Abstract
Anaphase in budding yeast is triggered by cleavage of the central subunit, Scc1, of the chromosomal cohesin complex by the protease separase. Here we show that separase also cleaves the kinetochore-associated protein Slk19 at anaphase onset. Separase activity is further required for proper localisation of a stable Slk19 cleavage product to the spindle midzone in anaphase. Slk19 cleavage and localisation are necessary to stabilise the anaphase spindle, and we show that a stable spindle is a prerequisite for timely exit from mitosis. This demonstrates separase cleavage of targets other than cohesin in the orchestration of high fidelity anaphase.
Introduction
Chromosomes in metaphase are under the tension of the mitotic spindle that attaches to their kinetochores, trying to separate sister chromatids. This tension is counteracted by cohesion between the sister chromatids, mediated by the DNA-binding multi-protein cohesin complex1-3. Anaphase onset is characterised by the sudden loss of cohesion between the sister chromatids and their poleward movement. In budding yeast, loss of cohesion is achieved by cleavage of the cohesin subunit Scc1 by the protease Esp1, called separase4-6. Scc1 cleavage destroys the cohesin complex and frees sister chromatids for segregation. After cleavage, the C-terminal Scc1 fragment is unstable, owing to its N-terminal arginine that is recognised by the N-end rule pathway of protein degradation7,8. Instability of the Scc1 cleavage fragment is vital for cells, possibly because a stable fragment interferes with cohesion in the next cell cycle9.
Once Scc1 is cleaved, the elongating mitotic spindle pulls the liberated sister chromatids into the daughter cells. The mitotic spindle is an assembly of microtubules originating at the spindle pole bodies (SPBs) where they are anchored with their minus ends. The dynamic microtubule plus ends reach out to capture the kinetochores on the chromosomes, and to form a region of overlap with the microtubules from the opposite SPB, the spindle midzone10,11. Spindle elongation in budding yeast is also thought to be the stimulus for the subsequent exit from mitosis. One SPB enters the daughter bud, and there the SPB-associated GTPase Tem1 is activated when it meets its exchange factor Lte1 (Ref. 12, 13). Lte1 is found concentrated at the cell cortex. It is not clear, however, whether to activate Tem1 it is sufficient for the SPB to enter the bud, or whether the SPB has to contact the cell cortex. Tem1 activation initiates a cascade, termed the mitotic exit network, that leads to completion of mitosis, including spindle breakdown, cyclin degradation and cytokinesis14.
The contribution of Scc1 cleavage to the anaphase programme has been analysed in budding yeast. Engineered Scc1 cleavage in metaphase, using TEV protease instead of separase, triggers complete sister chromatid separation and segregation to opposite cell poles by an elongating spindle5. This shows that the budding yeast metaphase spindle is prepared to enter anaphase, awaiting only sister separation by Scc1 cleavage. However, the spindle produced after TEV protease-triggered anaphase appears weak and breaks. In contrast, spindles produced after a normal separase-triggered anaphase are stable. Further, a striking localisation of separase to anaphase spindles has been described in both budding and fission yeast15-18.
Here, we address how separase stabilises the anaphase spindle and ask why yeast cells maintain a stable spindle even after chromosomes have segregated. We show that separase cleaves Slk19 in anaphase. Slk19 is found at kinetochores in metaphase and in addition at the spindle midzone in anaphase where it has been implicated in spindle stabilisation19. We find that a stable Slk19 cleavage product is present in anaphase, and that its absence leads to spindle breakage and a delay in mitotic exit. Separase has further roles in the organisation of a stable spindle midzone. This demonstrates a function of separase beyond cleavage of cohesin in the orchestration and execution of anaphase.
Slk19 is cleaved by separase during anaphase
We addressed whether the budding yeast separase Esp1 has a role in anaphase in addition to Scc1 cleavage by searching for potential additional cleavage targets. We aligned the four known separase cleavage sites in the cohesins Scc1 and Rec8 (Ref. 4, 20) to derive a consensus cleavage site motif (Fig. 1A). Only a few other proteins in the S. cerevisiae proteome contain this motif. Of these, Slk19 was of particular interest to us as it has previously been shown to be involved in spindle dynamics19.
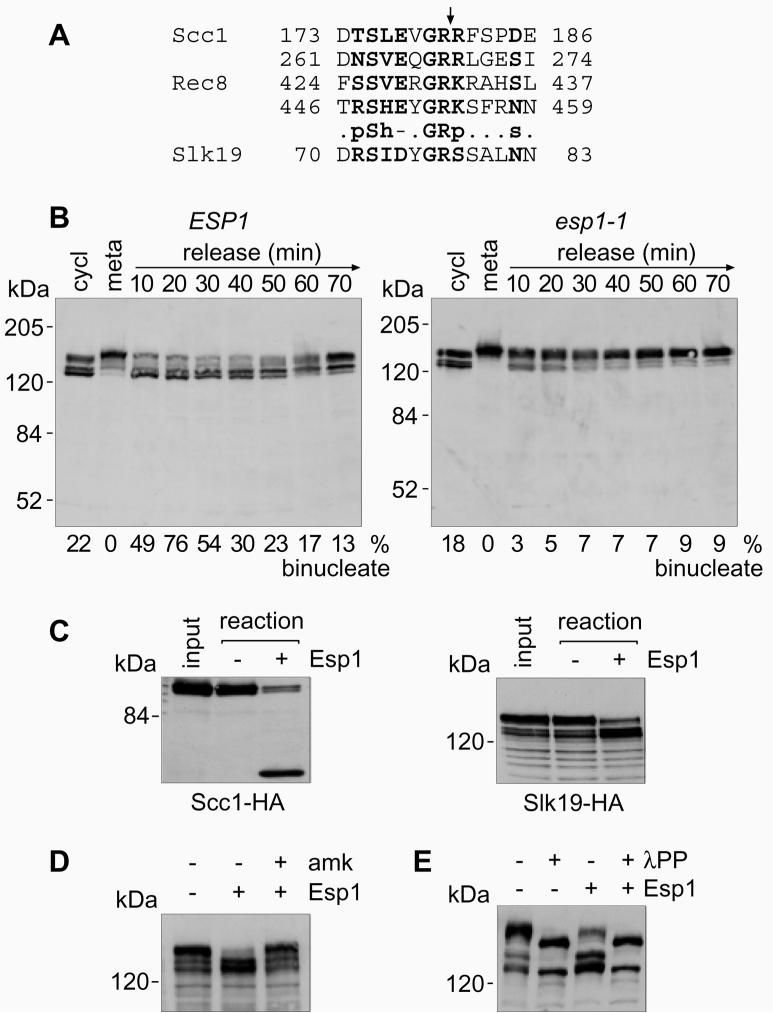
Figure 1. Slk19 is a cleavage target of separase.
A, Sequence alignment of the known separase cleavage sites in Scc1 and Rec8 (p: polar CDEHKNQRST, h: hydrophobic ACFGHILMVWY, -: negative charged DE, and s: small amino acids ADGNPSTV). The consensus was used to screen the S. cerevisiae proteome using the ISREC pattern finder (www.isrec.isb-sib.ch/software/PATFND_form.html). B, Metaphase arrest and release at 35.5°C of strains Y261 (MATa cdc20Δ GAL-CDC20 SLK19-HA6 SCC1-myc18) and Y254 (MATalpha esp1-1 cdc20Δ GAL-CDC20 SLK19-HA6). Protein extracts were analysed by Western blotting. Anaphase was monitored by DAPI staining. C, In vitro cleavage by separase of Scc1 and Slk19 in immunoprecipitates from strains K8869 (MATa SCC1-HA6) and Y138 (MATa SLK19-HA6). D, Specific inhibition of Slk19 cleavage by the separase inhibitor Bio-SVEQGR-amk. E, Cleavage of Slk19 requires its phosphorylation. Immunoprecipitates were treated with lambda protein phosphatase (λPP) before the cleavage reaction.
We first tested whether Slk19 is cleaved by separase at the metaphase to anaphase transition in vivo. To facilitate detection, C-terminal HA-epitopes were added to the SLK19 gene at its genomic locus. Western blot analysis of a logarithmically growing culture detected a number of protein bands around 120-140 kDa, close to the predicted molecular weight of Slk19 fused to the epitope tag (95 + 9 kDa). The status of Slk19 was examined in a culture that was arrested in metaphase and released into synchronous anaphase4 (Fig. 1B). In metaphase, Slk19 accumulated as a slower migrating form that was converted to faster migrating forms at the release into anaphase (Fig. 1B). The apparent doublet of bands produced in anaphase are phospho-isoforms (see below), and the difference in migration of Slk19 in metaphase and anaphase was estimated, using molecular weight standards, to be 13 kDa. This difference is consistent with cleavage at the predicted separase recognition site in Slk19, located 9 kDa from the N-terminus. In a strain containing a temperature sensitive mutation in separase, esp1-1, the processing of Slk19 during anaphase was strongly reduced at the restrictive temperature (Fig 1B), suggesting this processing might indeed be cleavage by separase. We noted that the Slk19 cleavage product produced in anaphase was stable and persisted in cells throughout the next cell cycle (Fig 1B). This is expected since the Slk19 cleavage product is predicted to carry an N-terminal serine, a stabilising residue in the N-end rule7,8.
To test more rigorously whether the change in Slk19 migration is due to cleavage by separase, we tested whether Slk19 is a substrate for separase cleavage in vitro. Slk19 was not stably bound to chromatin preparations that previously served as a substrate in separase cleavage assays4. We therefore established the use of immunoprecipitates of Scc1 and Slk19 from metaphase arrested cells as substrates (Fig. 1C). The source of separase in these reactions was yeast cell extract containing overproduced Esp1 (Ref. 4). Esp1-dependent proteolytic processing of Slk19 in vitro produced products of similar size as observed in anaphase in vivo (Fig. 1C). Importantly, this processing was prevented by addition of a specific peptide-based protease inhibitor against Esp1 (amk, Fig. 1D)5. This demonstrates that cleavage by separase is likely to be directly responsible for the mobility change of Slk19 during anaphase.
Cleavage of Scc1 by separase depends on its mitotic phosphorylation by polo-like kinase5,21. We addressed whether Slk19 is also phosphorylated in metaphase and whether phosphorylation of Slk19 contributes to its cleavage. When immunoprecipitates of Slk19 from metaphase arrested cells were treated with λ-protein phosphatase the multiple bands of Slk19 resolved into two sharp bands, corresponding to full-length and cleaved Slk19. Dephosphorylated Slk19 was no longer a substrate for cleavage by separase in vitro (Fig. 1E). This suggests that cleavage of Slk19, like Scc1, requires its phosphorylation.
Uncleavable Slk19 causes elevated chromosome loss
To address the role of Slk19 cleavage during anaphase we constructed a mutant protein in which the arginine preceding the proposed cleavage site was changed into glutamic acid (R77E). SLK19(R77E), as well as a wild type SLK19, were cloned under control of the SLK19 promoter and integrated at the TRP1 or URA3 locus into cells deleted for endogenous SLK19. Western blot analysis showed that Slk19(R77E) is no longer cleaved (Fig. 2A). Different integration multiplicities yielded transformants with varying expression levels. Two-fold integrants showed expression roughly comparable to expression from the endogenous SLK19 locus (Fig. 2A). Expression of uncleavable Slk19 had no obvious deleterious consequence for cell growth. We consider uncleavable Slk19 otherwise functional based on its localisation pattern that was indistinguishable from wild type protein (see below), and because it was able to correct for the metaphase spindle defect seen in the absence of Slk19 (Ref. 19, and data not shown).
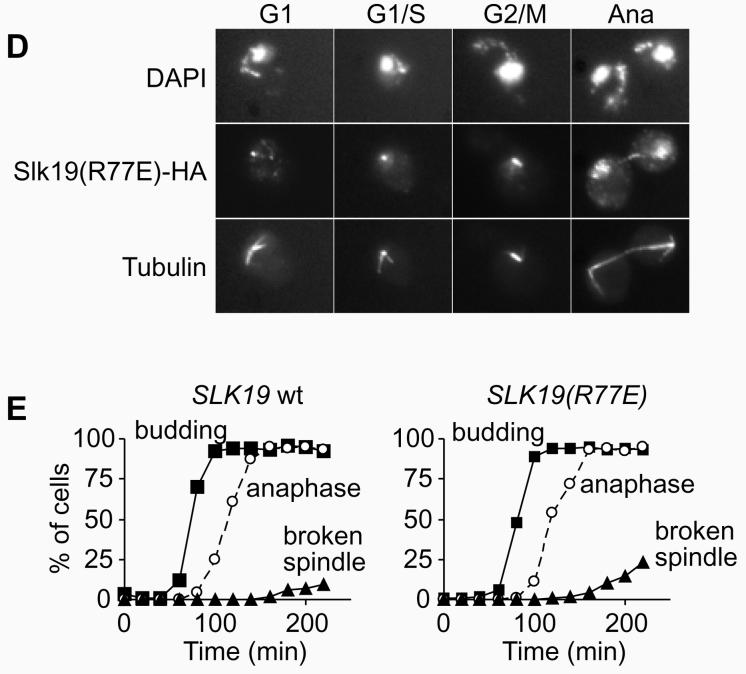
Figure 2. Phenotype of uncleavable Slk19.
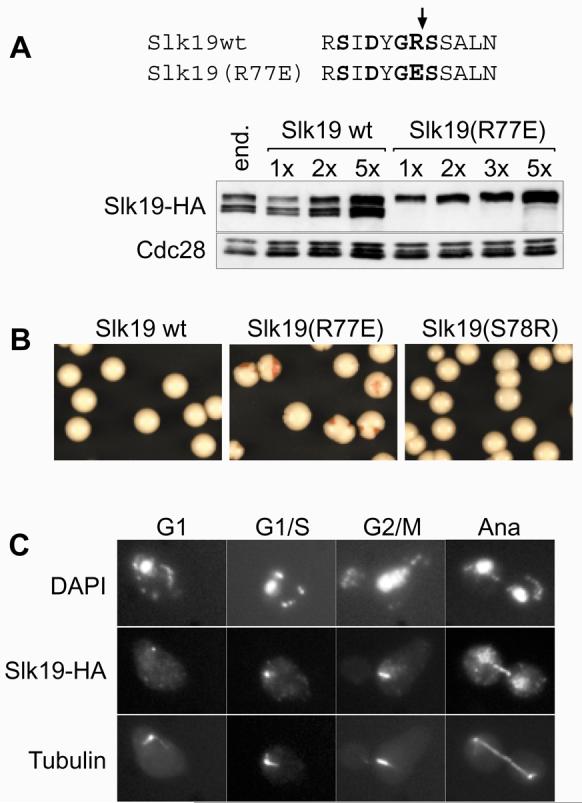
A, The R77E mutation in Slk19 prevents separase cleavage. Strains Y354 (MATa CFIII(CEN3.L.YPH278)URA3SUP11 SLK19-HA6), and (MATa CFIII(CEN3.L.YPH278)URA3SUP11 slk19Δ SLK19-HA6::TRP1xN) with the indicated integration multiplicities of SLK19 or SLK19(R77E) were analysed by Western blotting. B , Elevated expression of uncleavable Slk19 leads to increased levels of chromosome loss. Strains Y295 (MATa CFIII(CEN3.L.YPH278)URA3SUP11 slk19Δ SLK19-HA6x5), Y296 (MATa CFIII(CEN3.L.YPH278)URA3SUP11 slk19Δ SLK19(R77E)-HA6x5) and Y319 (MATa CFIII(CEN3.L.YPH278)URA3SUP11 slk19Δ SLK19(S78R)-HA6x5) were grown in medium lacking uracil and plated on rich medium. C, D, Strains Y305 (MATa cdc15-2 slk19Δ SLK19-HA6) and Y324 (MATa cdc15-2 slk19Δ SLK19(R77E)-HA6) were arrested by alpha factor treatment and released into telophase arrest at 37°C. Slk19-HA6 was stained with anti-HA monoclonal antibody 16B12 (Babco), and tubulin with monoclonal antibody YOL1/34 (Serotec). E, Quantitative analysis of the experiment in C and D. Spindles were scored as broken if there was no continuous tubulin signal visible between the spindle poles.
We next analysed the fidelity of chromosome segregation in these strains using a colony sectoring assay22. Uncleavable Slk19 caused a marked increase in the frequency of chromosome loss, however only when expressed at higher than endogenous levels in strains that contained five copies of the SLK19(R77E) gene (Fig. 2B). In such strains about half of the colonies showed red sectors indicative of chromosome loss, compared to less than 5% in controls. Expression of similar levels of wild type Slk19 did not cause an increased frequency of chromosome loss (Fig 2B). This suggests that although cleavage of Slk19 during anaphase is not essential, an accumulation of uncleaved Slk19 is incompatible with high fidelity chromosome segregation. An alternative explanation that uncleavable Slk19 competes for and inhibits separase cleavage of Scc1 was excluded: elevated levels of Esp1 could not reduce the chromosome loss. Esp1 expressed from a high copy number plasmid caused by itself a modest increase of chromosome loss (22% sectored colonies), presumably because of premature Scc1 cleavage. This effect was additive with chromosome loss caused by uncleavable Slk19 (63% sectored colonies, compared to SLK19(R77E) alone: 54%), indicating that both mechanisms of chromosome loss act independently.
We then analysed the localisation of Slk19 during the cell cycle and the stability of anaphase spindles, using strains expressing endogenous levels of wild type or uncleavable Slk19 (Fig 2C, D). For this, cells were synchronised by alpha factor block and release. After passing through a synchronous cycle, cells were arrested in telophase by the temperature sensitive cdc15-2 mutation23. In situ immunofluorescense showed wild type Slk19 as a weak single spot in G1 cells that increased in intensity at the G1/S boundary when synthesis of full-length Slk19 is transcriptionally upregulated (Fig. 2C). Uncleavable Slk19 presented a strong spot already in G1 cells that hardly increased in intensity during S-phase (Fig. 2D). The signal of both wild type and uncleavable Slk19 split towards metaphase and was found along the metaphase spindle, consistent with its localisation to kinetochores, as reported (Fig. 2C, D; and Ref. 19). In anaphase, both wild type and uncleavable Slk19 were found at the midzone of elongated anaphase spindles while the kinetochore staining was reduced or lost and a weak general nuclear staining became apparent. The loss of the kinetochore signal under our fixation conditions is in contrast to live cell imaging that shows persistent kinetochore localisation of Slk19 (Ref. 19, and data not shown), but it might indicate a reduced affinity of Slk19 for its binding partners at the kinetochore in anaphase. The elongated spindle in both cases remained initially stable. After longer times in telophase arrest spindles broke, and breakage was more pronounced in cells with uncleavable Slk19 (Fig. 2E). We conclude that cleavage of Slk19 is not essential for its association with the spindle midzone and has mild consequences on the stability of the anaphase spindle.
Destabilised Slk19 leads to spindle breakage
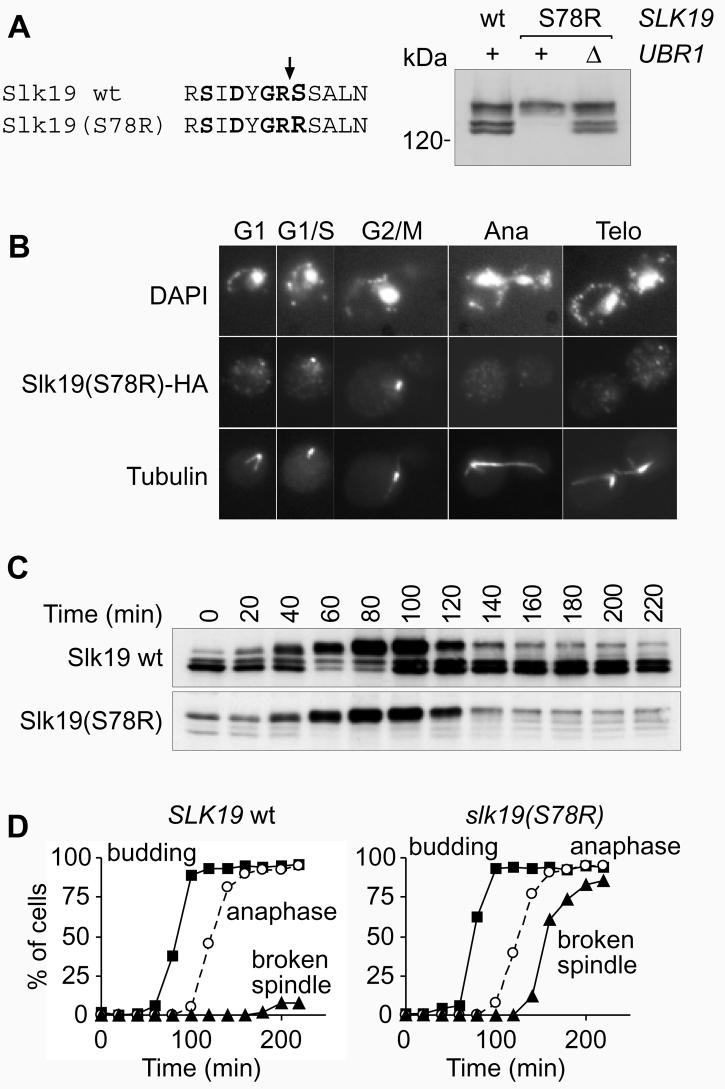
Most wild type Slk19 is cleaved at anaphase onset. Therefore not full-length Slk19, but the stable C-terminal Slk19 fragment that contains our epitope tag, is found on the anaphase spindle. To analyse more specifically the function of this Slk19 cleavage product we destabilised it, just as Scc1 cleavage products are unstable9, by making it a target for the N-end rule pathway. The P1′ residue in the Slk19 cleavage site, that becomes the N-terminus of the cleavage product, was changed from a serine into an arginine Slk19(S78R), i.e. from a stabilising into a destabilising residue in the N-end rule7,8. The cleavage product produced was now barely detectable, but could be stabilised again after inactivating the N-end rule pathway by deletion of UBR1 (Fig. 3A). As expected, Slk19(S78R) was present in cells from S-phase until in metaphase, but was no longer detectable in anaphase cells or in G1 (Fig. 3B).
Figure 3. The Slk19 cleavage product is required for spindle stability in anaphase.
A, Destabilisation of the Slk19 cleavage product. Western blot of extracts from strains Y284 (MATa slk19Δ SLK9-HA6), Y287 (MATa slk19Δ SLK19(S78R)-HA6), and Y323 (MATa slk19Δ SLK19(S78R)-HA6 ubr1Δ). B, Strains K1993 (MATa cdc15-2) and Y325 (MATa cdc15-2 slk19Δ SLK19(S78R)-HA6) were arrested with alpha factor and released into cdc15 arrest at 37°C. In situ immunofluorescense, as in Fig. 2, of strain Y325 at the indicated cell cycle stages. C, Strains Y305 and Y325 were arrested with alpha factor and released into cdc15 arrest at 37°C. D, Quantitative analysis of B. Spindles were scored as in figure 2E.
cdc15-2 cells containing wild type or destabilised Slk19 were again released from alpha-factor block and arrested in the following telophase. Western blot analysis showed that wild type and destabilised Slk19 were present in metaphase cells at similar levels, that both proteins were similarly cleaved in anaphase, but that the destabilised Slk19 cleavage fragment immediately disappeared (Fig 3C). This allowed us to specifically examine the role of the Slk19 cleavage product on the anaphase spindle. In cells containing destabilised Slk19 the spindle became thin, broke, and disappeared within 30 min after anaphase (Fig. 3B, D). This shows that after cleavage of Slk19 in anaphase the continued presence of the C-terminal cleavage product is crucial to maintain the spindle. Previous analysis of slk19Δ cells had indicated a role of Slk19 in spindle stabilisation in anaphase19. However, spindles in slk19Δ cells were abnormal already in metaphase making defects in anaphase difficult to interpret.
Uncleavable Slk19(R77E) had only a mild effect on anaphase spindle stability, but it caused increased chromosome loss if overproduced. If spindle instability was the reason for chromosome loss, then this effect should be even more pronounced in strains with destabilised Slk19 where spindles break much faster. The rate of chromosome loss, however, was not significantly increased over wild type controls (Fig. 2B). This indicates that spindle breakdown after anaphase is most likely not a reason for chromosome loss.
Spindle stability and the timing of mitotic exit
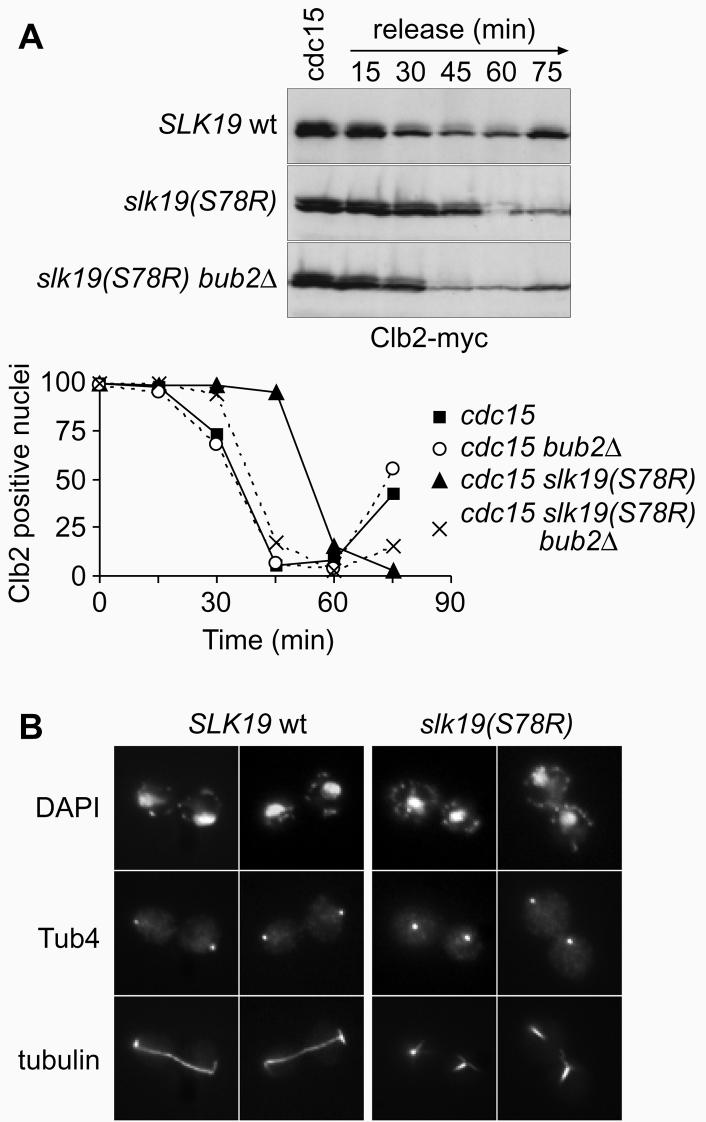
What is the reason for wild type cells to maintain a stable anaphase spindle? We noted that cells arrested in telophase using the cdc15-2 allele produced a bud elongation after longer times in the arrest, indicative that some aspects of the cell cycle had resumed. This failed to occur in similarly treated cells with destabilised Slk19 (data not shown). We therefore compared exit from mitosis after returning cells from cdc15 arrest to the permissive temperature. In cultures with destabilised Slk19 we observed a 30 min delay in the degradation of the mitotic cyclin Clb2 (Fig. 4A). This demonstrates a requirement for Slk19, and therefore possibly a stable spindle, for timely exit from mitosis.
Figure 4. Delay of mitotic exit after spindle breakdown.
A, Delayed degradation of Clb2 after release from telophase. Strains Y342 (MATa cdc15-2 slk19Δ SLK19-HA6 CLB2-myc9), Y344 (MATa cdc15-2 slk19Δ SLK19(S78R)-HA6 CLB2-myc9), Y450 (MATa cdc15-2 slk19Δ SLK19-HA6 CLB2-myc9 bub2Δ) and Y451 (MATa cdc15-2 slk19Δ SLK19(S78R)-HA6 CLB2-myc9 bub2Δ) were released from telophase arrest at 37°C into mitotic exit at 23°C. Protein extracts were prepared and analysed by Western blotting. Clb2 degradation in individual cells was seen by immunofluorescense. B, SPB mislocalisation in telophase arrested cells. Cells in the arrest from A were immunostained for tubulin and Tub4, a component of the spindle pole body (anti-Tub4 antibody was a kind gift of J. Kilmartin).
Because mitotic exit is initiated by the SPB-associated Tem1, we analysed the positioning of the SPBs in cdc15 arrested cells (Fig. 4B). Cells containing wild type Slk19 had intact spindles that brought the SPBs into close contact with the cell cortex. In contrast, after spindle breakage in cells with destabilised Slk19 the SPBs were found inside the cell, distant from the cortex. If the cause for the observed delay in mitotic exit is indeed lacking activation of Tem1 by Lte1 at the cell cortex, then removal of the negative regulator Bub2 should compensate for the delay. The delay of mitotic exit with destabilised Slk19 was almost completely suppressed by the deletion of Bub2 (Fig. 4A), indicating that the positioning of the SPB at the cell cortex might indeed contribute to the regulation of mitotic exit. Alternatively the integrity of the mitotic spindle or the presence of Slk19 might regulate mitotic exit by another yet unknown mechanism. We also analysed logarithmically growing populations of cells. The fraction of binucleates, i. e. cells after anaphase but before cytokinesis, was 10% in wild type but 21% in cells with destabilised Slk19. The majority of these binucleates (75%) stained positive for Clb2, indicating a delay before mitotic exit. Deletion of Bub2 in these cells reduced the fraction of binucleates again to 11%. This shows that a delay in mitotic exit caused by Slk19 destabilisation also affects progression through an otherwise undisturbed cell cycle.
Separase activity required for Slk19 localisation
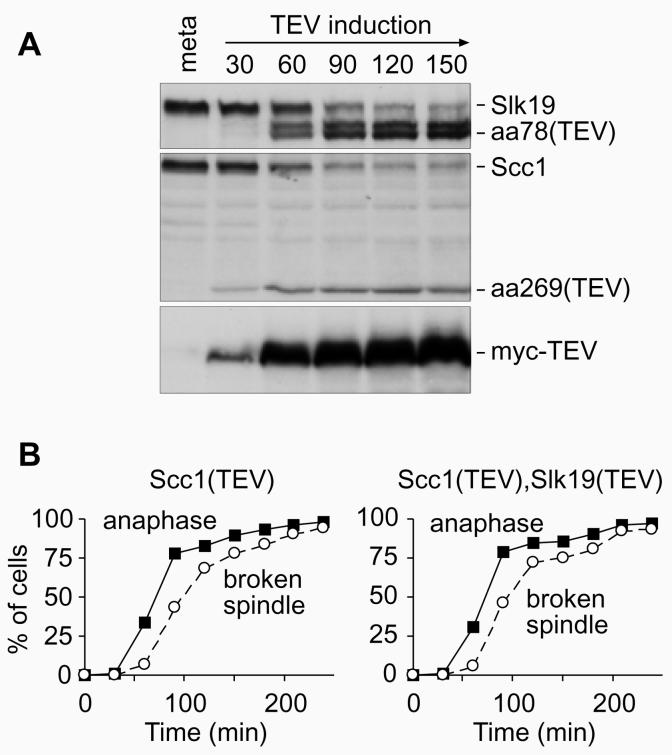
Spindles showed an increased tendency to break in cells containing uncleavable Slk19 (Fig. 2), but spindle instability was more severe if Slk19 was absent in anaphase (Fig. 3). How do these findings relate to the spindle instability after engineered, TEV-mediated, anaphase5? There, Slk19 was present just not cleaved. We therefore wondered whether cleavage of Slk19 together with Scc1 is sufficient to restore spindle stability after TEV-mediated anaphase. A TEV recognition sequence was engineered to replace the separase cleavage motif in Slk19, and this variant Slk19 was introduced into a strain that also contained TEV-cleavable Scc1. This strain was arrested in metaphase, and upon expression of TEV protease Slk19 was cleaved concomitant with Scc1 (Fig. 5A). We compared the spindle stability in this strain to a strain in which only Scc1 was cleaved. Anaphase efficiently proceeded in both strains, but also in both strains anaphase spindles appeared weak and broke within 30 min (Fig. 5B). This shows that cleavage of Slk19 is not alone sufficient to promote its function to stabilise the anaphase spindle.
Figure 5. Cleavage of Slk19 by TEV protease.
A, Cleavage o f Scc1 and Slk19 by TEV protease in vivo. Strain Y327 (MATa scc1Δ SCC1-TEV-HA3 slk19Δ SLK19-TEV-HA6 GAL-NLS-myc9-TEV-NLS2x10 MET3-CDC20) was arrested in metaphase by depletion of Cdc20, and TEV protease expression was induced. Protein extracts were prepared for Western blot analysis. B, As A, but strain Y116 (MATalpha scc1Δ SCC1-TEV-HA3 GAL-NLS-myc9-TEV-NLS2x10 MET3-CDC20) was used alongside. Samples were prepared for DNA staining with DAPI and immunostaining against tubulin.
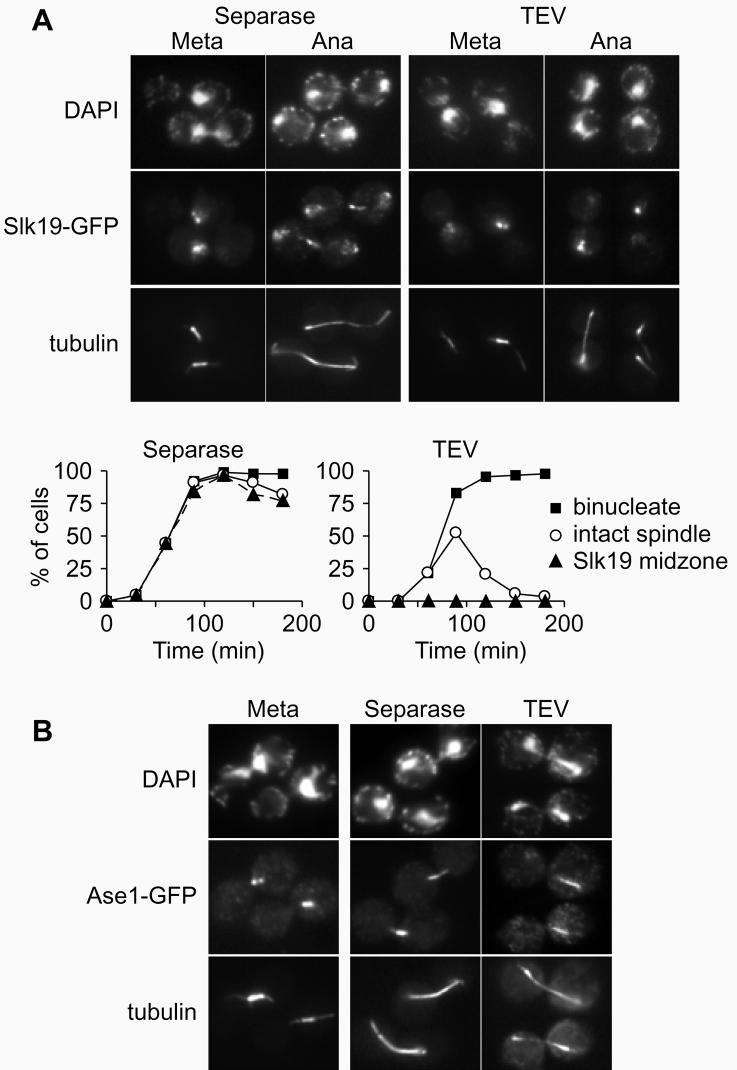
We therefore asked whether separase activity might be necessary not only to cleave Slk19 during anaphase but also for its proper localisation to the spindle midzone. We observed Slk19, tagged with GFP, in metaphase arrested cells when anaphase was triggered by ectopic expression of Esp1. As expected, the kinetochore signal of Slk19 segregated into the two daughter cells, and Slk19 was found decorating the stable spindle midzone in almost all of the cells (Fig. 6A). In contrast, when anaphase was triggered by TEV protease in a strain containing TEV-cleavable Scc1, no midzone staining was visible on any of the anaphase spindles that soon broke (Fig. 6A). Furthermore, Slk19 segregated unequally into the daughter cells, the signal on one side was much weaker than on the other (Fig. 6A). This shows that separase not only cleaves Slk19, but that it is also required for the localisation of Slk19 to the spindle midzone.
Figure 6. Separase activity in anaphase is required for Slk19 and Ase1 localisation.
A, Separase dependent localisation of Slk19 to the spindle midzone in anaphase. Strains Y359 (MATalpha scc1Δ Scc1-TEV-HA3 SLK19-GFP GAL-NLS-myc9-TEV-NLS2x10 MET3-CDC20) and Y406 (MATalpha SLK19-GFP GAL-ESP1x6 MET3-CDC20) were arrested in metaphase and expression of TEV protease or Esp1 was induced. Indirect immunofluorescense against tubulin and Slk19-GFP, using an anti-GFP antibody (a kind gift of K. Sawin), is shown. B, Quantitation of A. C, Separase dependent localisation of Ase1 to the spindle midzone in anaphase. As in A, but using strains Y442 (MATalpha ASE1-GFP GAL-ESP1x6 MET3-CDC20) and Y443 (MATalpha scc1Δ Scc1-TEV-HA3 ASE1-GFP GAL-NLS-myc9-TEV-NLS2x10 MET3-CDC20).
The failure of Slk19 to localise correctly might have been for two reasons. Separase might be required to bring Slk19 to the spindle midzone, or separase might be required for the formation of a proper spindle midzone to which Slk19 can bind. To differentiate between these possibilities we observed the behaviour of another protein, Ase1, that localises to the spindle midzone and is required for anaphase spindle stability24. In anaphase triggered by separase, Ase1 was found at the stable spindle midzone (Fig. 6B). In contrast, in TEV-mediated anaphase Ase1 staining on the spindle was weak and mislocalised to a region close to either one of the spindle pole bodies in most cells (86 out of 89 cells observed) (Fig. 6B), before the signal disappeared when the spindle broke. This was not a consequence of missing Slk19 at the spindle midzone, as Ase1 localisation appeared normal in a slk19Δ strain (93 of 100 anaphase spindles observed). It seems therefore that the spindle after TEV-mediated anaphase is fundamentally changed. This indicates that separase might act to reorganise spindle dynamics at anaphase onset.
Discussion
The separase Esp1 is a unique site-specific protease in budding yeast, and probably all eukaryotic cells, that is activated during anaphase. The only characterised targets for cleavage by separase have so far been subunits of the chromosomal cohesin complex whose cleavage dissolves the cohesion between sister chromatids. Further involvement of Esp1 during anaphase has been postulated, but the mechanistic basis for this has remained unclear18,25. We have now shown that the kinetochore and spindle associated protein Slk19 is cleaved by separase during anaphase. Unlike cohesins, whose C-terminal cleavage products are rapidly degraded via the N-end rule, the Slk19 cleavage product is stable. This is important for cleaved Slk19 to stabilise the anaphase spindle. Accumulation of uncleaved Slk19 is incompatible with high fidelity chromosome segregation, and consistently overexpression from the strong GAL1 promoter of full-length Slk19, but not of a fragment corresponding to cleaved Slk19, was toxic to cells (data not shown). The reason for this is still unclear. Nevertheless, this demonstrates that separase cleavage can not only destroy proteins, like the cohesins, but that cleavage can also generate new protein species with altered properties.
Furthermore, and independent of its cleavage, the localisation of Slk19 to the spindle midzone in anaphase depended on separase. Another protein at the spindle midzone, Ase1, also mislocalised when anaphase was triggered by TEV protease instead of separase. The spindle midzone is thought to be the region where microtubule plus ends from opposite SPBs overlap10, and Slk19 and Ase1 might be involved in protecting the plus ends from shrinkage thereby stabilising the anaphase spindle. A normal spindle midzone failed to form during anaphase without separase. Indeed, separase and Slk19 might act in the same pathway to stabilise the spindle, as the anaphase spindle after TEV-mediated anaphase broke with similar kinetics as when Slk19 was absent. The molecular difference between separase and TEV-mediated anaphase spindles, however, remains to be elucidated. How could separase participate in microtubule plus end stabilisation? During TEV protease-triggered anaphase, separase was present in the cells but bound by securin. Securin inhibits the proteolytic activity of separase4, so cleavage of another yet unidentified protein might be necessary to promote spindle stability. We can not exclude the possibility that another function of separase, independent of its proteolytic activity, is required for spindle stability and is inhibited by securin.
Cleavage of cohesin subunits by separase has apart from budding yeast also been demonstrated in fission yeast and in human cells26,27. Are there additional cleavage targets for separase also in these organisms? Cleavage of spindle associated proteins might be of even greater importance in higher eukaryotes. While in budding yeast cleavage of cohesin is in principle sufficient to trigger anaphase, this is not true in all organisms. The metaphase to anaphase transition at least in a Xenopus oocyte cell free system depends not only on destroying sister chromatid cohesion, but likewise on the APC-dependent degradation of the chromokinesin Xkid28. It seems conceivable that also the specific and regulated protease activity of separase is used at anaphase onset to restructure components of the spindle apparatus. An example might be CENP-E that acts at the interface between the kinetochore and spindle microtubules in vertebrates, which has been observed in two isoforms of distinctly different size but unknown origin29. Our finding that separase cleavage sites can be identified by sequence analysis should encourage similar studies also in other organisms.
Material and Methods
Plasmids and yeast strains
The SLK19 promoter and open reading frame was amplified from yeast genomic DNA using the polymerase chain reaction (PCR) and cloned into YIplac211 and YIplac204 (Ref. 30). At the C-terminus, a sequence encoding 6 copies of the HA epitope was inserted. PCR fragments containing the R77E and S78R point mutations, and the replacement of the separase cleavage site (amino acids 72-78) with the TEV cleavage consensus (ENLYFQG)31, were used to replace wild type sequences. All constructs were verified by DNA sequencing. Integration into the genome was analysed by Southern blotting. Epitope tagging of endogenous ASE1, CLB2, SCC1, and SLK19 (Ref. 32), and genomic deletions of BUB2, SCC1,SLK19, and UBR1 (Ref. 33) were performed using one-step PCR strategies. Replacement of the CDC20 promoter for the MET3 promoter was as described5.
Cell cycle analysis
Cells were grown in YEP medium34 containing 2% glucose, 2% raffinose, or 2% raffinose plus 2% galactose as the carbon source at 25°C if not otherwise stated. Cells were blocked in G1 by addition of 0.3 μg/ml mating pheromone alpha-factor, followed by further 0.2 μg/ml and 0.1 μg/ml after 1 and 2 hours respectively. After 2.5 hours cells were >95% unbudded and were released into medium lacking alpha factor. Arrest in metaphase and release into anaphase using GAL-CDC20 (Ref. 4), arrest and release from telophase using the temperature sensitive cdc15-2 mutation35, and arrest in metaphase using MET-CDC20 and induction of the TEV protease or Esp1 5 were as described. The colony sectoring assay for chromosome loss was performed as previously described22.
Cytology
Cells were fixed for indirect immunofluorescense in 3.7% formaldehyde for 1 hour at 30°C and processed according to standard procedures36.
Separase cleavage assay using immunoprecipitated substrates
200 ml of exponentially growing yeast cells (107 cells) were arrested in metaphase by addition of nocodazole (5 μg/ml) for 2.5 hours, washed in cold 50 mM HEPES/KOH pH7.5 and resuspended in 0.4 ml of buffer W1 (50 mM HEPES/KOH pH7.5, 70 mM KOAc, 20 mM β-glycerophosphate, 5 mM Mg(OAc)2, 10% w/v glycerol, 0.1 % Triton X-100, and protease inhibitors). 1 g of glass beads were added for cell disruption by agitation. The extract was cleared by centrifugation and 1 μl of 16B12 mouse anti-HA ascites fluid (Babco) was added. After 1 hour, immunocomplexes were bound to 30 μl of protein A-sepharose. Beads were washed, and aliquots used for the separase cleavage assay. For this assay the beads were resuspended in extract prepared from yeast cells overexpressing Esp1 (or a control extract) and incubated as described previously4. After incubation the beads were washed and resuspended in SDS-PAGE loading buffer for Western blot analysis. The peptide inhibitor Bio-SVEQGR-amk was used as described previously5.
Acknowledgements
We thank J. Kilmartin for his kind gift of the anti-Tub4 antibody, K. Sawin for the anti-GFP antibody, K. Nasmyth for yeast strains and for his support and encouragement, and J. Diffley and T. Toda for critical comments on the manuscript.
References:
- 1.Nasmyth K, Peters J-M, Uhlmann F. Splitting the chromosome: cutting the ties that bind sister chromatids. Science. 2000;288:1379–1384. doi: 10.1126/science.288.5470.1379. [DOI] [PubMed] [Google Scholar]
- 2.Hirano T. Chromosome Cohesion, Condensation, and Separation. Annu. Rev. Biochem. 2000;69:115–144. doi: 10.1146/annurev.biochem.69.1.115. [DOI] [PubMed] [Google Scholar]
- 3.Koshland DE, Guacci V. Sister chromatid cohesion: the beginning of a long and beautiful relationship. Curr. Opin. Cell Biol. 2000;12:297–301. doi: 10.1016/s0955-0674(00)00092-2. [DOI] [PubMed] [Google Scholar]
- 4.Uhlmann F, Lottspeich F, Nasmyth K. Sister-chromatid separation at anaphase onset is promoted by cleavage of the cohesin subunit Scc1. Nature. 1999;400:37–42. doi: 10.1038/21831. [DOI] [PubMed] [Google Scholar]
- 5.Uhlmann F, Wernic D, Poupart M-A, Koonin EV, Nasmyth K. Cleavage of cohesin by the CD clan protease separin triggers anaphase in yeast. Cell. 2000;103:375–386. doi: 10.1016/s0092-8674(00)00130-6. [DOI] [PubMed] [Google Scholar]
- 6.Uhlmann F. Secured cutting: Controlling separase at the metaphase to anaphase transition. EMBO Rep. 2001 doi: 10.1093/embo-reports/kve113. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bachmair A, Finley D, Varshavsky A. In vivo half-life of a protein is a function of its amino-terminal residue. Science. 1986;234:179–186. doi: 10.1126/science.3018930. [DOI] [PubMed] [Google Scholar]
- 8.Varshavsky A. The N-end rule: functions, mysteries, uses. Proc. Natl. Acad. Sci. USA. 1996;93:12142–12149. doi: 10.1073/pnas.93.22.12142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rao H, Uhlmann F, Nasmyth K, Varshavsky A. Degradation of a cohesin subunit by the N-end rule pathway is essential for chromosome stability. Nature. 2001;410:955–959. doi: 10.1038/35073627. [DOI] [PubMed] [Google Scholar]
- 10.Winey M, et al. Three-dimensional ultrastructural analysis of the Saccharomyces cerevisiae mitotic spindle. J. Cell Biol. 1995;129:1601–1615. doi: 10.1083/jcb.129.6.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Winey M, O'Toole ET. The spindle cycle in budding yeast. Nat. Cell Biol. 2001;3:E23–E27. doi: 10.1038/35050663. [DOI] [PubMed] [Google Scholar]
- 12.Bardin AJ, Visintin R, Amon A. A mechanism for coupling exit from mitosis to partitioning of the nucleus. Cell. 2000;102:21–31. doi: 10.1016/s0092-8674(00)00007-6. [DOI] [PubMed] [Google Scholar]
- 13.Pereira G, Höfken T, Grindlay J, Manson C, Schiebel E. The Bub2p spindle checkpoint links nuclear migration with mitotic exit. Mol. Cell. 2000;6:1–10. [PubMed] [Google Scholar]
- 14.McCollum D, Gould KL. Timing is everything: regulation of mitotic exit and cytokinesis by the MEN and SIN. Trends Cell Biol. 2001;11:89–95. doi: 10.1016/s0962-8924(00)01901-2. [DOI] [PubMed] [Google Scholar]
- 15.Funabiki H, Kumada K, Yanagida M. Fission yeast Cut1 and Cut2 are essential for sister chromatid separation, concentrate along the metaphase spindle and form large complexes. EMBO J. 1996;15:6617–6628. [PMC free article] [PubMed] [Google Scholar]
- 16.Ciosk R, et al. An Esp1/Pds1 complex regulates loss of sister chromatid cohesion at the metaphase to anaphase transition in yeast. Cell. 1998;93:1067–1076. doi: 10.1016/s0092-8674(00)81211-8. [DOI] [PubMed] [Google Scholar]
- 17.Kumada K, et al. Cut1 is loaded onto the spindle by binding to Cut2 and promotes anaphase spindle movement upon Cut2 proteolysis. Curr. Biol. 1998;8:633–641. doi: 10.1016/s0960-9822(98)70250-7. [DOI] [PubMed] [Google Scholar]
- 18.Jensen S, Segal M, Clarke DJ, Reed SI. A novel role of the budding yeast separin Esp1 in anaphase spindle elongation: evidence that proper spindle association of Esp1 is regulated by Pds1. J. Cell Biol. 2001;152:27–40. doi: 10.1083/jcb.152.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zeng X, et al. Slk19p is a centromere protein that functions to stabilize mitotic spindles. J. Cell Biol. 1999;146:415–425. doi: 10.1083/jcb.146.2.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Buonomo SBC, et al. Disjunction of homologous chromosomes in meiosis I depends on proteolytic cleavage of the meiotic cohesin Rec8 by separin. Cell. 2000;103:387–398. doi: 10.1016/s0092-8674(00)00131-8. [DOI] [PubMed] [Google Scholar]
- 21.Alexandru G, Uhlmann F, Poupart M-A, Mechtler K, Nasmyth K. Phosphorylation of the cohesin subunit Scc1 by Polo/Cdc5 kinase regulates sister chromatid separation in yeast. Cell. 2001;105:459–472. doi: 10.1016/s0092-8674(01)00362-2. [DOI] [PubMed] [Google Scholar]
- 22.Hieter P, Mann C, Snyder M, Davis RW. Mitotic stability of yeast chromosomes: a colony color assay that measures nondisjunction and chromosome loss. Cell. 1985;40:381–392. doi: 10.1016/0092-8674(85)90152-7. [DOI] [PubMed] [Google Scholar]
- 23.Fitch IT, et al. Characterization of four B-type cyclin genes of the budding yeast Saccharomyces cerevisiae. Mol. Biol. Cell. 1992;3:805–818. doi: 10.1091/mbc.3.7.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Juang Y-L, et al. APC-mediated proteolysis of Ase1 and the morphogenesis of the mitotic spindle. Science. 1997;275:1311–1314. doi: 10.1126/science.275.5304.1311. [DOI] [PubMed] [Google Scholar]
- 25.Tinker-Kulberg RL, Morgan DO. Pds1 and Esp1 control both anaphase and mitotic exit in normal cells and after DNA damage. Genes Dev. 1999;13:1936–1949. doi: 10.1101/gad.13.15.1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tomonaga T, et al. Characterization of fission yeast cohesin: essential anaphase proteolysis of Rad21 phosphorylated in the S phase. Genes Dev. 2000;14:2757–2770. doi: 10.1101/gad.832000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Waizenegger IC, Hauf S, Meinke A, Peters J-M. Two distinct pathways remove mammalian cohesin complexes from chromosome arms in prophase and from centromeres in anaphase. Cell. 2000;103:399–410. doi: 10.1016/s0092-8674(00)00132-x. [DOI] [PubMed] [Google Scholar]
- 28.Funabiki H, Murray AW. The Xenopus chromokinesin Xkid is essential for metaphase chromosome alignment and must be degraded to allow anaphase chromosome movement. Cell. 2000;102:411–424. doi: 10.1016/s0092-8674(00)00047-7. [DOI] [PubMed] [Google Scholar]
- 29.Abrieu A, Kahana JA, Wood KW, Cleveland DW. CENP-E as an essential component of the mitotic checkpoint in vitro. Cell. 2000;102:817–826. doi: 10.1016/s0092-8674(00)00070-2. [DOI] [PubMed] [Google Scholar]
- 30.Gietz RD, Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- 31.Dougherty WG, Cary SM, Parks TD. Molecular genetic analysis of a plant virus polyprotein cleavage site: a model. Virology. 1989;171:356–364. doi: 10.1016/0042-6822(89)90603-x. [DOI] [PubMed] [Google Scholar]
- 32.Knop M, et al. Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast. 1999;15:963–972. doi: 10.1002/(SICI)1097-0061(199907)15:10B<963::AID-YEA399>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 33.Wach A, Brachat A, Pöhlmann R, Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- 34.Rose MD, Winston F, Hieter P. Laboratory course manual for methods in yeast genetics. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1990. [Google Scholar]
- 35.Spellman PT, et al. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol. Biol. Cell. 1998;9:3273–3297. doi: 10.1091/mbc.9.12.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hagan IM, Ayscough KR. In: Protein localization by fluorescence microscopy. Allan VJ, editor. Oxford University Press; Oxford: 2000. pp. 179–208. [Google Scholar]