Abstract
We have built a detailed kinetic model of translation initiation in yeast and have used a novel approach to determine the flux controlling steps based on limited experimental data. An efficient parameter estimation method was adapted in order to fit the most uncertain parameters (rate constants) to in vivo measurements in yeast. However, it was found that there were many other sets of plausible parameter values that also gave a good fit of the model to the data. We therefore used random sampling of this uncertain parameter space to generate a large number of diverse fitted parameter sets. A compact characterization of these parameter sets was provided by considering flux control. In particular, we suggest that the rate of translation initiation is most strongly influenced by one of two reactions: either the guanine nucleotide exchange reaction involving initiation factors eIF2 and eIF2B or the assembly of the multifactor complex from its constituent protein/tRNA containing complexes. It is hoped that the approach presented in this paper will add to our understanding of translation initiation pathway and can be used to identify key system-level properties of other biochemical processes.
Keywords: translation initiation, parameter fitting, flux control
1. Introduction
Biological processes can exhibit complex behaviour. A full understanding of this behaviour is often only possible by constructing a mathematical model, which can then be used to investigate the relative importance each individual reaction and species has on the overall pathway (Di Ventura et al. 2006). However, obtaining a model that reliably reproduces experimental results is currently a considerable challenge in biology (Gutenkunst et al. 2007). The set of reactions in the model will no doubt be an approximation. The rate equations will depend on one or several parameters; for example, a mass-action rate equation will have a forward and reverse rate constant. The model will also depend on the initial concentrations that we set for each species. Several or even all of these values may be unknown and where parameter values have been measured in vitro, they may not correspond to their values within the cell.
An essential part of building the model is therefore to fit these unknown parameters so that the model reproduces experimental data. This issue is addressed here, particularly for the case where only a limited amount of experimental data are available. When faced with such a task, we ask what use can be made of an uncertain model and apply our techniques to study the translation initiation pathway of protein synthesis. We show that it is still possible to make definite conclusions and predictions about the flux control within the pathway, even when we have only a small amount of in vivo data.
There are three stages to the translation process (McCarthy 1998) of protein synthesis: initiation; elongation; and termination. In the initiation phase the ribosomal subunits 40S and 60S are recruited to the mRNA, a process that requires the binding of several initiation factors in a complex sequence of events. Once complete, the initiation factors dissociate leaving the ribosome attached to the mRNA. In the elongation phase, the ribosome moves along the mRNA while forming a polypeptide chain of amino acids that fold to produce the protein. In the termination step, the ribosome dissociates into its 40S and 60S subunits releasing the protein. The initiation phase of translation is believed to be rate limiting (Jacques & Dreyfus 1990). In this study, we therefore focus our attention on building a model for translation initiation in yeast and investigate the subdivision of control within this pathway.
Previous studies (Heinrich & Rapoport 1980; Basu & Chowdhury 2007; Skjøndal-Bar & Morris 2007; Zouridis & Hatzimanikatis 2007) have used mathematical models in an attempt to investigate how the rate of initiation affects the overall rate of translation. The translation initiation process has either been separated into a subset of reactions (Skjøndal-Bar & Morris 2007), or simply assumed to be constant (Basu & Chowdhury 2007; Zouridis & Hatzimanikatis 2007). The difference in our study is that we build a detailed model of translation initiation in terms of all the known initiation factors. We use the recent parameter fitting methodology, the proximate parameter tuning (PPT) algorithm (Wilkinson et al. 2008), adapt it by calculating the parameter bounds on the fly, apply it to this ODE model of translation initiation and use principal component analysis on the distribution of flux control coefficients to make clear conclusions that can be tested or at least are consistent with other findings. This study suggests that there are two reactions in the translation initiation pathway which have greatest control over the flux through the pathway. We compare this finding with what is known in the literature.
2. The model
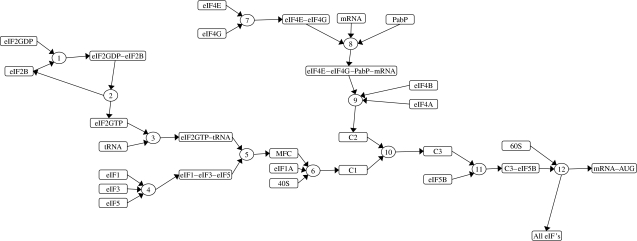
Figure 1 shows the sequence of binding events in the translation initiation pathway for our model. We approximate it as consisting of 12 key reactions and make the assumption that all eIF2 molecules are bound to GDP or GTP. The tRNA molecules enter the pathway by binding to the eIF2GTP complex. However, the initiation process hydrolyses this GTP molecule resulting in the eIF2GDP complex, and therefore the GDP molecule bound to eIF2 has to be replaced with GTP to prevent depletion of eIF2GTP. This event is catalysed by eIF2B (Pavitt et al. 1998; Manchester 2001). We reduce this mechanism to just two reactions. In reaction 1, eIF2GDP binds to eIF2B to form the eIF2GDP–eIF2B complex. In reaction 2, the GDP is replaced with GTP resulting in eIF2GTP and free eIF2B. The tRNA then binds to the eIF2GTP in reaction 3.
Figure 1.
The translation initiation pathway.
The initiation factors eIF1, eIF3 and eIF5 form the trimolecular complex eIF1–eIF3–eIF5 in reaction 4. This binds to the eIF2GTP–tRNA complex produced by reaction 3 resulting in the multifactor complex (MFC; Asano et al. 2000) in reaction 5. Then the MFC and the initiation factor eIF1A are recruited to the 40S ribosomal subunit in reaction 6 to form a complex that we have labelled C1.
The mRNA enters the pathway from a separate branch of the pathway. The eIF4E and eIF4G form the eIF4E–eIF4G complex in reaction 7. This complex binds to the capped end of the mRNA molecule along with the PabP initiation factor in reaction 8. The eIF4A and eIF4B initiation factors also bind to the mRNA complex in reaction 9 to form a complex that we have labelled C2. Then, in reaction 10, the tRNA complex C1 binds to the mRNA complex C2 to form the complex C3. The eIF5B initiation factor binds at reaction 11 and the 60S ribosomal subunit is finally recruited at reaction 12.
Reaction 12 actually covers a sequence of events where the 60S binds to the C3–eIF5B complex, which then moves along the mRNA until the AUG codon of the mRNA is reached. At this stage, the initiation complex dissociates leaving the ribosomal subunit attached to the mRNA from which the elongation phase of translation begins. We make the assumption that all initiation factors completely dissociate and re-enter the initiation pathway as free molecules.
All reactions were modelled using reversible mass-action kinetics, apart from reaction 12, which was assumed irreversible. Reactions 4, 6, 8 and 9 of the model involve the binding of three substrates to form a trimolecular complex. In these reactions, the order in which the substrates come together is uncertain. We therefore assume random ordering with mass-action kinetics, which allows the constituent bimolecular reactions to be lumped into a single trimolecular one, similar to the method of convenience kinetics (Liebermeister & Klipp 2006). This does not imply that these reactions are dependent on a coincident three-body collision between the molecules, but can be derived by considering a sequence of bimolecular reactions (see appendix A; Cornish-Bowden 1995). The rate equations for each reaction are displayed in table 1. and are the forward and reverse rate constants, respectively, for reaction i.
Table 1.
Rate equations in our model for translation initiation.
| reaction | rate equation |
|---|---|
| 1 | |
| 2 | |
| 3 | |
| 4 | |
| 5 | |
| 6 | |
| 7 | |
| 8 | |
| 9 | |
| 10 | |
| 11 | |
| 12 |
The steady-state concentrations of the initiation factors and the complexes they form are unknown. However, we do know the total number of molecules per cell, shown in table 2. In our model, the amount of each initiation factor is a conserved quantity (moiety). We can therefore set the initial concentration of each initiation factor equal to their total amount. The 40S subunit, 60S subunit, mRNA and tRNA form the end product of the translation initiation pathway as the mRNA–AUG complex of reaction 12. This complex is removed from the system and so the 40S subunit, 60S subunit, mRNA and tRNA concentrations are fixed at their initial concentrations, which are also given in table 2. All other intermediate species have an initial concentration of zero. Starting from these initial concentrations, the model can be brought to steady state by integrating the rate equations forward in time until all concentrations and the flux through the pathway reach their steady-state values.
Table 2.
Initial concentrations of each species used in the model.
| molecule | number per cell/105 | fixed? | reference |
|---|---|---|---|
| eIF2B | 0.3 | von der Haar & McCarthy (2002) | |
| eIF2GDP | 1.8 | von der Haar & McCarthy (2002) | |
| tRNA | 20 | yes | estimate |
| eIF1 | 2.5 | von der Haar & McCarthy (2002) | |
| eIF3 | 1 | von der Haar & McCarthy (2002) | |
| eIF5 | 0.483 | Ghaemmaghami et al. (2003) | |
| eIF1A | 0.5 | von der Haar & McCarthy (2002) | |
| 40S | 2 | yes | French et al. (2003) |
| eIF4E | 3.4 | von der Haar & McCarthy (2002) | |
| eIF4G | 0.175 | von der Haar & McCarthy (2002) | |
| mRNA | 0.15 | yes | Van Hoof et al. (2000) |
| PabP | 1.98 | Ghaemmaghami et al. (2003) | |
| eIF4A | 8 | von der Haar & McCarthy (2002) | |
| eIF4B | 1.55 | von der Haar & McCarthy (2002) | |
| eIF5B | 0.134 | Ghaemmaghami et al. (2003) | |
| 60S | 2 | yes | French et al. (2003) |
Unlike the initial concentrations, there is a large degree of uncertainty in the other parameter values required by the model, namely the reaction rate constants (table 3). Dealing with such uncertainty is a key concern of this paper and is discussed in more detail below.
Table 3.
Typical range for the rate constant values fitted to reproduce experimental results.
| reaction | units | lower | mean | upper |
|---|---|---|---|---|
| forward rate constant kf | ||||
| 1 | (μM s)−1 | 2.5 | 4.6 | 19.5 |
| 2 | s−1 | 4.1 | 8.4 | 13.7 |
| 3 | (μM s)−1 | 3.4 | 15.4 | 68.6 |
| 4 | (μM2 s)−1 | 3.8 | 13.4 | 36.6 |
| 5 | (μM s)−1 | 7.4 | 26.3 | 82.3 |
| 6 | (μM2 s)−1 | 1.7 | 6.0 | 16.9 |
| 7 | (μM s)−1 | 1.5 | 3.2 | 5.4 |
| 8 | (μM2 s)−1 | 2.9 | 7.8 | 23.7 |
| 9 | (μM2 s)−1 | 5.3 | 13.9 | 31.5 |
| 10 | (μM s)−1 | 30.1 | 56.5 | 85.6 |
| 11 | (μM s)−1 | 27.7 | 45.2 | 65.2 |
| 12 | (μM s)−1 | 26.1 | 56.8 | 86.3 |
| reverse rate constant kr | ||||
| 1 | s−1 | 55.3 | 122.6 | 199.4 |
| 2 | (μM s)−1 | 21.0 | 58.3 | 200.8 |
| 3 | s−1 | 19.2 | 68.9 | 209.0 |
| 4 | s−1 | 5.8 | 19.3 | 144.2 |
| 5 | s−1 | 21.8 | 87.0 | 446.1 |
| 6 | s−1 | 3.4 | 7.3 | 12.9 |
| 7 | s−1 | 50.0 | 109.2 | 198.4 |
| 8 | s−1 | 26.8 | 66.8 | 179.0 |
| 9 | s−1 | 10.0 | 23.3 | 108.1 |
| 10 | s−1 | 3.7 | 8.7 | 13.1 |
| 11 | s−1 | 2.3 | 7.6 | 13.8 |
3. Model uncertainty
We can divide the uncertainty in the model presented above into two categories: structural and parametric uncertainties.
Structural uncertainty relates to the stoichiometry of the biological reaction network. Many of the reactions described above may themselves consist of multiple steps, or else the order in which proteins and complexes assemble and disassemble may differ from that which is assumed. It could even be the case that completely alternative routes exist. We believe that the model presented above is sufficiently detailed to represent current knowledge about the structure of the network while avoiding the ‘combinatorial explosion’ of attempting to include all possible steps that may or may not exist.
Parametric uncertainty, on the other hand, refers to the values of the model parameters, which, for the mathematical model (based on ordinary differential equations), comprise initial concentrations and reaction rate constants. For translation initiation there are more data available on the concentrations of each species. Hence, the reaction rate constants are by far the most uncertain and we therefore focus our attention upon these in the parameter fitting process described in the next sections. Since there are no published in vivo values, we estimate loose lower and upper bounds on their values using a first principle approach (see §4).
4. Results
4.1 Fit to the experimental data
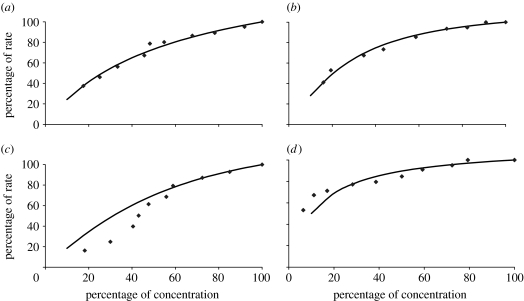
We take our experimental data from a recent study (Sangthong et al. 2007) in which the rate of protein synthesis was measured with respect to the concentration of initiation factors eIF1A, eIF4E, eIF4G and eIF5B. The rates and concentrations are given as a percentage of their naturally occurring values. The experimental results are shown in figure 2, along with the corresponding model-predicted values (MPVs) after the rate constants in our model had been fitted to reproduce these experimental data.
Figure 2.
Rate of protein synthesis versus initiation factor concentration for (a) eIF1A, (b) eIF4E, (c) eIF4G and (d) eIF5B. The graphs show the experimental data points (diamond) and model-predicted values (solid line).
It can be seen that good fits were achieved for each modulated elongation factor except eIF4G which has a sigmoid-shaped curve for eIF4G. In fact, we fitted the model only to the largest four experimental data points for eIF4G since this seemed to guide the PPT algorithm to the best overall fit across all elongation factors. Although this means that the model does not reproduce the eIF4G data for concentrations below 50%, it does ensure the behaviour is correct near conditions in which the yeast cells would naturally operate. A sigmoidal shape could arise from cooperative behaviour, but we have found no mechanisms involving the participation of multiple eIF4G proteins in the published literature. In this work, therefore, we adopted an unbiased strategy of assuming that all reactions follow mass-action kinetics with the kinetic order of each substrate being unity (table 1). However, the data for eIF4G hint at the possibility of more complex kinetics and our assumption will be relaxed in future work if additional data confirm this.
4.2 Efficiency of the parameter fitting
We found that multiple sets of parameter values gave fits to the measured data of a similar accuracy to that shown in figure 2. This problem is therefore an under-determined inverse problem (see §6). A family of models was therefore generated, each model having the same structure but a different set of parameters fitted to reproduce the experimental data, in order to extract important system-level properties of the translation initiation process (Kuepfer et al. 2007).
In order to perform a systematic evaluation of the nonlinear parameter space, we used a scaleable and effective parameter fitting algorithm called the PPT algorithm (Wilkinson et al. 2008; see §5). The PPT algorithm is a local optimization method and the computationally expensive part of this parameter fitting algorithm is in the calculation of the sensitivities of the steady-state flux with respect to changes in each parameter. We developed a computationally efficient implementation, whereby the sensitivities were calculated using the SBAOsenssim routine from the SBAOtoolbox, an extension of the Systems biology toolbox (Schmidt & Jirstrand 2006) for Matlab. This routine was able to calculate all sensitivities from a single time-course simulation to steady state. The speed and efficiency of the SBAOsenssim routine in calculating the sensitivities was the key feature that made it possible to obtain many sets of fitted parameters on which this study is based.
We found that the convergence of the PPT algorithm was highly dependent on the starting point used. Instabilities in the algorithm often occurred, perhaps due to sensitivities varying in sign and/or magnitude as the algorithm progressed. We developed a strategy to correct this pathological behaviour by limiting the amount by which any parameter could be changed to ±30% of its value in the previous iteration. This dramatically improved the probability of achieving a convergence to a good fit when using variable starting points. Implementing this additional constraint increased the chances of successfully fitting the parameters starting from a random set of initial values from less than 50% to around 90%. A successful fit was defined to have a sum of residuals value less than 1.0, where the residual is the absolute difference between the experimental data point and the MPV in logarithmic space.
4.3 Distribution of fitted parameter values
To investigate the behaviour of the model, initial values for the rate constants were randomly drawn from a uniform distribution and passed through the PPT algorithm to fit to the experimental data. For this fitting procedure, it was convenient to use model-specific units for the concentrations and reaction rates. The concentrations were expressed in units of 105 molecules per yeast cell, as expressed in table 2, making the concentrations of the order of unity. Each time-varying species is produced by one reaction and removed by another. At steady state all reaction rates must be equal and, as a first approximation, we therefore expect all forward rate constants to be of the same magnitude. Initial estimates of the reverse rate constants were also chosen to be of a similar size, the same tactic as employed in a previous study (Wilkinson et al. 2008), so that initially there would be no forward bias along the pathway. We were interested to see how the PPT algorithm would adjust these parameters in order to fit the model to the data. All initial values of the rate constants were drawn from a uniform distribution with a lower bound of 0 and an upper bound of 1000. The choice for the upper limit is essentially arbitrary as all rates were expressed relative to the steady-state flux when all initiation factors are present at their maximum value (figure 2).
Table 3 shows the typical range of the fitted rate constants converted to micromolar units for the concentration and seconds for the time. In making the conversion to these units, we have assumed that the volume of the yeast cell is 7×10−14 l (Sherman 1991), which contains a total of 50×106 protein molecules (Futcher et al. 1999) and has a doubling time of 2.23 hours, measured using the yeast strain in the in vivo study (Sangthong et al. 2007). Therefore, the wild-type protein synthesis rate is approximately 6200 protein molecules s−1. The typical deviation about the mean value of a fitted rate constant is characterized by lower and upper bounds. The lower bound is 1 standard deviation below its mean value, calculated from those rate constants whose values were below this mean. Similarly, the rate constants above the mean were used in the calculation of the upper bound.
Diffusion-limited protein–protein association rate constants are generally of the order of 0.1–1 (μM s)−1 (Schlosshauer & Baker 2004) but, in the presence of electrostatic steering forces, the association rates are larger. Our model estimates for the forward rate constants are in the range of 1–50 (μM s)−1 and are typical of such values where electrostatic steering forces are important. Dissociation rate constants can vary considerably but our model estimates for the backward rate constants are generally higher than the dissociation constants of protein complexes, which are usually of the order of 0.1 s−1 (Wilkinson et al. 2008). However, we point out that the model is a simplified version of the real situation in terms of its individual reactions, the overall order of events (structural uncertainty) and the assumption that the species were uniformly distributed across the cell. The rate constants presented here are fitted in order for this model to reproduce the in vivo experimental data. Not only are these values dependent on our model assumptions, but also on our estimates made for the volume of the yeast cell and the rate of protein synthesis and therefore the values may not necessarily correlate with experimentally measured values in vitro. As further experimental data become available, it should be possible to build a more detailed and comprehensive model of the translation initiation process. Rate constant values measured in vitro may then be incorporated into the model prior to parameter fitting and this will be investigated in future work.
We found that 90% of the initial sets of rate constants drawn from the uniform distribution resulted in acceptable fits (sum of residuals less than 1.0), when passed through the parameter fitting algorithm. The mean value of the sum-of-residuals was 0.89 with a standard deviation of 0.04, equal to 4.5% of the mean, showing that the fitted profiles we obtained followed a tight distribution around the measured data points. On the other hand, the large standard deviations for most of the fitted parameter values show that they are not uniquely identifiable from the available data. In other words, there are many different combinations of parameter values that give good fits. For this reason, we shift our focus away from the identification of unique parameter values and instead analyse the patterns of flux control. Below, we demonstrate that this system-level feature is more identifiable since we are able to formulate an exclusive disjunction on the most controlling reaction.
4.4 Control patterns of the fitted models
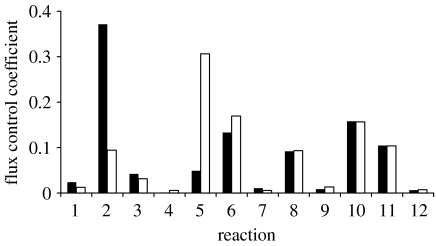
For each initial set of randomly drawn rate constant values, the resulting flux control coefficients were calculated for each reaction (see §5). The rate constants were then fitted to reproduce the experimental data and a fitted set of flux control coefficients were calculated. Figure 3 shows the mean value for each flux control coefficient, the average being taken over all 100 randomly selected and fitted parameter sets. The flux control for the randomly drawn parameters is centred firmly on reactions 11 and 12. Although this does not represent a physically realistic scenario, it does show that there is very little variation in the flux control across a large and randomly sampled parameter space. The effect of fitting to the experimental data is to dramatically modify this pattern of flux control whereby control is now distributed across six reactions (2, 5, 6, 8, 10 and 11). The remaining reactions (1, 3, 4, 7, 9 and 12) exert relatively little control over the rate of translation initiation.
Figure 3.
Average flux control coefficients for (a) the initial randomly selected rate constants and (b) the fitted rate constants.
Principal component analysis (see §5) gave further insight into the model results by projecting each set of 12 control coefficients onto the two largest principal components (figure 4). Within the plane of these two principal axes, 94% of the variance within the control coefficients is captured. The results suggest that the control coefficients have formed two clusters. Cluster 1 has a negative value on the first principal axis whereas for cluster 2 it is positive. The flux control coefficients were averaged over each separate cluster and are shown in figure 5.
Figure 4.
The flux control coefficients projected onto the plane defined by the two largest principal components here appear to be less than 100 points since many are coincident. (a) Cluster 1 and (b) cluster 2.
Figure 5.
Average flux control coefficients of each cluster identified by the principal component analysis. For each reaction, mean of the coefficient values weighted by the size of their respective cluster correspond to the values given in figure 3. Filled bars, cluster 1; open bars, cluster 2.
The flux control coefficients are essentially the same for both clusters except for reactions 2 and 5. For cluster 1, the flux control coefficient of reaction 2 is largest with a value of 0.37, while the flux control coefficient of reaction 5 is minimal. The opposite effect is found in cluster 2, reaction 5 having the larger flux control coefficient of 0.31.
4.5 Response coefficients
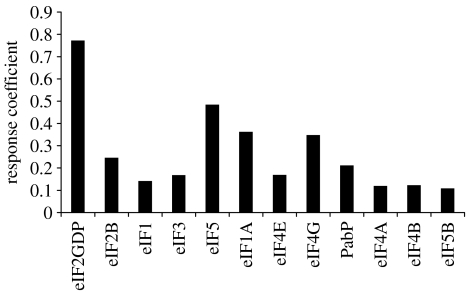
The response of the flux with respect to the initial concentrations of the initiation factors (see §5) was calculated for each set of fitted parameters. The averages of these response coefficients are plotted in figure 6, and presented in table 4 with their corresponding standard deviations.
Figure 6.
The average value of the response coefficients in terms of the initial concentration of each initiation factor.
Table 4.
The response coefficients averaged over the family of models with their standard deviations.
| initiation factor | mean | s.d. |
|---|---|---|
| eIF2GDP | 0.77 | 0.06 |
| eIF2B | 0.25 | 0.17 |
| eIF1 | 0.14 | 0.18 |
| eIF3 | 0.17 | 0.20 |
| eIF5 | 0.48 | 0.14 |
| eIF1A | 0.36 | 0.02 |
| eIF4E | 0.17 | 0.04 |
| eIF4G | 0.35 | 0.05 |
| PabP | 0.21 | 0.06 |
| eIF4A | 0.12 | 0.08 |
| eIF4B | 0.12 | 0.08 |
| eIF5B | 0.11 | 0.01 |
The results show that the initiation factor with the largest control over the flux is eIF2 with a response coefficient of 0.77, whereas eIF5B has the smallest response coefficient of 0.11. Note that one might not expect this by just considering the flux control coefficients. Reaction 1 has a very small flux control coefficient, yet eIF2 (a reactant of reaction 1) has the largest response coefficient. Likewise, reaction 11 has a relatively large flux control coefficient, but the response coefficient of eIF5B (a reactant of reaction 11) is small. Unlike the flux control coefficients that must sum to 1, the response coefficients need not sum to any particular value. Here we find that the average values for the response coefficients of all the initiation factors sum to 3.3±0.4. This says that if the concentrations of all the initiation factors were simultaneously increased by 1%, the flux through the pathway would increase by 3.3%.
From an experimental viewpoint these response coefficients are more informative than the flux control coefficients, as the response coefficients demonstrate how the flux through the pathway responds to a change in concentration that the cell itself may regulate and the standard deviations in the values of these response coefficients may be used in experimental design. Several initiation factors, such as eIF1 and eIF3, have relatively large standard deviations. The model, in conjunction with the experimental data, has therefore revealed a larger degree of uncertainty in these response coefficients compared with the others. The next set of experiments can therefore have greatest impact by removing this uncertainty, which suggests that these experiments should be in obtaining the rate versus concentration curves for the initiation factors eIF1 and eIF3.
5. Methods
5.1 Parameter fitting
Fitting the rate constants so that the model reproduces the experimental results was extensively used in this study. We have developed a quick and effective parameter fitting algorithm that can handle a vast and highly nonlinear parameter space, called the PPT (Wilkinson et al. 2008) algorithm. This algorithm fits m parameters using n experimental data points by minimizing the linear fitting function,
| (5.1) |
Δki is defined to be greater than or equal to the absolute difference in log space between the ith parameter value ki and an initial estimate , which we call the nominal value,
| (5.2) |
Δk∞ is the infinity norm of Δki, defined to be greater than or equal to the maximum Δki; therefore,
| (5.3) |
Ri is defined to be greater than or equal to the ith residual. The residual is the absolute difference in log space between an experimental data point, called the target value Ti, and the corresponding model-predicted value MPVi. Therefore,
| (5.4) |
R∞ is the infinity norm of Ri, defined to be greater than or equal to the maximum Ri; therefore,
| (5.5) |
Each term in the fitting function is multiplied by a weight: , , or . In this study, the weights were chosen to have a value of 10−5. All other weights had a value of 1.
The first term of the fitting function can be used to restrain the parameter values to their nominal values. In this study, the nominal values were set equal to the randomly drawn initial values. Since we did not want to bias any parameter towards any particular value, the weights were set to a negligible value of 10−5. When there are many parameters and little experimental data, an infinite number of points may exist that all identically minimize the fitting function at each iteration of the PPT algorithm. The second term of the fitting function breaks this symmetry ensuring that the algorithm moves to the closest point within the infinite set of possibilities. The third term of the fitting function is the main term that calculates the difference between the experimental data and the MPVs. However, to prevent the algorithm from fitting preferentially to some experimental data points at the expense of others, the fourth term of the fitting function penalizes the maximum residual value.
The fitting function of equation (5.1) is minimized, subject to equations (5.2)–(5.5), using a linear programming algorithm, such as the simplex method or interior-point method (Williams 1993). This method requires that constraints, like the fitting function, must be linear with respect to the parameters log(ki) and the variables Δki, Δk∞, Ri and R∞. The expression for the MPVs in equations (5.4) and (5.5) must therefore be expanded to first order to give the unknown value at iteration r+1 in terms of the known value at iteration r.
| (5.6) |
5.2 Control analysis
Flux control coefficients (Heinrich & Rapoport 1974; Kholondenko & Westerhoff 1993) show how each individual reaction within the network controls the steady-state flux through the network and are defined as
| (5.7) |
∂ ln vk is a change made to the rate of the kth reaction by perturbing both the forward and reverse rate constants of the reaction. ∂ ln J is the steady-state response of the flux through the network. The summation theorem for flux control coefficients states that all flux control coefficients within the network must sum to 1 (Kholondenko & Westerhoff 1993). So the value of the flux control coefficient for reaction k shows what control this reaction has on the steady-state flux. A value of zero would imply that it has no control whereas a value of one implies it has total control (all other reactions must have a zero flux control coefficient) and therefore would be rate limiting.
Response coefficients show how each individual species controls the steady-state flux through the network and are defined as
| (5.8) |
∂ ln [Xk] is a change made to the initial concentration of species Xk with ∂ ln J being the response of the steady-state flux as with the flux control coefficient above.
5.3 Principal component analysis
For each set of fitted rate constants, 12 flux control coefficients, corresponding to each reaction in the network, can be calculated. One can imagine that this set of 12 flux control coefficients occupies a point in a 12-dimensional space and when all 100 sets of fitted rate constants are taken into account we therefore have a distribution of points. The principal components of this distribution capture the greatest amount of variance and showed that 94% of the variance was captured by just two principal components. The distribution of points is essentially on a two-dimensional plane within the 12-dimensional space. The dimensionality of the flux control coefficients has therefore been reduced from 12 to 2.
Principal component analysis is an eigenvalue problem (Leach 2001; Allen et al. 2003). Suppose there are x sets of flux control coefficients, each set containing y values (in this study, x=100 and y=12). These data can be arranged in a matrix D containing x columns and y rows to calculate the variance–covariance matrix
| (5.9) |
The eigenvectors of Z are the principal components. The corresponding eigenvalues (λi) are used to calculate the variance captured by the eigenvector. The fraction of the total variance along the ith principal axis is
| (5.10) |
6. Discussion
Obtaining an accurate biological model of translation initiation is limited by the following problems.
Several reactions in our simplified model could be subdivided into individual steps. The apparent rate constants for each reaction are therefore hard to derive and in many cases unknown.
Experimental estimates of rate constants, where they exist, will have been performed in vitro and may therefore not correspond to the in vivo rate constants within a crowded cellular environment.
The order of the binding events within the model is based on our current understanding of the translation initiation process and will naturally be an approximation. Furthermore, valid alternative pathways may exist in parallel (Sangthong et al. 2007).
Our assumption that the initiation factors are uniformly distributed across the cell does not take into account spatial variance within the cell (Minton 2006; Sun & Weinstein 2007).
A major problem is then to select suitable values for the 23 rate constants in the model. However, even an accurate value for a binding constant is of little use if any of the above problems are significant. We therefore have to accept that the rate constants used in our model cannot easily be obtained by experiment or derived from physical reasoning. This forces us to take a different approach that focuses on the behaviour displayed by the model.
The parameter fitting methodology, the PPT algorithm (Wilkinson et al. 2008), was used to fit randomly selected rate constants to the limited experimental data. The probability of successfully fitting a set of random parameters to the experimental data was greatly improved by applying an additional constraint, which prevented each rate constant from changing beyond 30% of its value at the previous iteration. It should be emphasized that there are many combinations of parameter values that give fits as good as those shown in figure 2. This problem is therefore an under-determined one when viewed from a classic parameter estimation perspective. It is not possible to uniquely identify the values of most of the parameters in the model and this is an extremely common problem for this type of model given the limited experimental data.
The range of behaviour displayed by the model was investigated by considering the flux control coefficients obtained from each set of fitted parameters. The results demonstrate that there is no single reaction that is rate limiting, but the rate control is distributed among several of the reactions. However, principal component/clustering analysis on the flux control coefficients shows that the model predicts two distinct patterns of rate control that differ only in the flux control coefficient of two out of the 12 reactions. The first distribution identifies reaction 2 (eIF2B-catalysed reaction) as having the greatest control over the rate whereas the second distribution predicts reaction 5 (production of the MFC). Clearly, the two possibilities cannot both be correct. It is interesting to note, however, that other studies have proposed that the eIF2B-catalysed reaction is a major control point in protein synthesis (Welsh et al. 1996; Campbell et al. 2005; Pavitt 2005; Campbell & Ashe 2006) and is consistent with our model prediction that reaction 2 may have the largest control over the rate. The response coefficients calculated using our model also support this result, predicting that the initiation factor eIF2 exerts the largest control over the rate of initiation.
A recent study (Asano & Sachs 2008) proposes that the MFC controls translation via a cooperative set of interactions involving its constituents, the eIF2 and the tRNA. It is clear then that the role of eIF2 in the formation of the MFC is more complex than the simplified mass-action kinetics presented here. However, this study shows that it is still possible to extract meaningful results and make testable predictions, even when the model may be approximate and little experimental data are available.
7. Conclusion
In this paper, we have made a first attempt at quantitatively understanding the distribution of rate control in the translation initiation pathway. The key contribution of this paper is to use the available data to infer an important system-level property of the system that we hope can be experimentally tested. We found that generating a family of possible parameter sets, each one capable of reproducing the experimental data, was a powerful and effective approach; particularly where parameter values are unknown, the structure of the model is uncertain and the experimental data are limited.
It should be noted that the output from this exercise is not a particular model instance—that is, one with a specific set of parameter values that is able to accurately simulate translation initiation under a range of conditions. While this remains the ultimate goal of our efforts, the structural uncertainty and paucity of available experimental data completely preclude this at present. Nevertheless, we have been able to explore a diverse space of possible models to produce a subspace of fitted models that collectively suggest directions for further experimental effort. In particular, for eIF1 and eIF3, our results indicate that a more precise quantification of their influence on translation would be valuable. Furthermore, some degree of validation of our analysis has been provided by previous work on the importance of the eIF2 cycle.
Finally, we note that the problem of insufficient data addressed in this paper is a very common one and is becoming an issue of intense interest in systems biology. Can under-determined kinetic models still add value to the experimental process? It is hoped that our methodology can be extended into a general framework in which detailed models can enhance knowledge, even when unique estimation of their constituent parameter values is not possible.
Acknowledgments
R.J.D. acknowledges funding from BBSRC grant BBE0037291 awarded to J. E. McCarthy and H. V. Westerhoff. S.J.W. acknowledges funding from BBSRC grant BBD0190791 awarded to H. V. Westerhoff, D. B. Kell and S. Oliver.
Appendix A.
We now provide justification for our use of pseudo-third-order rate laws for some of the forward reactions, such as reaction 4 in our model, in which three initiation factors assemble into an intermediate complex. Consider the general case of three species A, B and C that can associate in a random order. We break this aggregate reaction into a set of subreactions, each conforming to mass-action kinetics. We assume rapid equilibration between the monomeric and the heterodimeric species via forward and reverse reactions. For the formation of the final complex, however, we consider only the forward reaction since we are only interested in an expression for the forward rate of the overall reaction—that is, the rate of production of the final complex. We can write the set of subreactions as
| (A1) |
| (A2) |
| (A3) |
Looking at the subreactions given by equation (A 1), we can write the rate of production of the heterodimer AB as
| (A4) |
We now assume that this heterodimer is close to steady state (the same assumption is made in the derivation of the Michaelis–Menten equation in which the concentration of the enzyme–substrate complex is assumed to be approximately steady). This yields an expression for the concentration of the heterodimer
| (A5) |
The expression for the rate of production of complex ABC in equation (A 1) is therefore,
| (A6) |
Looking at the denominator we now assume that —the rate at which the heterodimer dissociates is much more rapid than the rate at which it associates with species C. Here, we are implying that the first-order dissociation reaction (unlimited by diffusion) is rapid compared with the second-order complex association reaction, which is limited by the diffusion of the co-substrate C. This gives a final third-order rate equation for the overall forward rate of the subreaction given by equation (A 1),
| (A7) |
Similarly, the same form of expression can be derived for the other reactions (equations (A 2) and (A 3)) to give the final overall expression for the forward rate of complex formation.
| (A8) |
References
- Allen J.K, Davey H.M, Broadhurst D, Heald J.K, Rowland J.J, Oliver S.G, Kell D.B. High-throughput classification of yeast mutants for functional genomics using metabolic footprinting. Nat. Biotechnol. 2003;21:692–696. doi: 10.1038/nbt823. [DOI] [PubMed] [Google Scholar]
- Asano K, Sachs M.S. Translation factor control of ribosome conformation during start codon selection. Genes Dev. 2008;21:1280–1287. doi: 10.1101/gad.1562707. [DOI] [PubMed] [Google Scholar]
- Asano K, Clayton J, Shalev A, Hinnebusch A.G. A multifactor complex of eukaryotic initiation factors, eIF1, eIF3, eIF5, and initiator tRNAMet is an important translation initiation intermediate in vivo. Genes Dev. 2000;14:2534–2546. doi: 10.1101/gad.831800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu A, Chowdhury D. Traffic of interacting ribosomes: effects of single machine mechanochemistry on protein synthesis. Phys. Rev. E. 2007;75:021 902. doi: 10.1103/PhysRevE.75.021902. [DOI] [PubMed] [Google Scholar]
- Campbell S.G, Ashe M.P. Localization of the translational guanine nucleotide exchange factor eIF2B. Cell Cycle. 2006;5:678–680. doi: 10.4161/cc.5.7.2607. [DOI] [PubMed] [Google Scholar]
- Campbell S.G, Hoyle N.P, Ashe M.P. Dynamic cycling of eIF2 through a large eIF2B-containing cytoplasmic body: implications for translational control. J. Cell Biol. 2005;170:925–934. doi: 10.1083/jcb.200503162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornish-Bowden A. Portland Press; London, UK: 1995. Fundamentals of enzyme kinetics. [Google Scholar]
- Di Ventura B, Lemerle C, Michalodimitrakis K, Serrano L. From in vivo to in silico biology and back. Nature. 2006;443:527–533. doi: 10.1038/nature05127. [DOI] [PubMed] [Google Scholar]
- French S.L, Osheim Y.N, Cioci F, Nomura M, Beyer A.L. In exponentially growing saccharomyces cerevisiae cells, RNA synthesis is determined by the summed RNA polymerase I loading rate rather than by the number of active genes. Mol. Cell. Biol. 2003;23:1558–1568. doi: 10.1128/MCB.23.5.1558-1568.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futcher B, Latter G.I, Monardo P, McLaughlin C.S, Garrels J.I. A sampling of the yeast proteome. Mol. Cell. Biol. 1999;19:7357–7368. doi: 10.1128/mcb.19.11.7357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaemmaghami S, Huh W.K, Bower K, Howson R.W, Belle A, Dephoure N, O'Sheah E.K, Weissman J.S. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- Gutenkunst R.N, Waterfall J.J, Casey F.P, Brown K.S, Myers C.R, Sethna J.P. Universally sloppy parameter sensitivities in systems biology models. PLoS Comp. Biol. 2007;3:e189. doi: 10.1371/journal.pcbi.0030189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrich R, Rapoport T.A. A linear steady-state treatment of enzymatic chains. Eur. J. Biochem. 1974;42:89–95. doi: 10.1111/j.1432-1033.1974.tb03318.x. [DOI] [PubMed] [Google Scholar]
- Heinrich R, Rapoport T.A. Mathematical modelling of translation of mRNA in eukaryotes; steady-states, time dependent processes and application to reticulocytes. J. Theor. Biol. 1980;86:279–313. doi: 10.1016/0022-5193(80)90008-9. [DOI] [PubMed] [Google Scholar]
- Jacques N, Dreyfus M. Translation initiation in Escherichia coli: old and new questions. Mol. Micobiol. 1990;4:1063–1067. doi: 10.1111/j.1365-2958.1990.tb00679.x. [DOI] [PubMed] [Google Scholar]
- Kholondenko B.N, Westerhoff H.V. Metabolic channelling and control of the flux. FEBS Lett. 1993;320:71–74. doi: 10.1016/0014-5793(93)81660-R. [DOI] [PubMed] [Google Scholar]
- Kuepfer L, Peter M, Sauer U, Stelling J. Ensemble modelling for analysis of cell signalling dynamics. Nat. Biotechnol. 2007;25:1001–1006. doi: 10.1038/nbt1330. [DOI] [PubMed] [Google Scholar]
- Leach A.R. 2nd edn. Pearson; Harlow, UK: 2001. Molecular modelling: principles and applications. [Google Scholar]
- Liebermeister W, Klipp E. Bringing metabolic networks to life: convenience rate law and thermodynamic constraints. Theor. Biol. Med. Mod. 2006;3:41. doi: 10.1186/1742-4682-3-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manchester K.L. Catalysis of guanine-nucleotide exchange on eIF2 by eIF2B: can it be both a substituted enzyme and a sequential mechanism? Biochem. Biophys. Res. Commun. 2001;289:643–646. doi: 10.1006/bbrc.2001.6010. [DOI] [PubMed] [Google Scholar]
- McCarthy J. Posttranscriptional control of gene expression in yeast. Microbiol. Mol. Biol. Rev. 1998;62:1492–1553. doi: 10.1128/mmbr.62.4.1492-1553.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minton A.P. How can biochemical reactions within cells differ from those in test tubes? J. Cell Sci. 2006;119:2863–2869. doi: 10.1242/jcs.03063. [DOI] [PubMed] [Google Scholar]
- Pavitt G.D. eIF2B, a mediator of general and gene-specific translation control. Biochem. Soc. Trans. 2005;33:1487–1492. doi: 10.1042/BST20051487. [DOI] [PubMed] [Google Scholar]
- Pavitt G.D, Ramaiah K.V.A, Kimball S.R, Hinnebusch A.G. eIF2 independently binds two distinct eIF2B sub complexes that catalyze and regulate guanine-nucleotide exchange. Genes Dev. 1998;12:514–526. doi: 10.1101/gad.12.4.514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangthong P, Hughes J, McCarthy J.E.G. Distributed control for recruitment, scanning and subunit joining steps of translation initiation. Nucleic Acids Res. 2007;35:3573–3580. doi: 10.1093/nar/gkm283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlosshauer M, Baker D. Realistic protein-protein association rates from a simple diffusional model neglecting long-range interactions, free energy barriers, and landscape ruggedness. Protein Sci. 2004;13:1660–1669. doi: 10.1110/ps.03517304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt H, Jirstrand M. Systems biology toolbox for Matlab: a computational platform for research in systems biology. Bioinformatics. 2006;22:514–515. doi: 10.1093/bioinformatics/bti799. [DOI] [PubMed] [Google Scholar]
- Sherman F. Getting started with yeast. Methods Enzymol. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- Skjøndal-Bar N, Morris D.R. Dynamics model of the process of protein synthesis in eukaryotic cells. Bull. Math. Biol. 2007;69:361–393. doi: 10.1007/s11538-006-9128-2. [DOI] [PubMed] [Google Scholar]
- Sun J, Weinstein H. Toward realistic modelling of dynamics processes in cell signalling: quantification of macromolecular crowding effects. J. Chem. Phys. 2007;127:155 105. doi: 10.1063/1.2789434. [DOI] [PubMed] [Google Scholar]
- Van Hoof A, Lennertz P, Parker R. Yeast exosome mutants accumulate 3′-extended polyadenylated forms of U4 small nuclear RNA and small nuclear RNAs. Mol. Cell. Biol. 2000;20:441–452. doi: 10.1128/MCB.20.2.441-452.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von der Haar T, McCarthy J.E.G. Intracellular translation initiation factor levels in Saccharomyces cerevisiae and their role in cap-complex function. Mol. Microbiol. 2002;2:531–544. doi: 10.1046/j.1365-2958.2002.03172.x. [DOI] [PubMed] [Google Scholar]
- Welsh G.I, Miyamoto S, Price N.T, Safer B, Proud C.G. T-cell activation leads to rapid stimulation of translation initiation factor eIF2B and inactivation of glycogen synthase kinase-3. J. Biol. Chem. 1996;271:11 410–11 413. doi: 10.1074/jbc.271.4.2121. [DOI] [PubMed] [Google Scholar]
- Wilkinson S.J, Benson N, Kell B.K. Proximate parameter tuning for biochemical networks with uncertain kinetic parameters. Mol. Biosyst. 2008;4:74–97. doi: 10.1039/b707506e. [DOI] [PubMed] [Google Scholar]
- Williams H.P. Wiley; New York, NY: 1993. Model building in mathematical programming. [Google Scholar]
- Zouridis H, Hatzimanikatis V. A model for protein translation: polyzome self-organization leads to maximum protein synthesis rates. Biophys. J. 2007;92:717–730. doi: 10.1529/biophysj.106.087825. [DOI] [PMC free article] [PubMed] [Google Scholar]