 3 animals per group) viable cells. Pretreatment with cyclopamine completely inhibited tumor engraftment and growth, as all animals survived for the duration of the experiment, and microscopic analysis of the injection site failed to identify any tumor deposits (Fig. 5G, 5H). In contrast, massive infiltrative gliomas were present in all mice implanted with vehicle-treated cells, resulting in death as early as 2 months following injection (Fig. 5G).
Discussion
Because of their diffuse infiltration of the brain, complete surgical removal of malignant gliomas is generally not possible. Additional therapies will therefore be required to achieve a better outcome in patients affected by these aggressive tumors. Unfortunately, although a transient response to radiation is seen in most patients, and some chemotherapeutic agents also appear to reduce tumor mass, essentially all malignant gliomas, particularly GBM, rapidly recur. It has been postulated that the recurrence of GBM following current therapies is due to the persistent presence of a small group of stem-like cancer cells within tumors that are relatively resistant to cell death (reviewed in [48]). One possible mechanism for this resistance is the expression of ABC-type transporters in these cancer stem cells, which define a side population that can efflux chemotherapeutic agents. Significantly, it has been shown by two groups that small side populations are present in established glioma cell lines such as C6, U87-MG, and U373 and that the side population within the C6 and U373 lines appears to be uniquely capable of forming tumor xenografts [31, 49].
Our data suggest that the survival of stem-like side population cells in glioblastoma is dependent on ongoing Hh pathway activity. We first demonstrated that the components required to transduce the Hh signal are present in the majority of GBM samples and that mRNA expression of the pathway target Gli1 is relatively high in approximately one-quarter of cases. Forced differentiation of GBM neurospheres reduced both stem cell and Hh activity markers. Hh pathway blockade by cyclopamine reduced growth in glioma lines with elevated pathway activity but not in those with lower levels of Gli1, suggesting that when elevated Hh signaling is detected, it is required for ongoing tumor growth. Reduction of Gli levels using siRNA also reduced the viability and growth of glioma cells. Most significantly, cyclopamine therapy dramatically reduced the side and Aldefluor-positive populations present in GBM cells, resulting in cultures no longer able to form colonies in vitro or xenografts in vivo. We also show that the effects of cyclopamine and radiation on stem-like glioma cells are quite different, with only the former able to deplete this clinical critical population.
Our findings also point to expression of Shh ligand by GBM cells as a mechanism for Hh pathway activation. We detected Shh mRNA in many primary tumor samples, and the highest level of Shh expression was found in the tumor with the most elevated Gli1 level. In mRNA extracted from the primary tumor samples, it is not possible to determine whether the ligand is generated by tumor cells, entrapped non-neoplastic glia and neurons, or stromal elements such as vessels. It is therefore significant that we found that GBM cells grown in culture as neurospheres secreted even larger amounts of active Shh into their media than 3T3 cells genetically engineered to express the ligand. This suggests that some gliomas are capable of secreting their own ligand and are not dependent on exogenous sources.
Clement et al. have recently reported findings similar to ours in several ways [50]. They show that Hh signaling is active in both adherent and neurosphere-based glioma cultures and that pathway inhibition using either cyclopamine or siRNA reduced tumor growth, the self-renewal of glioma stem cells, and in vivo tumorigenicity. Our studies differ in the markers used to evaluate cancer stem cells, as they used CD133, whereas we examined side population and aldehyde dehydrogenase-expressing cells. We also demonstrate for the first time that malignant glioma cells are capable of secreting biologically active Shh ligand, suggesting antibody-mediated ligand depletion as a potential therapy. Also, our results are partially consistent with the report by Ehtesham et al., who found that Hedgehog activity is confined to progenitor cells within gliomas [51]. However, we detected Hedgehog pathway activity in grade IV gliomas, a finding that is in contrast to theirs. This discrepancy may be explained by the fact that gliomas are extremely heterogeneic, so differences in pathway activity may relate to sample location within the tumor. Taken together, our studies provide a strong rationale for testing Hh inhibitors in patients with malignant gliomas.
Acknowledgments
We thank Dr. Duncan Stearns for helping to derive GBM cell lines. Funding for this project was provided by the Brain Tumor Funders Collaborative.
Footnotes
Disclosure of Potential Conflicts of Interest
The authors indicate no potential conflicts of interest.
References
- 1.Holland EC. Gliomagenesis: Genetic alterations and mouse models. Nat Rev Genet. 2001;2:120–129. doi: 10.1038/35052535. [DOI] [PubMed] [Google Scholar]
- 2.Maher EA, Furnari FB, Bachoo RM, et al. Malignant glioma: Genetics and biology of a grave matter. Genes Dev. 2001;15:1311–1333. doi: 10.1101/gad.891601. [DOI] [PubMed] [Google Scholar]
- 3.Shih AH, Holland EC. Notch signaling enhances nestin expression in gliomas. Neoplasia. 2006;8:1072–1082. doi: 10.1593/neo.06526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vescovi AL, Galli R, Reynolds BA. Brain tumour stem cells. Nat Rev Cancer. 2006;6:425–436. doi: 10.1038/nrc1889. [DOI] [PubMed] [Google Scholar]
- 5.Zhu Y, Guignard F, Zhao D, et al. Early inactivation of p53 tumor suppressor gene cooperating with NF1 loss induces malignant astrocytoma. Cancer Cell. 2005;8:119–130. doi: 10.1016/j.ccr.2005.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dahmane N, Ruiz i Altaba A. Sonic hedgehog regulates the growth and patterning of the cerebellum. Development. 1999;126:3089–3100. doi: 10.1242/dev.126.14.3089. [DOI] [PubMed] [Google Scholar]
- 7.Wallace VA. Purkinje-cell-derived Sonic hedgehog regulates granule neuron precursor cell proliferation in the developing mouse cerebellum. Curr Biol. 1999;9:445–448. doi: 10.1016/s0960-9822(99)80195-x. [DOI] [PubMed] [Google Scholar]
- 8.Wechsler-Reya RJ, Scott MP. Control of neuronal precursor proliferation in the cerebellum by Sonic Hedgehog. Neuron. 1999;22:103–114. doi: 10.1016/s0896-6273(00)80682-0. [DOI] [PubMed] [Google Scholar]
- 9.Dahmane N, Sanchez P, Gitton Y, et al. The Sonic Hedgehog-Gli pathway regulates dorsal brain growth and tumorigenesis. Development. 2001;128:5201–5212. doi: 10.1242/dev.128.24.5201. [DOI] [PubMed] [Google Scholar]
- 10.Ahn S, Joyner AL. In vivo analysis of quiescent adult neural stem cells responding to Sonic hedgehog. Nature. 2005;437:894–897. doi: 10.1038/nature03994. [DOI] [PubMed] [Google Scholar]
- 11.Kinzler KW, Bigner SH, Bigner DD, et al. Identification of an amplified, highly expressed gene in a human glioma. Science. 1987;236:70–73. doi: 10.1126/science.3563490. [DOI] [PubMed] [Google Scholar]
- 12.Katayam M, Yoshida K, Ishimori H, et al. Patched and smoothened mRNA expression in human astrocytic tumors inversely correlates with histological malignancy. J Neurooncol. 2002;59:107–115. doi: 10.1023/a:1019660421216. [DOI] [PubMed] [Google Scholar]
- 13.Hooper JE, Scott MP. Communicating with Hedgehogs. Nat Rev Mol Cell Biol. 2005;6:306–317. doi: 10.1038/nrm1622. [DOI] [PubMed] [Google Scholar]
- 14.Lum L, Beachy PA. The Hedgehog response network: Sensors, switches, and routers. Science. 2004;304:1755–1759. doi: 10.1126/science.1098020. [DOI] [PubMed] [Google Scholar]
- 15.Mullor JL, Sanchez P, Altaba AR. Pathways and consequences: Hedgehog signaling in human disease. Trends Cell Biol. 2002;12:562–569. doi: 10.1016/s0962-8924(02)02405-4. [DOI] [PubMed] [Google Scholar]
- 16.Ruiz i Altaba A. Gli proteins and Hedgehog signaling: Development and cancer. Trends Genet. 1999;15:418–425. doi: 10.1016/s0168-9525(99)01840-5. [DOI] [PubMed] [Google Scholar]
- 17.Berman DM, Karhadkar SS, Hallahan AR, et al. Medulloblastoma growth inhibition by hedgehog pathway blockade. Science. 2002;297:1559–1561. doi: 10.1126/science.1073733. [DOI] [PubMed] [Google Scholar]
- 18.Berman DM, Karhadkar SS, Maitra A, et al. Widespread requirement for Hedgehog ligand stimulation in growth of digestive tract tumours. Nature. 2003;425:846–851. doi: 10.1038/nature01972. [DOI] [PubMed] [Google Scholar]
- 19.Karhadkar SS, Bova GS, Abdallah N, et al. Hedgehog signalling in prostate regeneration, neoplasia and metastasis. Nature. 2004;431:707–712. doi: 10.1038/nature02962. [DOI] [PubMed] [Google Scholar]
- 20.Thayer SP, di Magliano MP, Heiser PW, et al. Hedgehog is an early and late mediator of pancreatic cancer tumorigenesis. Nature. 2003;425:851–856. doi: 10.1038/nature02009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Watkins DN, Berman DM, Burkholder SG, et al. Hedgehog signalling within airway epithelial progenitors and in small-cell lung cancer. Nature. 2003;422:313–317. doi: 10.1038/nature01493. [DOI] [PubMed] [Google Scholar]
- 22.Al-Hajj M, Becker MW, Wicha M, et al. Therapeutic implications of cancer stem cells. Curr Opin Genet Dev. 2004;14:43–47. doi: 10.1016/j.gde.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 23.Ehtesham M, Stevenson CB, Thompson RC. Stem cell therapies for malignant glioma. Neurosurg Focus. 2005;19:E5. doi: 10.3171/foc.2005.19.3.6. [DOI] [PubMed] [Google Scholar]
- 24.Lam JS, Reiter RE. Stem cells in prostate and prostate cancer development. Urol Oncol. 2006;24:131–140. doi: 10.1016/j.urolonc.2005.11.038. [DOI] [PubMed] [Google Scholar]
- 25.Perez-Caro M, Sanchez-Garcia I. Killing time for cancer stem cells (CSC): Discovery and development of selective CSC inhibitors. Curr Med Chem. 2006;13:1719–1725. doi: 10.2174/092986706777452533. [DOI] [PubMed] [Google Scholar]
- 26.Schulenburg A, Ulrich-Pur H, Thurnher D, et al. Neoplastic stem cells: A novel therapeutic target in clinical oncology. Cancer. 2006;107:2512–2520. doi: 10.1002/cncr.22277. [DOI] [PubMed] [Google Scholar]
- 27.Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med. 1997;3:730–737. doi: 10.1038/nm0797-730. [DOI] [PubMed] [Google Scholar]
- 28.Reya T, Morrison SJ, Clarke MF, et al. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 29.Florek M, Haase M, Marzesco AM, et al. Prominin-1/CD133, a neural and hematopoietic stem cell marker, is expressed in adult human differentiated cells and certain types of kidney cancer. Cell Tissue Res. 2005;319:15–26. doi: 10.1007/s00441-004-1018-z. [DOI] [PubMed] [Google Scholar]
- 30.Kobari L, Giarratana MC, Pflumio F, et al. CD133+ cell selection is an alternative to CD34+ cell selection for ex vivo expansion of hematopoietic stem cells. J Hematother Stem Cell Res. 2001;10:273–281. doi: 10.1089/15258160151134980. [DOI] [PubMed] [Google Scholar]
- 31.Kondo T, Setoguchi T, Taga T. Persistence of a small subpopulation of cancer stem-like cells in the C6 glioma cell line. Proc Natl Acad Sci U S A. 2004;101:781–786. doi: 10.1073/pnas.0307618100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Richardson GD, Robson CN, Lang SH, et al. CD133, a novel marker for human prostatic epithelial stem cells. J Cell Sci. 2004;117:3539–3545. doi: 10.1242/jcs.01222. [DOI] [PubMed] [Google Scholar]
- 33.Singh SK, Hawkins C, Clarke ID, et al. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 34.Galli R, Binda E, Orfanelli U, et al. Isolation and characterization of tumorigenic, stem-like neural precursors from human glioblastoma. Cancer Res. 2004;64:7011–7021. doi: 10.1158/0008-5472.CAN-04-1364. [DOI] [PubMed] [Google Scholar]
- 35.Dunaeva M, Michelson P, Kogerman P, et al. Characterization of the physical interaction of Gli proteins with SUFU proteins. J Biol Chem. 2003;278:5116–5122. doi: 10.1074/jbc.M209492200. [DOI] [PubMed] [Google Scholar]
- 36.Ingham PW, McMahon AP. Hedgehog signaling in animal development: Paradigms and principles. Genes Dev. 2001;15:3059–3087. doi: 10.1101/gad.938601. [DOI] [PubMed] [Google Scholar]
- 37.Karlstrom RO, Tyurina OV, Kawakami A, et al. Genetic analysis of zebrafish gli1 and gli2 reveals divergent requirements for gli genes in vertebrate development. Development. 2003;130:1549–1564. doi: 10.1242/dev.00364. [DOI] [PubMed] [Google Scholar]
- 38.Purow BW, Haque RM, Noel MW, et al. Expression of Notch-1 and its ligands, Delta-like-1 and Jagged-1, is critical for glioma cell survival and proliferation. Cancer Res. 2005;65:2353–2363. doi: 10.1158/0008-5472.CAN-04-1890. [DOI] [PubMed] [Google Scholar]
- 39.Lee J, Kotliarova S, Kotliarov Y, et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell. 2006;9:391–403. doi: 10.1016/j.ccr.2006.03.030. [DOI] [PubMed] [Google Scholar]
- 40.Chen JK, Taipale J, Young KE, et al. Small molecule modulation of Smoothened activity. Proc Natl Acad Sci U S A. 2002;99:14071–14076. doi: 10.1073/pnas.182542899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bao S, Wu Q, McLendon RE, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444:756–760. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- 42.Hemmati HD, Nakano I, Lazareff JA, et al. Cancerous stem cells can arise from pediatric brain tumors. Proc Natl Acad Sci U S A. 2003;100:15178–15183. doi: 10.1073/pnas.2036535100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Leemhuis T, Yoder MC, Grigsby S, et al. Isolation of primitive human bone marrow hematopoietic progenitor cells using Hoechst 33342 and Rhodamine 123. Exp Hematol. 1996;24:1215–1224. [PubMed] [Google Scholar]
- 44.McAlister I, Wolf NS, Pietrzyk ME, et al. Transplantation of hematopoietic stem cells obtained by a combined dye method fractionation of murine bone marrow. Blood. 1990;75:1240–1246. [PubMed] [Google Scholar]
- 45.Wolf NS, Kone A, Priestley GV, et al. In vivo and in vitro characterization of long-term repopulating primitive hematopoietic cells isolated by sequential Hoechst 33342-rhodamine 123 FACS selection. Exp Hematol. 1993;21:614–622. [PubMed] [Google Scholar]
- 46.Armstrong L, Stojkovic M, Dimmick I, et al. Phenotypic characterization of murine primitive hematopoietic progenitor cells isolated on basis of aldehyde dehydrogenase activity. Stem Cells. 2004;22:1142–1151. doi: 10.1634/stemcells.2004-0170. [DOI] [PubMed] [Google Scholar]
- 47.Corti S, Locatelli F, Papadimitriou D, et al. Identification of a primitive brain-derived neural stem cell population based on aldehyde dehydrogenase activity. Stem Cells. 2006;24:975–985. doi: 10.1634/stemcells.2005-0217. [DOI] [PubMed] [Google Scholar]
- 48.Dean M, Fojo T, Bates S. Tumour stem cells and drug resistance. Nat Rev Cancer. 2005;5:275–284. doi: 10.1038/nrc1590. [DOI] [PubMed] [Google Scholar]
- 49.Patrawala L, Calhoun T, Schneider-Broussard R, et al. Side population is enriched in tumorigenic, stem-like cancer cells, whereas ABCG2+ and ABCG2- cancer cells are similarly tumorigenic. Cancer Res. 2005;65:6207–6219. doi: 10.1158/0008-5472.CAN-05-0592. [DOI] [PubMed] [Google Scholar]
- 50.Clement V, Sanchez P, de Tribolet N, et al. HEDGEHOG-GLI1 signaling regulates human glioma growth, cancer stem cell self-renewal, and tumorigenicity. Curr Biol. 2007;17:165–172. doi: 10.1016/j.cub.2006.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ehtesham M, Sarangi A, Valadez JG, et al. Ligand-dependent activation of the hedgehog pathway in glioma progenitor cells. Oncogene. 2007 doi: 10.1038/sj.onc.1210359. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 52.Bar EE, Chaudhry A, Farah MH, et al. Hedgehog signaling promotes medulloblastoma survival via Bc/II. Am J Pathol. 2007;170:347–355. doi: 10.2353/ajpath.2007.060066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kleinschmidt-DeMasters BK, Orr EA, Savelieva E, et al. Paucity of retinoic acid receptor alpha (RAR alpha) nuclear immunostaining in gliomas and inability of retinoic acid to influence neural cell adhesion molecule (NCAM) expression. J Neurooncol. 1999;41:31–42. doi: 10.1023/a:1006162211296. [DOI] [PubMed] [Google Scholar]
- 54.Vescovi AL, Parati EA, Gritti A, et al. Isolation and cloning of multipotential stem cells from the embryonic human CNS and establishment of transplantable human neural stem cell lines by epigenetic stimulation. Exp Neurol. 1999;156:71–83. doi: 10.1006/exnr.1998.6998. [DOI] [PubMed] [Google Scholar]
- 55.Taipale J, Chen JK, Cooper MK, et al. Effects of oncogenic mutations in Smoothened and Patched can be reversed by cyclopamine. Nature. 2000;406:1005–1009. doi: 10.1038/35023008. [DOI] [PubMed] [Google Scholar]
- 56.Stewart SA, Dykxhoorn DM, Palliser D, et al. Lentivirus-delivered stable gene silencing by RNAi in primary cells. RNA. 2003;9:493–501. doi: 10.1261/rna.2192803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fan X, Matsui W, Khaki L, et al. Notch pathway inhibition depletes stem-like cells and blocks engraftment in embryonal brain tumors. Cancer Res. 2006;66:7445–7452. doi: 10.1158/0008-5472.CAN-06-0858. [DOI] [PubMed] [Google Scholar]
- 58.Stearns D, Chaudhry A, Abel TW, et al. c-myc overexpression causes anaplasia in medulloblastoma. Cancer Res. 2006;66:673–681. doi: 10.1158/0008-5472.CAN-05-1580. [DOI] [PubMed] [Google Scholar]