Abstract
The present study focuses on potential agents of chromoblastomycosis and other endemic diseases in the state of Paraná, Southern Brazil. Using a highly selective protocol for chaetothyrialean black yeasts and relatives, environmental samples from the living area of symptomatic patients were analysed. Additional strains were isolated from creosote-treated wood and hydrocarbon-polluted environments, as such polluted sites have been supposed to enhance black yeast prevalence. Isolates showed morphologies compatible with the traditional etiological agents of chromoblastomycosis, e.g. Fonsecaea pedrosoi and Phialophora verrucosa, and of agents of subcutaneous or systemic infections like Cladophialophora bantiana and Exophiala jeanselmei. Some agents of mild disease were indeed encountered. However, molecular analysis proved that most environmental strains differed from known etiologic agents of pronounced disease syndromes: they belonged to the same order, but mostly were undescribed species. Agents of chromoblastomycosis and systemic disease thus far are prevalent on the human host. The hydrocarbon-polluted environments yielded yet another spectrum of chaetothyrialean fungi. These observations are of great relevance because they allow us to distinguish between categories of opportunists, indicating possible differences in pathogenicity and virulence.
Keywords: Black yeasts, Chaetothyriales, chromoblastomycosis, enrichment, environmental isolation, opportunists, phaeohyphomycosis, virulence
INTRODUCTION
Knowledge of natural ecology and evolution is essential for a better understanding of pathogenicity and opportunism. Members of different fungal orders and families tend to be differentially involved in human mycoses. Among melanised fungi, for example, etiologies of members of Dothideaceae and Herpotrichiellaceae show basic differences (de Hoog 1993, 1997). Of these families, only species belonging to the Herpotrichiellaceae (black yeasts and relatives) are associated with recurrent, clearly defined disease entities such as chromoblastomycosis and neurotropic dissemination in immunocompetent individuals. In contrast, members of Dothideaceae show coincidental opportunism, whereby the infection is largely dependent on the portal of entry and the immune status of the host.
Among the diseases caused by chaetothyrialean fungi (teleomorph family Herpotrichiellaceae), chromoblastomycosis and other traumatic skin disorders are the most frequent (Attili et al. 1998, Zeng et al. 2007). Although the agents are supposed to originate from the environment, their isolation from nature is difficult. This is probably due to their oligotrophic nature, low competitive ability, and in general insufficient data on their natural habitat. Several selective techniques have been developed enabling recovery of these fungi (de Hoog et al. 2005; Dixon et al. 1980, Prenafeta-Boldú et al. 2006, Satow et al. 2008, Zhao et al. 2008, Sudhadham et al. 2008). These investigations indicated that opportunism of these fungi must be explained from the perspective of unexpected environments such as rock, creosote-treated wood, hydrocarbon-polluted soil, and hyperparasitism of fungi and lichens (Sterflinger et al. 1999, Wang & Zabel 1997, Lutzoni et al. 2001).
In the present study we tried to find recover chaetothyrialean fungi from the natural environment in the State of Paraná, Southern Brazil, where chromoblastomycosis and phaeohyphomycosis are frequent in endemic areas. In addition, human-made substrates like creosote-treated wood and hydrocarbon-polluted soil were sampled. Strains morphologically similar to etiological agents of chromoblastomycosis, such as Fonsecaea pedrosoi and Phialophora verrucosa, and to agents of subcutaneous and systemic infections, such as Exophiala jeanselmei and Cladophialophora bantiana, were selected. The aim of this investigation was to clarify whether these fungi were identical to known etiologic agents of disease. Isolates were compared with clinical reference strains on the basis morphological, physiological and molecular parameters.
MATERIALS AND METHODS
Study area and strains
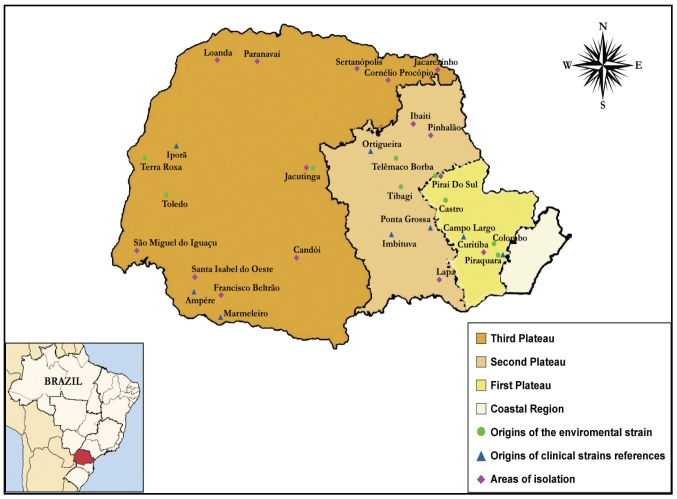
Samples were obtained from 28 localities belonging to three different geographical regions in the State of Paraná, Southern Brazil (Fig. 1). Locations were chosen on the basis of known records of chromoblastomycosis in the “Hospital de Clínicas” of the Paraná Federal University (HC-UFPR). The climate in the region is subtropical, with relatively regular rainfall throughout the yr (average 12500 mm/yr). The isolates obtained were compared to clinical isolates from the hospital of the Universidade Federal do Paraná, as well as to reference strains from the Centraalbureau voor Schimmelcultures (CBS, The Netherlands; Table 1). Samples from Eucalyptus wood in the “Navarro de Andrade” forest and creosote-treated railway ties near Rio Claro, and from hydrocarbon-polluted soil from the oil refinery of Paulínia, Paulínia state of São Paulo, Brazil, were also investigated (Table 1).
Fig. 1.
Sampling locations in the State of Paraná, S.P., Brazil.
Table 1.
Isolates from clinical and environmental sources in southern Brazil.
| Morphological ID | Final ITS ID | CBS | dH | Vicente / Attili | Origin | Source |
|---|---|---|---|---|---|---|
| Fonsecaea pedrosoi | Fonsecaea pedrosoi | 18223 | Fp28III | Marmeleiro | Chromoblastomycosis | |
| Fonsecaea pedrosoi | Fonsecaea pedrosoi | 18331 | Queiros | São Paulo | Subcutaneous, compromised | |
| Fonsecaea pedrosoi | Fonsecaea pedrosoi | 102244 | 11608 | Fp37 | Ipora | Chromoblastomycosis |
| Fonsecaea pedrosoi | Fonsecaea pedrosoi | 102245 | 11610 | Fp36I | Ampere | Chromoblastomycosis |
| Fonsecaea pedrosoi | Fonsecaea monophora | 102248 | 11613 | Fp82 | Piraquara | Chromoblastomycosis |
| Fonsecaea pedrosoi | Fonsecaea monophora | 102246 | 11611 | Fp65 | Campo Largo | Chromoblastomycosis |
| Fonsecaea pedrosoi | Fonsecaea monophora | 102243 | 11607 | Fp31I | Ibituva | Chromoblastomycosis |
| Cladophialophora bantiana1 | Cladophialophora immunda | 102227 | 11588 | FE9 = 9EMB | Colombo | Stem palm (Syagrum roman zoffianum) |
| Cladophialophora bantiana | Cladophialophora sp. 3 | 102231 | 11592 | FE1IIA | Colombo | Rotten Gochnatia polymorpha stem |
| Cladophialophora devriesii1 | Cladophialophora saturnica4 | 102230 | 11591 | FP4IIB | Piraquara | Plant litter |
| Cladophialophora sp. | Cladophialophora saturnica | 102228 | 11589 | FP4IIA | Piraquara | Rotten wood |
| Cladophialophora sp. | Fonsecaea monophora5 | 102229 | 11590 | FP8D = 8DPIRA | Piraquara | Plant litter |
| Cladophialophora sp. | Cladophialophora sp. 2 | 102236 | 11600 | F11PLA | Telêmaco Borba | Plant litter |
| Cladophialophora sp. | Cladophialophora immunda | 102237 | 11601 | F10PLA | Telêmaco Borba | Plant litter |
| Exophiala lecanii-corni2 | Exophiala xenobiotica4 | 102232 | 11594 | FE4IIB | Colombo | Rotten wood |
| Exophiala jeanselmei2 | Exophiala bergeri4 | 102241 | 11605 | F14PL | Cianorte | Soil under coffee tree |
| Exophiala sp. | Capronia semi-immersa | 102233 | 11595 | FE6IIB | Colombo | Rotten Araucaria trunk |
| Fonsecaea pedrosoi3 | Fonsecaea sp. 3 | 102223 | 11583 | FCL2 | Castro | Rotten root |
| Fonsecaea pedrosoi | Fonsecaea sp. 1 | 102224 | 11584 | F9PRA | Terra Roxa | Grevillea robusta wood |
| Fonsecaea pedrosoi | Fonsecaea monophora5 | 102225 | 11585 | FE5P4 | Colombo | Rotten wood (Gochnatia polymorpha) |
| Fonsecaea pedrosoi | Fonsecaea sp. 1 | 102254 | 11619 | FE5P6 | Colombo | Rotten wood |
| Fonsecaea pedrosoi | Fonsecaea sp. 2 | 102226 | 11587 | FE5II | Colombo | Rotten Araucaria trunk |
| Phialophora verrucosa3 | Phialophora sp. 1 | 102234 | 11596 | FE3 | Colombo | Lantana camara rhizosphere |
| Rhinocladiella sp. | Rhinocladiella sp. 1 | 102235 | 11597 | F9PR | Terra Roxa | Grevillea robusta wood |
| Rhinocladiella sp. | Cladophialophora immunda | 102249 | 11614 | F10PLB | Sarandi | Rotten Cinnamomum trunk |
| Rhinocladiella sp. | Cladophialophora chaetospira | 102250 | 11615 | F3PLB | Sertanópolis | Plant litter |
| Rhinocladiella sp. | Exophiala xenobiotica4 | 102251 | 11616 | F3PLC | Sertanópolis | Plant litter |
| Rhinocladiella sp. | Fonsecaea sp. 3 | 102239 | 11603 | FE1IIB | Colombo | Rotten Lantana camara stem |
| Rhinocladiella sp. | Fonsecaea sp. 3 | 102252 | 11617 | FE10IIB | Colombo | Plant litter |
| Rhinocladiella sp. | Rhinocladiella sp. 1 | 11618 | FE10IIB1 | Colombo | Plant litter | |
| Rhinocladiella sp. | Rhinocladiella sp. 1 | 102240 | 11604 | F9PRC | Terra Roxa | Podocarpos lambertii branch |
| Rhinocladiella sp. | Exophiala xenobiotica4 | 102255 | 11621 | F20PR3 | Jacutinga | Soil |
| Rhinocladiella sp. | Fonsecaea monophora5 | 102238 | 11602 | F1PLE | Rio Tibagi | Soil |
| Exophiala sp. | Exophiala bergeri4 | 122844 | 18627 | D0009 | Rio Claro | Railway tie treated with creosote 15 yr ago |
| Exophiala sp. | Exophiala bergeri4 | 122843 | 18629 | D0020 | Rio Claro | Railway tie treated with creosote 15 yr ago |
| Exophiala sp. | Exophiala bergeri4 | 122842 | 18636 | D0035 | Rio Claro | Railway tie treated with creosote 15 yr ago |
| Exophiala sp. | Exophiala bergeri4 | 122841 | 18643 | D0201 | Rio Claro | Railway tie treated with creosote 15 yr ago |
| Exophiala sp. | Exophiala bergeri4 | 122840 | 18654 | D0213 | Rio Claro | Eucalyptus wood |
| Exophiala sp. | Exophiala dermatitidis6 | 122839 | 18635 | D0029b | Rio Claro | Eucalyptus wood |
| Exophiala sp. | Exophiala dermatitidis6 | 122838 | 18646 | D0204a | Rio Claro | Eucalyptus wood |
| Exophiala sp. | Exophiala dermatitidis6 | 122837 | 18651 | D0210 | Rio Claro | Eucalyptus wood |
| Exophiala sp. | Exophiala dermatitidis6 | 122836 | 18648 | D0206 | Rio Claro | Eucalyptus wood |
| Exophiala sp. | Exophiala dermatitidis6 | 122835 | 18652 | D0211 | Rio Clarov | Eucalyptus wood |
| Exophiala sp. | Exophiala dermatitidis6 | 122834 | 18653 | D0212 | Rio Claro | Eucalyptus wood |
| Exophiala sp. | Exophiala dermatitidis6 | 122833 | 18656 | D0215 | Rio Claro | Eucalyptus wood |
| Exophiala sp. | Exophiala dermatitidis (mel- mut) | 122830 | 18650 | D0209 | Rio Claro | Railway tie treated with creosote 16 yr ago |
| Exophiala sp. | Exophiala xenobiotica4 | 122832 | 18647 | D0205 | Rio Claro | Eucalyptus wood |
| Exophiala sp. | Exophiala xenobiotica4 | 122910 | 18638 | D0044 | Rio Claro | Eucalyptus wood |
| Exophiala sp. | Exophiala xenobiotica4 | 122831 | 18655 | D0214 | Rio Claro | Eucalyptus wood |
| Exophiala sp. | Exophiala xenobiotica4 | 122829 | 18631 | D0023 | Rio Claro | Railway tie treated with creosote 15 yr ago |
| Exophiala sp. | Exophiala xenobiotica4 | 122846 | 18632 | D0024 | Rio Claro | Railway tie treated with creosote 15 yr ago |
| Exophiala sp. | Exophiala xenobiotica4 | 122828 | 18637 | D0037 | Rio Claro | Railway tie treated with creosote 15 yr ago |
| Black fungus sp. | Veronaea botryosa4 | 122826 | 18639 | D0045 | Rio Claro | Railway tie treated with creosote 20 yr ago |
| Black fungus sp. | Veronaea botryosa4 | 122824 | 18640 | D0047 | Rio Claro | Railway tie treated with creosote 20 yr ago |
| Black fungus sp. | Veronaea botryosa4 | 122822 | 18641 | D0060 | Rio Claro | Railway tie treated with creosote 20 yr ago |
| Black fungus sp. | Veronaea botryosa4 | 122825 | 18642 | D0063 | Rio Claro | Railway tie treated with creosote 20 yr ago |
| Black fungus sp. | Veronaea botryosa4 | 122823 | 18628 | D0017 | Rio Claro | Railway tie treated with creosote 20 yr ago |
| Aureobasidium sp. | Aureobasidium pullulans4 | 122827 | 18657 | D0216 | Paulinia | Polluted soil, Replan Co. |
Abbreviations used: CBS = Centraalbureau voor Schimmelcultures; dH = G.S. de Hoog working collection.
Known agent of: 1systemic and disseminated disease; 2mycetoma; 3chromoblastomycosis; 4mild cutaneous disease; 6systemic disease and pulmonary colonization 5Known opportunistic agent (including chromoblastomycosis).
Fungal isolation
In each location, fragments of litter and decaying wood showing the presence of black spots, as well as soil samples, were randomly collected. Approximately 20 g from each sample were processed for fungal isolation, with 10 replicates per sample. Each sample was incubated at room temperature for 30 min in 100 mL sterile saline solution containing 200 U penicillin, 200 μg/L streptomycin, 200 μg/L chloramphenicol and 500 μg/L cycloheximide (Fig. 2C). After the initial incubation, 20 mL of sterile mineral oil was added to the solution, followed by vigorous 5 min shaking and the flasks were left to settle for 20 min. The oil-water interphase was carefully collected, inoculated onto Mycosel agar (Difco) and incubated for 4 wks at 36 °C (Dixon & Shadomy 1980, Iwatsu et al. 1981). The grown dark colonies were then isolated and stored on Mycosel agar (Fig. 2D).
Fig. 2.
A. Sampling location in the area of “FE” series of samples in the first plateau at Colombo (Paraná) with native species (arrows) dominated by cambara tree (Gochnathia polymorpha) and stem palm (Syagrus romanzoffiana); B. Sampling location of FE5P4 of Fonsecaea monophora, from decaying cambara wood; C. Sample incubated at room temperature in sterile saline solution, containing antibiotics added with sterile mineral oil after vigorous shaking; D. Black yeast colonies on Mycosel medium.
Morphology
Preliminary identification was carried out based on macro- and microscopic features of the colonies after slide culturing on Sabouraud's dextrose agar at room temperature (de Hoog et al. 2000a). In addition, vacuum-dried samples were mounted on carbon tape and sputtered with gold for 180 s for SEM. Observations were done in a Zeiss DSM 940 A microscope, operated at 5 kV.
Nutritional physiology
Some isolates with cultural and morphological similarity to known agents of disease were selected for physiological testing. Growth and fermentative abilities were tested in duplicate, negative controls were added. The fungi were incubated at 28 and 36 °C on the following culture media: Mycosel, Potato Dextrose Agar (PDA), Minimal Medium (MM), Complete Medium (CM), and Malt Extract Agar (MEA). Assimilation and fermentation tests were carried out in liquid medium according to de Hoog et al. (1995,1995). Halotolerance was tested in a liquid medium at 2.5, 5 and 10 % (w/v) NaCl and MgCl2. Cycloheximide tolerance was determined in liquid medium at 0.01, 0.05 and 0.1 % (w/v).
DNA extraction
About 1 cm2 mycelium of 20 to 30-d-old cultures was transferred to a 2 mL Eppendorf tube containing 300 μL CTAB (cetyltrimethylammonium bromide) buffer [CTAB 2% (w/v), NaCl 1.4 M, Tris-HCl 100 mM, pH 8.0; EDTA 20 mM, b-mercaptoethanol 0.2 % (v/v)] and about 80 mg of a silica mixture (silica gel H, Merck 7736, Darmstadt, Germany / Kieselguhr Celite 545, Machery, Düren, Germany, 2:1, w/w). Cells were disrupted manually with a sterile pestle for approximately 5 min. Subsequently 200 μL CTAB buffer was added, the mixture was vortexed and incubated for 10 min at 65 °C. After addition of 500 μL chloroform, the solution was mixed and centrifuged for 5 min at 20 500 g and the supernatant transferred to a new tube with 2 vols of ice cold 96 % ethanol. DNA was allowed to precipitate for 30 min at –20°C and then centrifuged again for 5 min at 20 500 g. Subsequently the pellet was washed with cold 70 % ethanol. After drying at room temperature it was resuspended in 97.5 μL TE-buffer plus 2.5 μL RNAse 20 U.mL–1 and incubated for 5 min at 37 °C, before storage at –20 °C (Gerrits van den Ende & de Hoog 1999).
Sequencing
rDNA Internal Transcribed Spacer (ITS) was amplified using primers V9G and LS266 (Gerrits van den Ende & de Hoog 1999) and sequenced with ITS1 and ITS4 (White et al. 1990). Amplicons were cleaned with GFX PCR DNA purification kit (GE Healthcare, U.K.). Sequencing was performed on an ABI 3730XL automatic sequencer. Sequences were edited using the Seqman package (DNAStar, Madison, U.S.A.) and aligned using BioNumerics v.4.61 (Applied Maths, Kortrijk, Belgium). Sequences were compared in a research data database of black fungi maintained at CBS, validated by ex-type strains of all known species.
RESULTS
Eighty-one isolates from a total of 540 showed morphologies compatible with the traditional etiological agents of chromoblastomycosis and phaeohyphomycosis. Twenty -six strains were selected and processed for taxonomic studies and listed in Table 1 with additional strains from hydrocarbon-polluted soil and wood (natural and creosote-treated).
Isolate FE9 was morphologically very similar to Cladophialophora bantiana. Physiological testing demonstrated ability to assimilate ethanol, lactose and citrate, but it was unable to grow at 40 °C (Table 2). Sequence data proved identity to C. immunda (Table 1). Strain F10PLB was physiologically similar to FE9 of C. immunda (Table 2), which was confirmed by molecular data (Table 1). F10PLA showed physiological characteristics close to the FE9, differing only by growth in the presence of creatine and creatinine (Table 2); also this strain was identified by ITS sequence data as C. immunda. The isolate FP4IIB was capable of growing with 0.1 % cycloheximide, showed reduced growth in the presence of ethanol and had a maximum growth temperature of 37 °C (Table 2). It presented ellipsoidal to fusiform conidia originating from denticles, consistent with Cladophialophora devriesii. However, molecular data identified the strain as C. saturnica (Table 1). FP4IIA, phenetically identified as Cladophialophora sp. and physiologically similar to FP4IIB was identified as C. saturnica by ITS sequencing (Table 1). FE1IIA and F11PLA had fusiform conidia in chains. FE1IIA was unable to assimilate galacticol, developed poorly in the presence of D-glucoronate, but was able to grow in a medium with ethanol; F11PLA assimilated glucoronate having a weak development in the presence of ethanol (Table 2). With ITS sequencing two undescribed Cladophialophora species appeared to be concerned (Table 1).
Table 2.
Physiological test results of Brazilian isolates.
| FE3 Phialophora sp. 1 | FE9 C. immunda | F10PLA C. immunda | FE1IIA Clad. sp. 3 | FP4IIB C. saturnica | FP4IIA C. saturnica | F10PLB C. immunda | F11PLA Clad. sp. 2 | F14PL E. bergeri | FE4IIB E. xenobiotica | FE6IIB Cap. semiimmersa | F20PR3 E. xenobiotica | F3PLC E. xenobiotica | F9PR Rhin. sp. 1 | F9PRC Rhin. sp. 1 | FE10IIB1 Rhin. sp. 1 | Fp82 F. monophora | Fp65 F. monophora | FE5P4 F. monophora | FP8D F. monophora | F1PLE F. monophora | F9PRA Fonsecaea sp. 1 | FE10IIB Fonsecaea sp. 3 | FE1IIB Fonsecaea sp. 3 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 | 24 | |
| D-Glucose | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | w/+ | + | + |
| D-Galactose | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| L-Sorbose | + | w | + | w | + | + | + | + | + | w | + | - | w | + | w | w | w | + | + | w | w/+ | w | w | w |
| D-Glucosamine | w/+ | w | w | + | w | w | w | w | w/+ | + | w | + | + | w | w/- | - | w | + | w | w | w | w/- | + | + |
| D-Ribose | - | + | + | + | + | + | + | + | + | + | w | + | + | w | - | w/- | w/- | w/- | w/- | w/- | - | - | + | + |
| D-Xylose | + | + | + | + | + | + | + | w | + | + | + | + | + | + | w/+ | + | w/+ | + | + | w/+ | w/+ | + | + | + |
| L-Arabinose | + | + | + | + | + | + | + | + | + | + | + | + | + | + | w/+ | + | + | + | + | + | w/+ | + | + | + |
| D-Arabinose | + | w | + | + | w | w | + | w | + | w/+ | w | + | + | w | - | - | - | + | w | - | w/- | - | + | + |
| L-Rhamnose | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Sucrose | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | w/+ | + | + | w/+ | + | + | + | + |
| Maltose | + | + | + | + | + | + | + | + | + | + | + | + | + | + | w/+ | + | w/+ | + | + | w/+ | w/+ | + | + | + |
| α,α-Trehalose | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Methyl-α-D-Glucoside | w | + | + | - | + | + | + | + | - | - | + | w | - | + | + | + | w/- | + | w | w/- | w | + | - | - |
| Cellobiose | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | w | w/+ | + | w | w/+ | w/+ | + | + |
| Salicin | + | + | + | + | + | + | + | + | w/- | + | + | + | + | + | + | + | + | + | + | + | + | w | + | + |
| Arbutin | + | + | + | + | + | + | + | + | w/+ | + | + | + | + | + | w/+ | + | + | + | + | + | + | + | + | + |
| Melibiose | - | + | + | + | w | w | + | + | - | + | + | + | + | + | w/+ | + | + | + | + | + | w/+ | w/+ | + | + |
| Lactose | - | + | + | - | + | + | + | + | w/- | - | - | - | w | + | + | + | w/- | + | w | w/- | w/- | w/+ | - | - |
| Raffinose | + | + | + | + | + | + | + | + | w | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Melezitose | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Inulin | - | - | - | w | - | - | - | - | - | w/- | w | w | w | - | - | - | - | - | - | - | - | - | w | w |
| Sol. starch | w | w | w | w/+ | - | - | w | - | - | w/- | - | w/+ | w/+ | - | w/- | w/- | w/- | w/- | - | w/- | w/- | w/- | w/+ | w/+ |
| Glycerol | + | + | + | + | + | + | + | w | + | + | w | + | + | w | + | + | w | + | w | w | w | + | + | + |
| meso-Erythritol | + | + | + | + | + | + | + | + | w/- | + | + | + | + | + | + | + | w/- | + | w | w/- | w | + | + | + |
| Ribitol | + | w | + | + | + | + | + | + | + | + | + | + | + | w | w | w/+ | w/- | + | w | w/- | w | w/- | + | + |
| Xylitol | - | + | + | + | + | + | + | + | + | + | w | + | + | + | + | + | - | + | w | - | w/- | + | + | + |
| L-Arabinitol | w | + | + | w/+ | w | w | + | + | + | + | w | w/+ | w/+ | w | w | w/+ | w/+ | + | + | w/+ | + | w/+ | w/+ | w/+ |
| D-Glucitol | + | + | + | + | w | w | + | + | + | + | + | + | + | + | w | w/+ | w/+ | + | + | w/+ | w/+ | w/+ | + | + |
| D-Mannitol | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | w/+ | + | + | + |
| Galactitol | w | + | + | - | w | w | + | w | - | - | w | - | - | w | + | + | w/- | + | w | w/- | w | + | - | - |
| myo-Inositol | + | + | + | + | + | + | + | + | + | + | w | + | + | - | - | + | - | w/+ | + | - | w | - | + | + |
| Glucono-d-Lactone | + | + | + | + | + | + | + | w | w/+ | w/+ | w | + | + | + | w | w | w | + | + | w | w | w | + | + |
| D-Gluconate | + | w | w | w | + | + | w | w | - | w | + | w | - | w | w | w | w | + | + | w | w | w/- | w | w |
| D-Glucuronate | + | w | w | w | + | + | w | + | w | w | w | w | w | w | w | w | w | + | + | w | w/- | w/- | w | w |
| D-Galacturonate | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | w/- | w/- | w | w/- | w | w/+ | + | + |
| DL-Lactate | + | + | + | + | w | w | + | w | + | + | w | + | + | w | + | + | w | - | w | w | w/+ | w | + | + |
| Succinate | + | w | + | + | + | + | + | + | w | w/+ | w | + | + | + | w | w | w | w/+ | w | w | w | w | + | + |
| Citrate | + | w | w | - | w/- | w/- | w | + | - | - | - | - | - | w | w/- | w/- | w | w | w | w | w | w/- | - | - |
| Methanol | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| Ethanol | w/+ | w/+ | + | + | w | w | + | w | + | w/+ | w | + | + | w | + | w/+ | w/+ | + | w | w/+ | w/+ | + | + | + |
| Nitrate | + | + | + | + | + | + | + | + | w/+ | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Nitrite | + | w/- | + | + | w | w | + | w | + | + | + | + | + | w | + | w | + | + | w | + | + | w/+ | + | + |
| Ethylamine | + | + | w/+ | w/+ | w | w | w/+ | + | w/+ | + | w | + | w/+ | w | w/+ | w/+ | w | w | w | w/+ | w | w/+ | w/+ | w/+ |
| L-Lysine | + | w | w/+ | w | - | - | w/+ | w | w/- | w/+ | - | - | w | - | w/+ | w/+ | + | + | w | + | w/+ | w/+ | w | w |
| Cadaverine | + | + | + | + | + | + | + | w | + | + | + | - | + | + | + | + | + | + | + | w/+ | + | + | + | + |
| Creatine | + | + | w | + | - | - | w/+ | w | - | + | w | + | + | w | + | w | w/+ | w/+ | w | + | w/+ | w/+ | + | + |
| Creatinine | + | + | w | + | - | - | w/+ | w/- | - | + | w | + | + | w | + | - | w/+ | w/+ | w | w/+ | w | w | + | + |
| 2.5% MgCl2 | + | + | + | + | + | + | + | w/+ | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| 5.0% MgCl2 | + | + | w/+ | + | w/+ | w/+ | w/+ | w/+ | + | + | + | + | + | + | w/+ | + | + | + | + | + | + | + | + | + |
| 10% MgCl2 | w | + | w/+ | + | w/+ | w/+ | w/+ | w/+ | + | + | + | + | + | w/+ | w/+ | + | w/+ | + | + | w/+ | + | w/+ | + | + |
| 2.5% NaCl | + | + | + | + | + | + | + | w/+ | + | + | + | + | + | + | + | + | w/+ | + | + | w/+ | + | + | + | + |
| 5.0% NaCl | w | + | w | + | w/+ | w/+ | w | w/+ | + | + | + | + | + | + | w/+ | + | - | + | + | - | + | + | + | + |
| 10% NaCl | - | - | - | - | - | - | - | - | + | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| 0.1% Cycloheximide | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| 0.05% Cycloheximide | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| 0.01% Cycloheximide | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Urease | - | - | - | + | - | - | - | - | - | - | - | + | + | w | w/+ | + | + | - | + | + | + | - | + | + |
| 30°C | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| 37°C | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| 40°C | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| Fermentation | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| Acid production | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| Mycosel | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Gelatine | - | - | - | - | w | w | - | w | - | - | w | - | - | w | - | - | - | - | w | - | w | - | - | - |
| DNAse | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| Arbutin | ? | ? | w/+ | w/- | w/+ | w/+ | w/+ | w/+ | - | w/+ | - | w/- | - | - | - | - | - | - | - | - | - | - | w/- | w/- |
Abbreviations used: + = growth; w = weak growth; - = no growth; ? = ambiguous or unknown.
C. = Cladophialophora; Cap = Capronia; E = Exophiala; Phial = Phialophora; Rhin = Rhinocladiella. 1Phialophora sp. 1; 2-8 Cladophialophora species; 9-13 Exophiala species; 14 -16 Rhinocladiella. sp; 17-18 Clinical strain: (m) Fonsecaea monophora and (p) F. pedrosoi; 19-21 (m) Fonsecaea monophora; 22-24 (sp) Fonsecaea sp. (molecular identifications partly according to Najafzadeh et al. 2008)
Strain FE5P4 was isolated from decaying cambara wood (Fig. 2B) in an area of native species (Fig. 2A) dominated by cambara trees (Gochnathia polymorpha) and stem palm (Syagrus romanzoffiana) near Colombo city (Fig. 1). This isolate was morphologically identified as Fonsecaea pedrosoi. Physiologically it differed from F. pedrosoi by assimilation of L-sorbose, melibiose, ribitol, xylitol, myo-inositol, glucono-б-lactone, D- and L-lactate, succinate, nitrite, urease and tolerance to 5% NaCl (Table 2). This physiological profile was similar to that of clinical strains FP65 and FP82 (Table 2) originating from symptomatic patients of the same geographic region (first plateau, Fig. 1). With ITS sequencing FE5P4 was identified as Fonsecaea monophora. Environmental isolate FP8D morphologically was cladophialophora-like but was identified as F. monophora based on molecular data. It had physiological similarity with clinical strain FP82 of F. monophora (Table 2) and was isolated from the same location where the patient, a carrier of chromoblastomycosis, had acquired his infection (Piraquara city, Fig. 1). All strains grew at 37 °C but not at 40 °C, similar to known Fonsecaea species (de Hoog et al. 2004). Isolate F1PLE was recovered from soil, located on the second plateau (Fig. 1). It showed similar morphology to Rhinocladiella but through molecular data it was identified as F. monophora (Table 1). Strains FE5P6, FE5II and FCL2 strains appeared to represent undescribed species of the genus Fonsecaea (Table 1).
In the same region isolate (FE3) was recovered which was morphologically identified as Phialophora verrucosa on the basis of pronounced funnel-shaped collarettes from which the conidia were released. The isolate did not assimilate glucose, ribose and inulin but was capable of L-lysine assimilation (Table 2), a result that is consistent with the physiological characteristics of the P. verrucosa reference strain (de Hoog et al. 1999). With molecular ID a hitherto undescribed Phialophora species was found (Table 1).
Strain FE6IIB, morphologically identified as Exophiala species (Table 1) physiologically differed from reference strains of Exophiala (de Hoog et al. 2000a) by positive responses to lactose, L-arabinose, myo-inositol, D-gluconate and DL-lactate, by being able to assimilate D-ribose, and also presenting weak assimilation of glucoronate. By molecular identification was identified as the anamorph of Capronia semi-immersa. Isolate F14PL was preponderantly yeast-like and was provisionally identified as Exophiala jeanselmei, but ITS sequence data suggested E. bergeri (Table 1). Isolate FE4IIB showed morphological similarity to E. lecanii-corni, but differed from reference strains (de Hoog et al. 2000a) by positive assimilation of lactose, L-arabinose, myo-inositol, D-glucuronate and DL-lactate and by being able to assimilate D-ribose and D-glucoronate. Molecular data suggested identity with E. xenobiotica.
Isolates F20PR3 and F3PLC, morphologically having the appearance of Rhinocladiella species were identified as Exophiala xenobiotica by ITS data. These strains were unable to ferment glucose, to assimilate methanol, to grow at 40 °C and were citrate negative. None of the strains analysed produced extracellular DNAse. Rhinocladiella-like strains F9PR and F9PRC were physiologically similar, differing only in assimilation of glycerol and L-lysine (Table 2). Using molecular data, they were identified as an undescribed Rhinocladiella species (Table 1). Strains FE10IIB and FE10IIB1 were initially thought to be Rhinocladiella- or fonsecaea-like species. FE10IIB1 did not assimilate inulin and was physiologically similar to Rhinocladiella atrovirens (CBS 264.49 and CBS 380.59). No close molecular match was found for either of these strains (Table 1).
DISCUSSION
Chaetothyrialean black yeasts and relatives are interesting microorganisms from ecological as well as clinical points of view. The recurrent and consistent infections cause by many representatives of the order indicates a possible adaptation of the fungi to the human host. In the environment they occupy specific micro-habitats, probably due to their low competitive ability towards co-occurring microorganisms. Their oligotrophism (Satow et al. 2008) enables them to thrive and maintain at low density in adverse substrates where common saprobes are absent (de Hoog 1993, 1997). An eventual potential as an environmental pathogen may involve a composite life cycle of the fungi concerned. However, the invasive potential is polyphyletic and differs significantly between species (Badali et al. 2008). Recurrent, consistently identifiable diseases are caused by relatively few species, which may be morphologically very similar to environmental counterparts which in many cases seem to be undescribed (Table 1). Therefore a reliable taxonomic system is mandatory to obtain better understanding of the link between clinical disease and environmental ecology.
The state of Paraná in southern Brazil is an endemic region for chromoblastomycosis. Fonsecaea pedrosoi is supposed to be responsible for more than 95% of the clinical cases, mainly infecting agricultural laborers (Queiroz-Telles 1997). This species is now known to comprise two cryptic entities, causing the same disease but seemingly differing in virulence (de Hoog et al. 2004). Out of five clinical strains tested from Paraná, two appeared to be F. monophora (Table 1). Our extensive environmental sampling in 56 locations in the state of Paraná showed that Fonsecaea pedrosoi was not isolated from nature, but instead we repeatedly encountered F. monophora. The natural source and route of infection of F. pedrosoi therefore still remains a mystery.
Several chaetothyrialean opportunists were isolated which are known to be associated with mild disorders, such as the cutaneous species Cladophialophora saturnica (Badali et al. 2008) and Exophiala xenobiotica (de Hoog et al. 2006). None of the systemic pathogens, such as Cladophialophora bantiana, were found. Several species listed in Table 1 concern hitherto undescribed, apparently saprobic representatives of the order Chaetothyriales that have never been reported as agents of human or animal disorders. The discrepancy of molecular identification and morphological and physiological results that were validated by analysis of ex-type strains of chaetothyrialean fungi (de Hoog et al. 1995,1995) indicated that a vast number of saprobic species still awaits discovery and description.
The hydrocarbon-polluted environments yielded another spectrum of chaetothyrialean fungi. Exophiala dermatitidis is a fairly common opportunist, occasionally causing fatal, systemic disease. Exophiala bergeri, E. xenobiotica, E. angulospora and Veronaea botryosa are exceptional and/or low-virulent opportunists. Exophiala bergeri has thus far rarely been reported as an agent of disease, but was abundantly isolated when monoaromatic hydrocarbons were used for enrichment. The presence of aromatic compounds in the sample increases colony density and diversity of black yeasts. The ecological and physiological patterns of species concerned suggests an evolutionary connection between the ability to develop on alkylbenzenes and the ability to cause diseases in humans and animals (Prenafeta-Boldú et al. 2006).
The present study was an attempt to verify whether infections caused by Fonsecaea pedrosoi and other agents of human mycosis are likely to be initiated by traumatic inoculation of environmental strains, and, more in general, to find the source of infection of invasive black yeasts-like fungi. Our results showed that this link is complex: environmental strains cannot always be linked directly to clinical cases. This is illustrated above by the genus Fonsecaea, known from two clinically relevant species. Mostly F. monophora or unknown Fonsecaea species were isolated. The apparently more virulent species F. pedrosoi is likely to require special, hitherto unknown parameters for isolation, such as the use of an animal bait (Dixon et al. 1980, Gezuele et al. 1972). Thus far it only has been encountered on the human patient, always causing chromoblastomycosis when the host is immunocompetent. In contrast, F. monophora can be isolated from the environment without an animal bait, and is a less specific opportunist (Surash et al. 2006). In general, pathogenicity and virulence of chaetothyrialean black yeasts may differ between closely related species. The group can be divided in three ecological groups, as follows. (1) Saprobes not known from vertebrate disorders, such as the majority of undescribed strains reported in Table 1; (2) Low-virulent opportunists that can directly be isolated from the environment, such as F. monophora, and (3) Highly specific pathogens that cannot be isolated from the environment directly but require a living mammal bait, resp. a human host. This suggests that isolation efficiencies differing between species reflect different pathogenic tendencies in pathogenic adaptation of the species.
Acknowledgments
This study was supported by CAPES/Ministério da Educação, Brazil and CNPq.
References
- Attili DS, Hoog GS de, Pizzirani-Kleiner AA (1998). rDNA-RFLP and ITS1 sequencing of species of the genus Fonsecaea, agents of chromoblastomycosis. Medical Mycology 36: 219–225. [PubMed] [Google Scholar]
- Badali H, Gueidan C, Najafzadeh MJ, Bonifaz A, Gerrits van den Ende AHG, Hoog GS de (2008). Biodiversity of the genus Cladophialophora. Studies in Mycology 61: 175–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon DM, Shadomy HJ, Shadomy S (1980). Dematiaceous fungal pathogens isolated from nature. Mycopathologia 70: 153–161. [DOI] [PubMed] [Google Scholar]
- Dixon DM, Shadomy HJ (1980). Taxonomy and morphology of dematiaceous fungi isolated from nature. Mycopathologia 70: 139–144. [DOI] [PubMed] [Google Scholar]
- Gerrits van den Ende AHG, Hoog GS de (1999). Variability and molecular diagnostics of the neurotropic species Cladophialophora bantiana. Studies in Mycology 43: 151–162. [Google Scholar]
- Gezuele E, Mackinnon JE, Conti-Diaz IA (1972). The frequent isolation of Phialophora verrucosa and Fonsecaea pedrosoi from natural sources. Sabouraudia 10: 266–273. [PubMed] [Google Scholar]
- Hoog GS de (1993). Evolution of black yeasts: possible adaptation to the human host. Antonie van Leeuwenhoek 63: 105–109. [DOI] [PubMed] [Google Scholar]
- Hoog GS de (1997). Significance of fungal evolution for the understanding of their pathogenicity, illustrated with agents of phaeohyphomycosis. Mycoses 40: 5–8. [DOI] [PubMed] [Google Scholar]
- Hoog GS de, Gerrits van den Ende AHG, Uijthof JMJ, Untereiner WA (1995). Nutritional physiology of type isolates of currently accepted species of Exophiala and Phaeococcomyces. Antonie van Leeuwenhoek 68: 43–49. [DOI] [PubMed] [Google Scholar]
- Hoog GS de, Guarro J, Gené J, Figueras MJ (2000a). Atlas of Clinical Fungi, ed. 2. Centraalbureau voor Schimmelcultures / Universitat Rovira i Virgili, Utrecht / Reus.
- Hoog GS de, Guého E, Masclaux F, Gerrits van den Ende AHG, Kwon-Chung KJ, McGinnis MR (1995). Nutritional physiology and taxonomy of human-pathogenic Cladosporium-Xylohypha species. Journal of Medical and Veterinary Mycology 33: 339–347. [PubMed] [Google Scholar]
- Hoog GS de, Matos T, Sudhadham M, Luijsterburg KF, Haase G (2005). Intestinal prevalence of the neurotropic black yeast Exophiala (Wangiella) dermatitidis in healthy and impaired individuals. Mycoses 48: 142–145. [DOI] [PubMed] [Google Scholar]
- Hoog GS de, Queiroz Telles F, Haase G, Fernandez-Zeppenfeldt G, Attili-Angelis D, Gerrits van den Ende AHG, Matos T, Peltroche-Llacsahuanga H, Pizzirani-Kleiner AA, Rainer J, Richard-Yegres N, Vicente VA, Yegres F (2000b). Black fungi: clinical and pathogenic approaches. Medical Mycology 38: 24–250. [PubMed] [Google Scholar]
- Hoog GS de, Zeng JS, Harrak MJ, Sutton DA (2006). Exophiala xenobiotica sp. nov., an opportunistic black yeast inhabiting environments rich in hydrocarbons Antonie van Leeuwenhoek 90: 257–268. [DOI] [PubMed] [Google Scholar]
- Hoog GS de. Attili-Angelis D, Vicente VA, Gerrits van den Ende AHG, Queiroz Telles F (2004). Molecular ecology and pathogenic potential of Fonsecaea species. Medical Mycology 42: 405–416. [DOI] [PubMed] [Google Scholar]
- Hoog GS, Weenink XO, Gerrits van de Ende AHG (1999). Taxonomy of the Phialophora verrucosa complex with the description of two new species Studies in Mycology 43: 107–122. [Google Scholar]
- Iwatsu T, Miyaji M, Okmoto S (1981). Isolation of Phialophora verrucosa and Fonsecaea pedrosoi from nature in Japan. Mycopathologia 75: 149–158. [Google Scholar]
- Lutzoni F, Pagel, M, Reeb V (2001). Major fungal lineages are derived from lichen symbiotic ancestors. Nature 411: 937–940. [DOI] [PubMed] [Google Scholar]
- Prenafeta-Boldú FX, Summerbell RC, Hoog GS de (2006). Fungi growing on aromatic hydrocarbons: biotechnology's unexpected encounter with biohazard. FEMS Microbiological Reviews 30: 109–130. [DOI] [PubMed] [Google Scholar]
- Queiroz Telles F (1997). A cromoblastomicose no estado do Paraná: etiologia, epidemiologia, clínica e terapêutica com itraconazol. Revista da Sociedade Brasileira de Medicina Tropical 30: 345–346. [Google Scholar]
- Satow MM, Attili-Angelis D, Hoog GS de, Angelis DF, Vicente VA (2008). Selective factors involved in oil flotation isolation of black yeasts from the environment Studies in Mycology 61: 157–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterflinger K, Hoog GS de, Haase G (1999). Phylogeny and ecology of meristematic Ascomycetes. Studies in Mycology 43: 5–22. [Google Scholar]
- Sudhadham M, Dorrestein G, Prakitsin S, Sivichai S, Chaiwat R, Hoog GS de (2008a). Two genotypes of the human-brain-infecting black yeast Exophiala dermatitidis indicate a possible origin of in the tropical rain forest. Studies in Mycology 61: 145–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surash S, Tyagi A, Hoog GS de, Zeng JS, Barton RC, Hobson RP (2005). Cerebral phaeohyphomycosis caused by Fonsecaea monophora. Medical Mycology 43: 465–472. [DOI] [PubMed] [Google Scholar]
- Wang CJK, Zabel RA (1990). Identification manual for fungi from utility poles in the Eastern United States. Allen Press, Lawrence, U.S.A.
- White TJ, Bruns T, Lee S, Taylor J (1990). Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: PCR Protocols A guide to methods and applications. (MA Innis, DA Gelfand JJ, Sninsky. TJ White eds) Academic Press, San Diego, U.S.A.
- Zeng JS, Sutton DA, Fothergill WA, Rinaldi MR, Harrak MJ, Hoog GS de (2007). Spectrum of clinically relevant Exophiala species in the U.S.A. Journal of Clinical Microbiology 45: 3713–3720. [DOI] [PMC free article] [PubMed] [Google Scholar]