Abstract
There is some evidence to suggest that peptide segments that are found rarely or never in the host proteome play a role in the immune response to disease-related proteins, both those derived from microbes and those derived from the host itself. We conjecture that this pattern may extend to human proto-oncoproteins. Our hypothesis in this study is that the frequency of rare peptide segments in sets of human proto-oncoproteins is significantly higher than in sets of control proteins, and we show that this is the case. Possible immunological implications of this observation are discussed.
Keywords: proto-oncoproteins, self–non-self discrimination, rare peptides, tyrosine kinases
1. Introduction
Studies employing computational analysis of cancer-related sequence data have focused mainly on modelling and predicting the effect of mutations, usually at the level of individual nucleotides or codons in a single proto-oncogene or family of proto-oncogenes (Wang et al. 2005). At the protein sequence level, bioinformatics and proteomics methods have been employed mainly to identify biomarkers for various types of cancers (Cho 2007). However, the number of bioinformatics studies that have focused on the protein products of proto-oncogenes has been limited.
The general sequence features of the protein products that are relevant to oncogenesis have not yet been established. One promising sequence feature, which has recently been suggested as important in disease-related proteins (derived from either pathogens or the host itself), is the presence of short peptide segments that are found rarely or never in the host proteome. In a broad-based analysis, Capone et al. (2008) found that such rare peptide sequences are found unexpectedly often in proteins implicated in autoimmunity and cancer, and Amela et al. (2007) discovered that there exists surprisingly little sequence similarity between human proteins and B-cell epitopes derived from pathogens. In a more narrowly defined study, Rolland et al. (2007) studied peptides derived from HIV-1, and discovered a negative correlation between similarity to the human proteome and frequency of immune recognition. Similarly, Polimeno et al. (2008) described immunodominant epitopes from hepatitis C virus that are characterized by a low level of similarity to the human proteome. Finally, short peptide segments—particularly 5-mers—have been suggested as a fundamental unit of immunological recognition (Lucchese et al. 2007).
In this study, we investigated whether proto-oncoproteins contain a significantly different proportion of short peptide segments that are found rarely or never in the human proteome, when compared with control proteins. If a statistically significant difference is found, it may indicate that the presence of rare peptides plays a role in oncogenesis.
2. Procedure
2.1 Comparisons performed and proteins used
In this study, 30 comparisons were performed, which were divided into three different categories. In each comparison, two distinct sets of proteins were used: a ‘proto-oncoprotein set’, which consisted of proteins that are known to have oncogenic potential, and a ‘control set’, which consisted of proteins not known to be proto-oncogenic.
2.1.1 Category 1: proto-oncoproteins compared with housekeeping proteins
Category 1 consisted of just a single comparison. The proto-oncoprotein set for this comparison contained 20 proto-oncoproteins, each of which is described as proto-oncogenic in its Swiss-Prot annotation. The control set consisted of 20 housekeeping proteins that perform basic cellular functions, such as glycolysis and cellular respiration. Table 1 in the electronic supplementary material contains the complete list of proteins in both sets.
2.1.2 Category 2: proto-oncoproteins compared with diverse sets of other proteins
Category 2 consisted of multiple comparisons. The category 2 comparisons had the common objective of expanding upon the category 1 comparison by incorporating more diverse sets of control proteins. To acquire such sets, the gene ontology (GO; Ashburner et al. 2000) terms were gathered for each housekeeping protein used in the category 1 comparison. GO terms are simply standardized descriptions of a protein, and fall into three broad classes: ‘biological process’; ‘molecular function’; and ‘cellular component’. GO terms in the biological process class (e.g. ‘striated muscle contraction’) generally facilitate categorization of a corresponding protein as housekeeping or non-housekeeping, while GO terms in the molecular function class (e.g. ‘protein binding’) and the cellular component class (e.g. ‘extrinsic to membrane’) do not; thus, only GO terms in the biological process class were used. For each of these GO terms, all of the human proteins having that GO term were downloaded from the Swiss-Prot database. The number of human proteins associated with each GO term varied widely, from just one protein to over 400 proteins. The initial number of GO terms was 40. To ensure meaningful results, GO terms associated with fewer than 10 human proteins were discarded. After this filtering step, 28 sets of proteins remained, with every protein in a given set being described by a specific GO term. Each of these sets was compared separately with the same proto-oncoprotein set as was used in the category 1 comparison; thus, there were 28 comparisons in category 2. Table 2 in the electronic supplementary material lists all 40 GO terms, along with the number of proteins that correspond to each. The full list of proteins corresponding to each of the 40 GO terms is included in the electronic supplementary material.
2.1.3 Comparison 3: proto-oncogenic tyrosine kinases compared with tyrosine kinases not known to be oncogenic
As with category 1, category 3 consisted of just a single comparison. Should differences in the above sets be found, one possible (though unlikely) explanation is that rare peptides are simply more likely to be found in proteins involved in cellular differentiation than in housekeeping proteins. This motivated a third comparison involving proteins from a single functional family. As many proto-oncoproteins are tyrosine kinases, the proto-oncoprotein and control sets for the category 3 comparison comprised human tyrosine kinases derived from KinBase (Manning et al. 2002; http://kinase.com/kinbase/index.html). Tyrosine kinases (as opposed to some other group of kinases) were selected because they form the largest group of human protein kinases given in KinBase. For each tyrosine kinase in KinBase, a Blast (Altschul et al. 1997) search was used to find the corresponding record at Swiss-Prot. For some proteins, the Swiss-Prot sequence was slightly different from the KinBase sequence, but the discrepancies were minor (all but seven proteins had 99% or more identity, and all had 95% or more identity). The actual sequences used in the comparisons were from Swiss-Prot. This list of all human tyrosine kinases was separated into two groups: known proto-oncoproteins (the proto-oncoprotein set) and proteins that have not been identified as proto-oncogenic (the control set). To accomplish this, the gene summary and GeneRIFs (‘gene reference into function’) from the Entrez Gene record for each tyrosine kinase were used to classify each kinase into one of the two categories. For some tyrosine kinases, the Entrez Gene summary and GeneRIFs were ambiguous as to the potential oncogenicity of the kinase; where no definitive statements were found characterizing the protein as proto-oncogenic, it was placed in the control set. Table 3 in the electronic supplementary material lists the proteins in the proto-oncoprotein and control sets for the category 3 comparison.
2.2 Comparing the similarity of two protein sets with the human proteome
The human proteome was downloaded from Integr8/UniProtKB (www.ebi.ac.uk/integr8/) on 30 April 2007, and contained 38 009 sequences. This set of proteins was denoted as ‘proteome A’. To reduce sequence redundancy, all possible pairs of protein sequences from proteome A were examined for instances in which both sequences were exactly the same, or in which one protein was a fragment of the other. In the former case, one protein record was arbitrarily chosen for deletion, and in the latter case, the shorter protein (the fragment) was removed. The resulting proteome, denoted ‘proteome B’, contained 36 014 sequences.
For each of the comparisons described in §2.1, we wished to determine how similar the proteins in each set (the proto-oncoprotein and control sets) were to the rest of the human proteome. Segments of five amino acid residues in length (5-mers) have been identified as a basic unit of recognition for functional protein–protein interactions, particularly in the immune system (Lucchese et al. 2007); thus, our primary measure of similarity was taken to be how often each 5-mer in the proteins in each set is found in the rest of the human proteome, and the methodology described below is specific for 5-mers. For completeness, however, the same analysis was also performed for 6- and 7-mers.
One potential problem with this approach is that the proteins in one set may have more homologues in the human proteome than those in the other set, and close homologues are likely to share many 5-mers. To address this problem, Blast searches were used to identify proteins in proteome B that were homologous to any protein in one of the two sets. Each protein in each set was used as a query sequence to Blast when searching the database of proteins in proteome B. For each of these queries, any protein in proteome B for which the E-value was less than 10−3 was deleted from the proteome, creating a new proteome called proteome C. This procedure was performed separately for each comparison; thus, 30 different versions of proteome C were created.
For each comparison, a graph was drawn where a given point (x,y) indicates that y per cent of the 5-mers in a given protein set (the proto-oncoprotein or control set) are found x times in that comparison's proteome C. Thus, 5-mers that are found often in the human proteome are found on the right-hand side of the graph, while those occurring rarely are found on the left-hand side. For instance, if the point (1,5.5) exists on the graph, then 5.5 per cent of the 5-mers in that protein set are found exactly once in proteome C. Note that a given protein in either comparison set may contain a 5-mer that is found only once in proteome B (i.e. the 5-mer occurs only in its source protein). Since this protein would have been deleted in the proteome B→proteome C filtering process, a point with x=0 is valid on the graph.
In this paper, a ‘rare 5-mer’ is defined as one that occurs zero times in proteome C. To get an idea of the range of times that a given 5-mer may occur, consider that EEEEE (the most frequent 5-mer) occurs 3454 times in the human proteome, while KPGSR occurs just eight times and ACNEW never occurs. We desired to determine whether there was a statistically significant difference in the occurrence of rare 5-mers in the proto-oncoprotein set versus the control set for each comparison. The number of 5-mers that occur zero times, as well as the number that occur one or more times, was computed for both sets, and a chi-squared test was performed to ascertain statistical significance. p<0.05 was considered significant (not corrected to account for multiple comparisons). Because our definition of a rare 5-mer is somewhat arbitrary, we have also computed p-values for two alternative definitions of a rare 5-mer—namely, one that occurs two or fewer times in proteome C, and one that occurs five or fewer times in proteome C.
3. Results
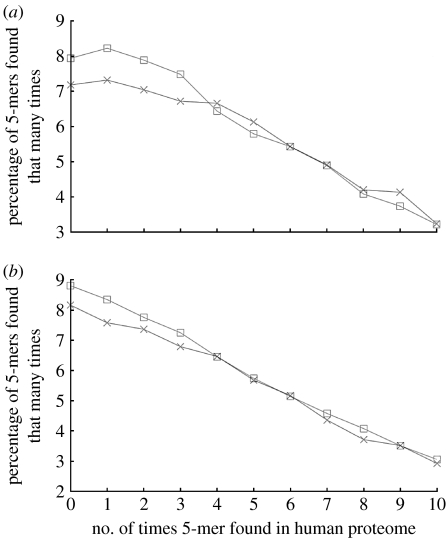
The results of the category 1 and 3 comparisons for 5-mers are shown in figure 1a,b. The most notable feature of both graphs is that the proportion of rare 5-mers (those found zero times in proteome C) is greater in the proto-oncoprotein set than in the control set. The chi-squared test indicates that the difference in occurrence is statistically significant: the proto-oncoprotein set had a significantly greater proportion of rare 5-mers in both the category 1 comparison (p=0.017) and the category 3 comparison (p=0.0014).
Figure 1.
(a) Percentage of 5-mers in the proto-oncoprotein set (squares) and the housekeeping set (crosses) that are found a given number of times in the human proteome. A point (x,y) indicates that y% of the 5-mers in that protein set are found x times in the human proteome. (b) Percentage of 5-mers in proto-oncogenic tyrosine kinases (squares) and non-proto-oncogenic tyrosine kinases (crosses) that are found a given number of times in the human proteome.
The difference in rare 5-mer frequency between the proto-oncoprotein and control sets is even more significant if the alternative definitions of a rare 5-mer are used. If a rare 5-mer is defined as one that occurs two or fewer times in proteome C, the p-value for the difference in rare 5-mer frequency between the proto-oncoprotein and control sets is 5.78×10−7 for the category 1 comparison and 5.16×10−9 for the category 3 comparison. Similarly, if a rare 5-mer is defined as one occurring five or fewer times in proteome C, then comparison 1 is significant with p=3.85×10−6, and comparison 3 is significant with p=8.89×10−11.
If other lengths of peptide are considered, the results are similar—for both the category 1 and 3 comparisons, the proto-oncoprotein sets contain a higher percentage of rare k-mers (for k=5, 6 and 7) for all three definitions of a rare k-mer. Full data for the category 1 and 3 comparisons are given in Tables 4 and 8 in the electronic supplementary material, respectively.
The results of the category 2 comparisons are summarized in table 1. For all three lengths of peptide considered (5-, 6- and 7-mers), the majority of the GO protein sets contained a significantly smaller percentage of rare k-mers than the proto-oncoprotein set. This was true for each of our possible definitions of a rare k-mer. Full data for the category 2 comparisons are given in tables 5–7 in the electronic supplementary material (for 5-, 6- and 7-mers, respectively). As described in §2.1, GO terms for which there were fewer than 10 corresponding proteins were not used in the analysis. However, the majority of the GO protein sets that were not included for this reason do still have a lower proportion of rare k-mers than the proto-oncoprotein set, with this difference being statistically significant (data not shown).
Table 1.
Percentage of the category 2 comparisons in which the proto-oncoprotein set had a significantly greater frequency of rare k-mers (p), the control set (i.e. the GO proteins) had a significantly greater frequency of rare k-mers (C) or the difference in rare k-mer frequency between the two sets of proteins was not statistically significant (n.s.). (p<0.05 was considered significant. These data are shown for k=5, 6 and 7, and for different definitions of a rare k-mer (found zero times in proteome C, found two or fewer times in proteome C or found five or fewer times in proteome C). Percentages may not sum to 100 due to rounding.)
| 5-mers | 6-mers | 7-mers | |||||||
|---|---|---|---|---|---|---|---|---|---|
| no. of occurrences | p (%) | C (%) | n.s. (%) | p (%) | C (%) | n.s. (%) | p (%) | C (%) | n.s. (%) |
| 0 | 71 | 4 | 25 | 71 | 7 | 21 | 82 | 7 | 11 |
| ≤2 | 75 | 4 | 21 | 93 | 7 | 0 | 86 | 4 | 11 |
| ≤5 | 71 | 4 | 25 | 93 | 4 | 4 | 71 | 4 | 25 |
It was suggested to us that the length of the proteins in each set may bias the results. The rationale for this is that evolutionary forces favouring an increase in the proportion of rare k-mers may be more constrained with shorter proteins compared with longer proteins due to the presence of other evolutionary forces acting on the smaller proteins. While the proto-oncoproteins in the category 1 and 2 comparisons do have an average length that is longer than most of the control sets, there were two category 2 comparisons having control sets with an average length longer than that of the proto-oncoprotein set (corresponding to the GO accession numbers GO0006941 and GO0030048), and both of these sets contain significantly fewer rare k-mers than the proto-oncoprotein set.
4. Discussion
In this paper, we have shown that k-mers occurring rarely in the human proteome are found in significantly greater proportions in proto-oncoproteins compared with control proteins. Although several explanations for this phenomenon are possible, we hypothesize that reduced self-similarity may have evolved in proto-oncoproteins in order to facilitate recognition of cancerous cells. The rationale for this hypothesis involves two major facts: first, T and B lymphocytes are targeted for apoptosis if they interact strongly to self-antigen (Kishimoto & Sprent 1997), and second, short peptides—particularly 5-mers—have been identified as a fundamental unit of antigenic recognition (Lucchese et al. 2007). As such, a reasonable conjecture is that the immune system is more likely to recognize k-mers that are rare in the human proteome than it is to recognize k-mers found frequently in the human proteome, because in the former case there would be fewer opportunities for negative selection to take place. Thus, a high proportion of rarely occurring k-mers in a particular protein may make that protein a better target for the organism's immune system. This may have a particularly strong effect on the organism's immune response in the cases where the oncogenicity of a particular protein is due to its overexpression, as opposed to being due to a somatic gene mutation.
Another interesting possibility is that low similarity might control/regulate crucial cell cycle functions. If rare peptide sequences represent peptides difficult to be synthesized, then their presence in proto-oncogenes (i.e. in molecular entities potentially dangerous when deregulated) would represent an additional device for controlling the cell cycle. On the other hand, peptide sequences with a high similarity level would fit with proteins performing basic, routine, ‘non-proliferative’ functions such as those performed by housekeeping proteins.
While the trend of proto-oncoproteins generally having a greater number of rare k-mers than control proteins was consistent for all three values of k, it is likely that only the differences at the 5- and 6-mer levels are biologically significant. With respect to 7-mers, the proportion of 7-mers found zero times in proteome C was very high—approximately 95 per cent—for all protein sets examined, making it unlikely that any differences observed would have biological significance. Further supporting this contention is that in most of the individual comparisons, the percentage of 7-mers in the control set found zero times in proteome C differed from that of the proto-oncoprotein set by less than 1 percentage point. If 95 per cent of the 7-mers in a protein are unique in that organism's proteome, then it seems unlikely that an additional 1 per cent would make a difference in terms of the organism's ability to recognize the overexpression or mutation of the protein. Thus, the differences in rare 7-mer frequency, while statistically significant due to the large number of 7-mers in each protein set, are unlikely to be biologically significant. As longer peptides repeat even less often than 7-mers, we have not analysed peptides longer than seven amino acids in this study. In addition, we have not also included an analysis of 4-mers, because they repeat too often (just 0.14% of all possible 4-mers are never found in the unfiltered human proteome, and almost 98% of all possible 4-mers are found more than five times), making it unlikely that any analysis performed using 4-mers would be biologically meaningful.
On the other hand, it is entirely plausible that the differences at the 5- and 6-mer levels are biologically significant. The proportion of 5-mers found zero times in proteome C for a given set of proteins is approximately 7–9 per cent; for a given comparison, the control set typically had a value between 0.5 and 2 percentage points lower than that of the proto-oncoprotein set. In nearly all comparisons, the magnitudes of the differences become greater if the definition of a rare 5-mer is extended to include those found two or fewer times in proteome C, or five or fewer times in proteome C. In the case of 6-mers, the proportion found zero times in proteome C is approximately 60–70 per cent for a given protein set, with the control set in an individual comparison typically having a value between 1 and 5 percentage points lower than that of the proto-oncoprotein set. Even a 5 percentage point difference in the proportion of rare 6-mers may seem unimportant, given that 60–70 per cent of 6-mers are rare in both the control and proto-oncoprotein sets. However, immune responses against any given target are generally limited to a small set of immunodominant epitopes (Rolland et al. 2007), and so a small increase in the number of potential immunodominant epitopes may create a large difference in the ability of the organism to mount an immune response. Furthermore, if our hypothesis is correct, it is possible that selective pressure may be concentrated in specific areas in the protein most amenable to immunological recognition—specifically, on the surface of the protein (for recognition by antibodies), and in areas of the protein likely to be recognized by T cells, such as those sites containing favourable motifs for proteasomal cleavage, major histocompatibility complex binding and so on. Future work could examine solved structures for various proto-oncoproteins to determine whether there is a bias in rare k-mer frequency in these regions.
The difference in rare 5-mer frequency between the proto-oncoprotein and control sets is lower in the category 3 comparison than in the category 1 comparison, as well as in most of the category 2 comparisons. This may be due to the ease in which the proteins can be classified into a proto-oncoprotein and a control set for each comparison. In the category 1 and 2 comparisons, the classifications can be assumed to be very accurate: all of the proteins in the proto-oncoprotein set have been well characterized as having oncogenic potential, while it is likely that few, if any, of the proteins in the control sets of the category 1 and 2 comparisons have oncogenic potential. By contrast, the tyrosine kinases used in comparison 3 were more difficult to correctly classify, and it is certainly possible that some of our classifications were erroneous. As more tyrosine kinases become well characterized, this classification task can become much more accurate.
With respect to the category 2 comparisons, the only GO term for which the corresponding proteins consistently contained a significantly greater percentage of rare k-mers was ‘carbohydrate metabolic process’. There is a growing body of knowledge (reviewed in, for example, Fuster & Esko (2005) and Vollmers & Brandlein (2007)) concerning the role of carbohydrates in cancer-related immunity, and it is possible that the rare k-mer frequency in proteins related to carbohydrate metabolism is somehow involved. Future work will attempt to characterize the role of these proteins, if any, in oncogenesis.
The data presented in this paper are applicable to both basic and clinical immunology. Given the premise that, for the most part, tumour-associated antigens are represented by proto-oncogenes that degenerate into uncontrollable oncogenes, low-similarity peptides from cell-cycle-associated oncoproteins might have strong implications for the rational development of peptide-based treatments for cancer diseases. From a clinical point of view, the most attractive feature of the similarity concept would be the guarantee of the highest specificity and the lowest cross-reactivity when designing effective and safe immunotherapeutic tools (Kanduc et al. 2007).
Acknowledgments
Funding for this work was provided in part by the Natural Sciences and Engineering Research Council of Canada (NSERC).
Supplementary Material
Supplementary tables that list the proteins used in the category 1 and 3 comparisons, as well as the GO terms used in the category 2 comparisons. Also included are tables showing the results for all 30 comparisons
Each row contains a GO accession number, followed by a comma-separated list of proteins corresponding to that GO term
References
- Altschul S.F, Madden T.L, Shaffer A.A, Zhang J, Zhang Z, Miller W, Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amela I, Cedano J, Querol E. Pathogen proteins eliciting antibodies do not share epitopes with host proteins: a bioinformatics approach. PLoS One. 2007;2:e512. doi: 10.1371/journal.pone.0000512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, et al. Gene ontology: tool for the unification of biology. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capone G, De Marinis A, Simone S, Kusalik A, Kanduc D. Mapping the human proteome for non-redundant peptide islands. Amino Acids. 2008;35:209–216. doi: 10.1007/s00726-007-0563-7. [DOI] [PubMed] [Google Scholar]
- Cho W.C.S. Contribution of oncoproteomics to cancer biomarker discovery. Mol. Cancer. 2007;6:25. doi: 10.1186/1476-4598-6-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuster M.M, Esko J.D. The sweet and sour of cancer: glycans as novel therapeutic targets. Nat. Rev. Cancer. 2005;5:526–542. doi: 10.1038/nrc1649. [DOI] [PubMed] [Google Scholar]
- Kanduc D, Lucchese A, Mittelman A. Non-redundant peptidomes from DAPs: towards “the vaccine”? Autoimmun. Rev. 2007;6:290–294. doi: 10.1016/j.autrev.2006.09.004. [DOI] [PubMed] [Google Scholar]
- Kishimoto H, Sprent J. Negative selection in the thymus includes semimature T cells. J. Exp. Med. 1997;185:263–271. doi: 10.1084/jem.185.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucchese G, Stufano A, Trost B, Kusalik A, Kanduc D. Peptidology: short amino acid modules in cell biology and immunology. Amino Acids. 2007;33:703–707. doi: 10.1007/s00726-006-0458-z. [DOI] [PubMed] [Google Scholar]
- Manning G, Whyte D.B, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- Polimeno L, Mittleman A, Gennero L, Ponzetto A, Lucchese G, Stufano A, Kusalik A, Kanduc D. Sub-epitopic dissection of HCV El315–328HRMAWDMMMNWSPT sequence by similarity analysis. Amino Acids. 2008;34:479–484. doi: 10.1007/s00726-007-0539-7. [DOI] [PubMed] [Google Scholar]
- Rolland M, et al. Recognition of HIV-1 peptides by host CTL is related to HIV-1 similarity to human proteins. PLoS One. 2007;2:e823. doi: 10.1371/journal.pone.0000823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollmers H.P, Brandlein S. Tumors: too sweet to remember? Mol. Cancer. 2007;6:78. doi: 10.1186/1476-4598-6-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Wang Y, You M. Identification of genetic polymorphisms through comparative DNA sequence analysis on the K-ras gene: implications for lung tumor susceptibility. Exp. Lung Res. 2005;31:165–177. doi: 10.1080/01902140490495543. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary tables that list the proteins used in the category 1 and 3 comparisons, as well as the GO terms used in the category 2 comparisons. Also included are tables showing the results for all 30 comparisons
Each row contains a GO accession number, followed by a comma-separated list of proteins corresponding to that GO term