Abstract
Adult tissue stem cells (SCs) share functional properties regardless of their tissue of residence. It had been thought that SCs might also share expression of certain “stemness” genes, though early investigations for such genes were unsuccessful. Here, we show that SCs from diverse tissues do preferentially express certain types of genes and that SCs resemble other SCs in terms of global gene expression more than they resemble the differentiated cells (DCs) of the tissues that they supply. Genes associated with nuclear function and RNA binding were over-represented in SCs. In contrast, DCs from diverse tissues shared enrichment in genes associated with extracellular space, signal transduction, and the plasma membrane. Further analysis showed that transit amplifying cells could be distinguished from both SCs and DCs by heightened expression of cell division and DNA repair genes and decreased expression of apoptosis-related genes. This transit-amplifying cell specific signature was confirmed by de novo generation of a global expression profile of a cell population highly enriched for transit amplifying cells: colonic crypt-base columnar cells responding to mucosal injury. Thus, progenitor cells preferentially express intracellular or biosynthetic genes, and differentiation correlates with increased expression of genes for interacting with other cells or the microenvironment. The higher-order, GO-term based analysis we use to distinguish SC- and DC-associated gene expression patterns can also be used to identify intermediate differentiation states (e.g., that of transit amplifying cells) and, potentially, any biological state that is reflected in changes in global gene expression patterns.
Keywords: Adult Stem Cells, Tissue-specific Stem Cells, Genomics, Gene Expression Profiling, Progenitor Cells
Introduction
The source of the constant stream of new differentiated cells (DCs) in continuously renewing adult tissues, such as skin and the gastrointestinal tract, is a cohort of resident stem cells (SCs). Because SCs from diverse tissues share properties such as the capacity for self-renewal and maintenance of an undifferentiated state, it has been argued that they may also share expression of a subset of specific genes 1, 2. However, others have argued that there is no genetic program of "stemness" and that the genes SCs express are as diverse as the tissues they supply 3–5. Within the past year, two studies have shown that certain patterns (modules) of gene espression that are enriched in embryonic SCs are also enriched in certain tissue SCs and in tumor cells 6, 7. Those studies suggest that there may be similarities in higher-order patterns of gene expression among diverse SCs, even if no specific genes are universal markers of the stem cell state.
Here, we address this controversy using a novel approach to analyze 45 different gene expression profiles from various tissues and sources. We show that SCs from diverse tissues do preferentially express certain types of genes and that SCs from one tissue resemble SCs from another tissue more than they resemble DCs from the same tissue. In addition, our results provide a working definition for the process of differentiation at the molecular level, where differentiation is the transition from expression of genes regulating intracellular processes to those required for cell communication and signaling.
The earliest comprehensive surveys of gene expression in adult tissue SCs were described several years ago.1, 2, 8 Since then, there has been a steady increase in published expression profiles of SCs 9–13, and the functional properties of diverse tissue-specific SCs have been examined in more detail 14–17. Global gene expression profiles have also been performed on progenitor and DCs in tissues such as the gastric and intestinal epithelium, where stem cells have been morphologically – more than functionally or molecularly – characterized 18–22.
Recently, we and others have developed algorithms that can determine overlap in patterns of gene expression among large expression profile datasets. These new algorithms compare expression profiles based not only on the individual genes within each profile but on the biological functions and properties those genes encode 23–26. For example, we have previously developed the GOurmet software to allow investigators to systematically condense a gene expression profile into a quantitative, parsable expression of its inherent functions and interrelationships (Fig. 1A). These re-expressed profiles can then be quantitatively compared (e.g., by hierarchical clustering) with multiple other profiles so that broad, shared patterns in gene expression can be determined. An advantage of this higher-order approach toward identifying shared patterns of gene expression is that data from multiple laboratories derived from multiple technologies (e.g., GeneChip, EST libraries), each with their own standards for measuring expression levels of individual genes, can be compared on equal footing 23, hus minimizing errors known to be introduced by cross-laboratory comparisons 27.
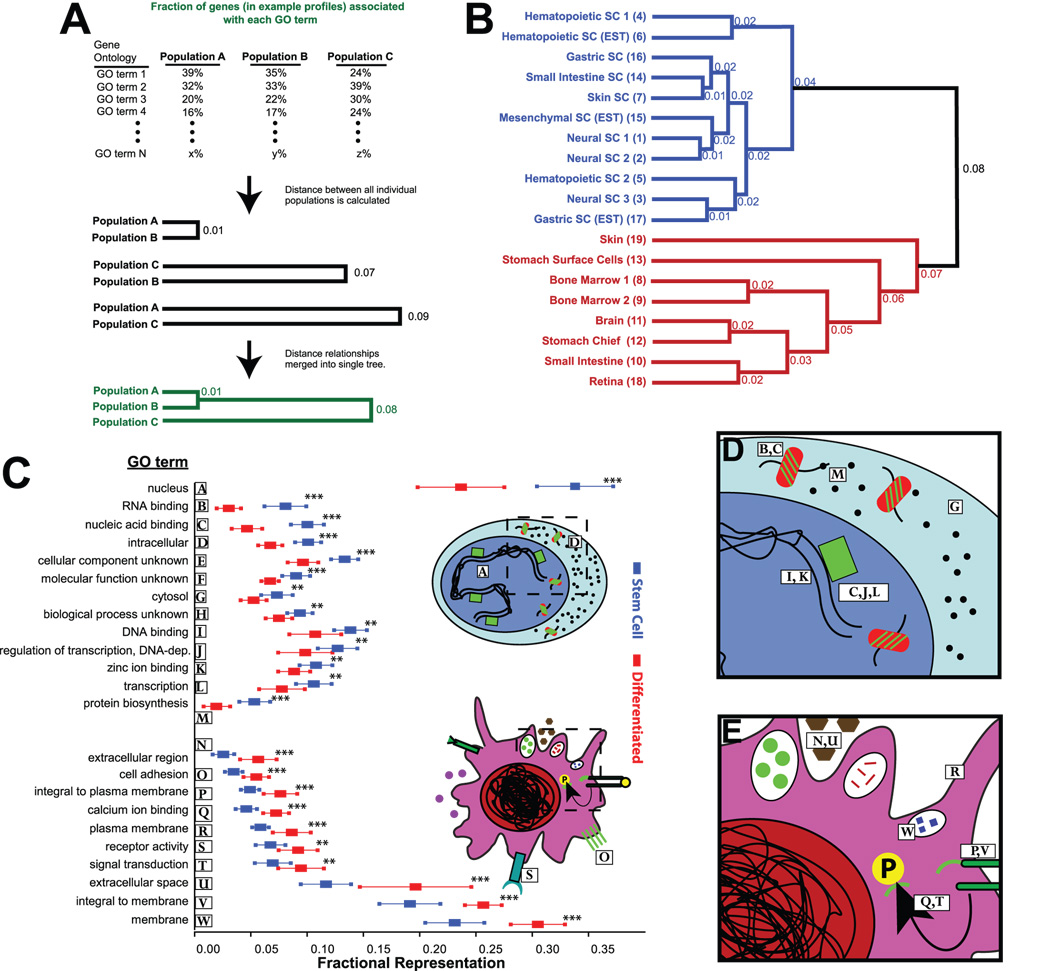
Fig. 1. Gene Ontology (GO) terms can be used to determine higher-order patterns of gene expression among diverse gene expression profiles.
A) Gene expression profiles from given cell populations (i.e., a list of genes preferentially expressed in, e.g., “Population A”) can be re-expressed as a function of the GO terms with which the genes in the profile are associated. The relative frequency of each GO term can be compared across multiple cell populations, and the overall similarity of each profile to every other profile can be determined based on the inherent distribution of GO terms. Finally, profile similarities can be plotted as a dendrogram. B) Dendrogram calculated as for panel A, from a dataset of stem/progenitor cells and differentiated progeny. Numbers (calculated as 1−Pearson’s coefficient of similarity) represent dissimilarity between the two profiles at the branch depicted. C) Table of GO terms determined to be statistically different when comparing SCs and DCs. ** – p of 0.01 to 0.001, ***– p<0.001. Note that those terms marked *** also meet the Bonferoni post hoc correction for significance when testing multiple hypotheses, where n=38, the number of GO terms representing on average ≥5% of the genes in either SC or DC profiles. Depicted are arithmetic means and standard deviations for each GO term in both groups. Inset - Illustrations depicting enriched characteristics by GO term (numbered as in table), in SCs and DCs. D,E) Higher magnifications of cartoons in inset of panel C.
Materials and Methods
Collection and preparation of expression profiles
In this study, the term “expression profile” denotes a cell-population-specific list of genes and the distribution of GO terms associated with those genes. In all cases, expression profiles were generated by first determining the genes preferentially expressed in a given cell population and then using the GOurmet software to determine the fractional representation of all the GO terms associated with that list of genes. The lists of genes expressed preferentially in each cell population were determined by several means and assembled from numerous published and unpublished sources, and, in one case, the list was generated specifically for the current manuscript (see below, Supplemental Experimental Procedures, and Tables 1 and 2 for details).
Preparation of GeneChips from dividing colonic crypt cells
All experiments involving animals were performed according to protocols approved by the Washington University School of Medicine Animal Studies Committee. Genotypes used were 1) conventionally raised Rag1−/− (containing TA cells), 2) germ-free wildtype, and 3) conventionally raised Myd88−/− mice (all C57BL/6 background; n=5,000 cells/mouse; n=3 mice/group). Highly proliferating, crypt base epithelial cells (TA cells) were laser-capture microdissected from Rag1−/− mice in the region of the descending colon bordering ulcers induced by one-week administration of 2.5% DSS in the drinking water. Control, non-stimulated crypt base epithelial cells were captured from the equivalent region of DSS-treated germ-free and Myd88−/− mouse controls. For each group, total cellular RNA was extracted, purified (PicoPure RNA isolation kit, Arcturus; Mountain View, CA), pooled and split into two samples. Duplicate samples for each group were individually amplified (RiboAmp HS RNA Amplification Kit Arcturus) to generate labeled cRNA probes to hybridize to Affymetrix MOE430A GeneChips. To identify a TA-specific gene list, Rag1−/− GeneChips were subtracted sequentially from each of the controls (wildtype germ free and conventionally raised Myd88−/−).
Conversion of cell-specific gene lists into GO distributions
Lists of genes were converted to Gene Ontology (GO) profiles using GOurmet Vocabulary of the GOurmet package 23. Clustering and relation of GO profiles in dendrogram format, using 1−Pearson’s coefficient as a metric, were performed using GOurmet Cartography. On this scale, a score of 0 means the two compared expression profiles have the same fractional representations for every GO term, and a score of 1 means the two profiles show no similarity. Additional analysis was performed using Spotfire Decisionsite 9.0 (Spotfire, Sommerville, MA) to generate relational trees using UPGMA (Unweighted Pair Group Method with Arithmetic mean) of Euclidean distances between the different GO profiles. We noted that there were no changes in the relative distribution of the various expression profiles using the two methods (data not shown).
Statistical analyses
Statistical analyses were performed in GraphPad Prism with verification of selected analyses using Stata10 software. GeneChips were analyzed with dChip threshold set at ≥1.3 fold lower bound of 90% confidence interval for each comparison and a difference intensity ≥100. The specific comparisons used to generate the expression profiles for each cell population can be found in Supplemental Table 3.
To analyze GO terms that distinguished SCs from DCs in the initial dataset (Fig. 1B, Supplemental Table 1), we calculated the mean fractional representation of all GO terms across all SC profiles and across all DC profiles. Rare GO terms (i.e., those associated with <5% of the genes in both the SC and DC profiles) were excluded from further analysis (however, see also Supplemental Experimental Procedures). Thirty-eight GO terms met the 5% threshold. To assess statistically significant differences, independent, two-tailed Student’s t-tests, assuming unequal variance, were performed analyzing the SC vs. DC means in pairwise fashion for all 38 GO terms. Twenty-three GO terms (all plotted in Fig. 1C) distinguished SCs from DCs (p<0.05). See supplemental procedures for additional statistical validation of this approach. We followed a similar approach to analyze TA-enriched GO terms, but set the GO term fractional representation at 1% and, to filter terms with only slight, but statistically significant changes, we further required that GO term representation be either increased or decreased by 0.7-fold on a log2 scale (Supplemental Table 4).
Results and Discussion
To test the hypothesis that all adult tissue stem cells might share certain genetic programs or patterns of gene expression, we generated lists of genes enriched in multiple adult stem/progenitor cell populations (SCs) and DC populations (DC) derived from the same tissues (Supplemental Table 1). SC- or DC-enrichment was determined by bioinformatic subtraction from a reference population; in most cases, SCs were referenced to DCs from the same tissue and vice versa (see Supplemental Experimental Methods and Supplemental Table 3 for details). We then determined the distributions of GO terms associated with the gene lists for each population and performed unbiased hierarchical clustering of all the SC and DC expression profiles using these GO term distributions (see Supplemental Workbook for an example of how gene expression profiles were converted to GO term distributions). With this approach, all the proposed SC profiles clustered together, and all the DC profiles clustered together (Fig. 1B). Thus, SCs from multiple tissues share higher level patterns of gene expression that transcended the laboratory and methods used, and profiles from functionally less well characterized SC populations, such as stomach and small intestine, cluster definitively with well defined SC profiles.
Our results suggested that specific genetic processes and functions may help maintain cells in an undifferentiated (stem) cell state. To identify those shared genetic functions, we determined which GO terms were correlated with the SC expression profiles relative to those from DCs. This analysis showed that of the 38 GO terms associated with ≥5% of genes on average in SCs and/or DCs, 13 correlated with the SC state with p<0.02 (Figure 1C; significance in this case determined by p test with two tails, assuming unequal variance; see Supplemental Experimental Procedures and Supplemental Figs. 2 and 3). “RNA binding” and its parent within the GO hierarchy, “nucleic acid binding”, were more than twice as common on average in SCs relative to DCs (for RNA binding: 8.1±1.8% vs. 3.1±1.1%, for Nucleic acid binding: 10.0±1.4% vs. 4.8±1.3%; Fig. 1C). Some GO terms representing a large fraction of the genes in both stem and DCs did not exhibit any discriminating potential (“protein binding”: 31.7±3.8% vs. 31.6±2.0%).
Surprisingly, though DCs must use widely divergent individual genes to perform functions unique to each tissue, there were certain higher-order patterns of gene expression that they all shared, regardless of their tissue of residence. For example, the term “membrane” was highly and consistently overrepresented in DCs relative to SCs (30.2±2.2% vs 23.1±2.5%, p<4×10−6). The capacity for self-renewal in part distinguishes SCs from DCs, which are either entirely post-mitotic or tend to proliferate more rarely. However, proliferation does not appear to account for the global differences in gene expression between SCs and DCs, because GO terms associated with proliferation, e.g., “cell cycle” (4.5±1.5% in SCs vs. 3.6±2.7%), “regulation of progression through cell cycle” (3.9±0.8% vs. 3.1±1.0%) , and “cell division” (2.2±1.1% vs. 1.9±1.9%) showed no statistically significant differences. Such GO terms are more important in defining highly proliferative populations such as transit amplifying cells (see below) and not SCs, which are only facultatively proliferative.
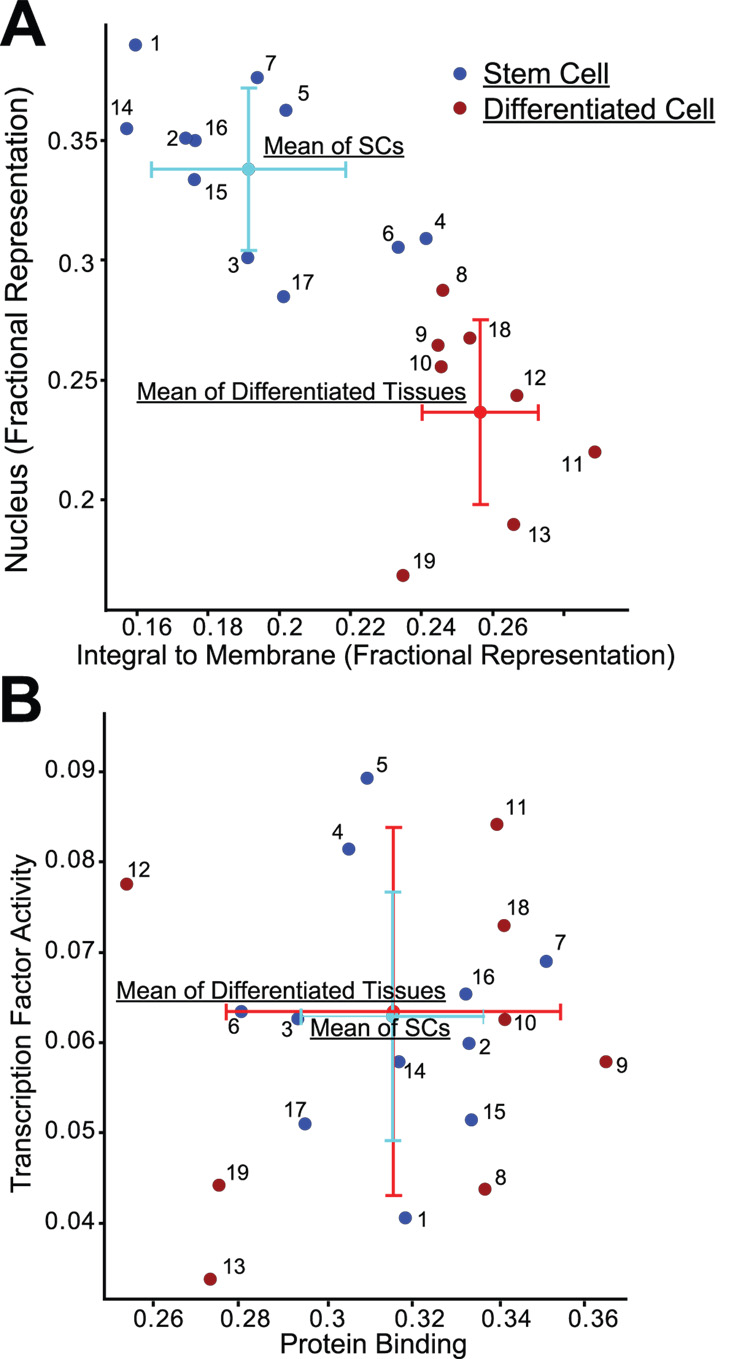
Given that certain GO terms were statistically significantly associated with SC expression profiles and others with DC expression profiles, we reasoned that plotting expression profiles on axes defined by a single DC-enriched GO term and a single SC-enriched GO term would be sufficient to distinguish all SC profiles from all DC profiles. Figure 2A shows how plotting the fractional representation of the terms “nucleus” vs. “integral to membrane” in all the lists completely distinguished SC from DC profiles with no profiles overlapping. Other pairs of GO terms have similar properties: “RNA binding” vs. “Calcium ion binding”, for example (data not shown). In contrast, plotting “transcription factor activity” vs. “protein binding” gave an entirely different pattern with no differentiation-dependent clustering of gene expression, showing that not all categories of gene function distinguish stem cells from their progeny (Fig. 2B).
Fig. 2. Stem and differentiated cell expression profiles can be categorized using fractional representation of only two GO terms.
A,B) Points representing profiles from SCs (blue) and DCs (red) are plotted along with the means and standard deviations of the entire SC and DC populations; expression profiles identified by number in Supplemental Table 1. A) GO terms showing statistically significant capacity to distinguish SCs from DCs. B) GO terms showing no statistically significant difference between progenitor and DCs.
Our results showing consistent higher-order patterns of gene expression can be explained biologically. In general, GO terms enriched in stem/progenitor cells describe intracellular processes, especially those associated with regulation of transcription and translation/biosynthesis: for example, “nucleus”, “intracellular”, “RNA binding”, “protein biosynthesis” (Fig. 1C–E). On the other hand, DCs are enriched for cell-cell communication and extracellular processes, e.g.: “membrane”, “extracellular space”, “receptor activity”, “signal transduction”. Thus, regardless of tissue, differentiation might be defined in molecular terms as the process of decreasing expression of genes involved in maintaining or building the intracellular state and increasing those for communicating with or modifying the extracellular environment. In other words, differentiation may represent a cell’s transformation from introversion to extroversion.
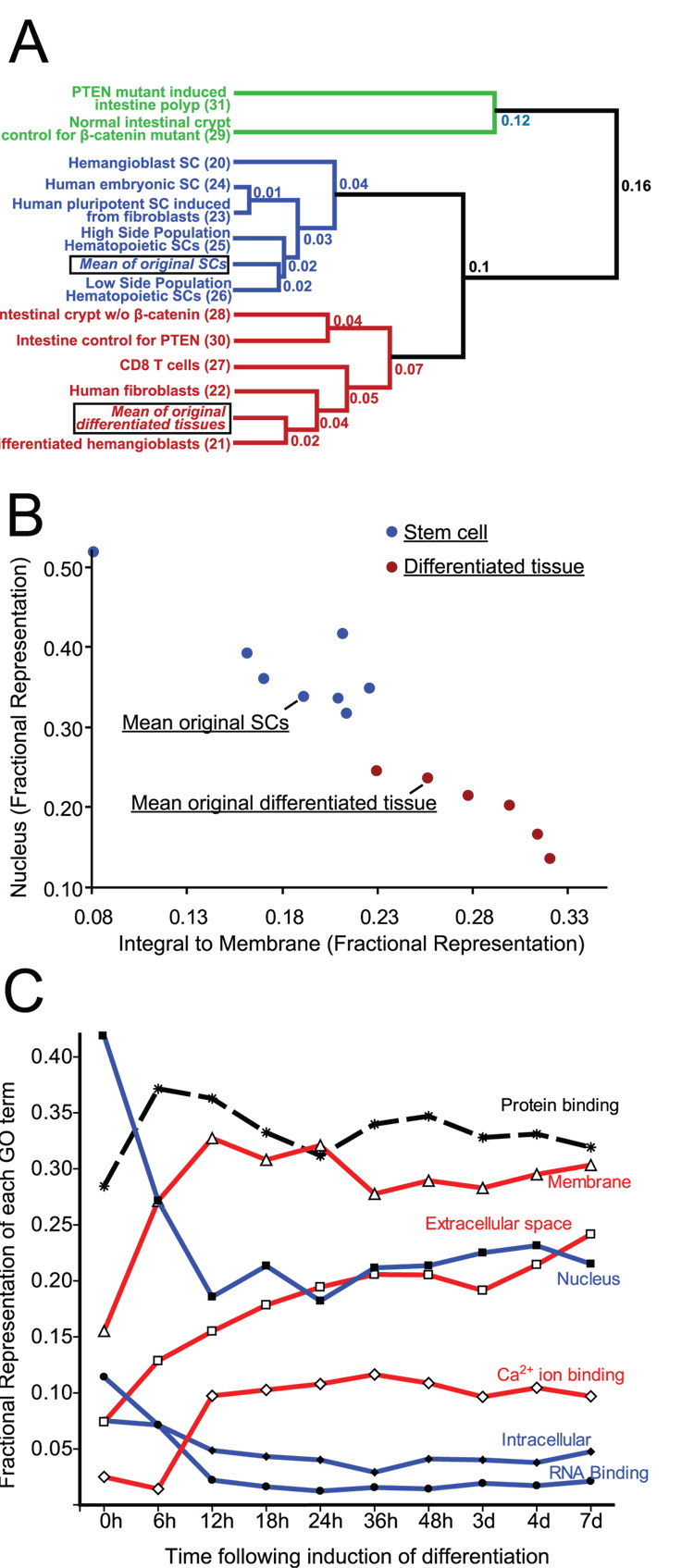
Given how tightly associated certain GO terms are with differentiation state, we reasoned that GO term distributions of gene expression profiles would aid in interpretation of future functional genomic results. For example, using the approach in Fig. 2A, fractional representation of only a few key GO terms such as “integral to membrane”, “nucleus”, and “RNA binding” within a gene expression profile from an unknown cell type might be useful as a shorthand to rapidly classify a cell population whose differentiation state is unknown as either a progenitor or differentiated cell. To test this hypothesis, we acquired multiple additional datasets from various tissues (Supplemental Table 2). From the GEO repository, we analyzed mouse expression profiles of: hematopoietic SCs isolated by Hoechst dye efflux (low and high side populations) using CD8 T cells as a DC reference population 28; hemangioblasts induced from embryonic SCs referenced to their differentiated progeny 29; intestinal polyps (i.e., hyperproliferative cells) induced by genetic deletion of PTEN and normal intestinal control 30; and laser capture microdissected intestinal crypt cells and profoundly hypoproliferative cells microdissected from β-catenin-deleted crypts 31. We also acquired data from an experiment where differentiated human fibroblasts were induced to de-differentiate into multipotent SCs by forced expression of Oct4, Sox2, Klf4, and Myc 32; from that study, we analyzed expression profiles of differentiated fibroblasts, induced pluripotent SCs and control embryonic SCs.
Fig. 3A shows that all the new profiles of DCs clustered together, and that this clustering was remarkably independent of species (human vs. mouse) using this GO term-based clustering analysis. Fig. 3B shows how most of the new SC and differentiated populations were even more divergent from each other than the previous set, using only “nucleus” and “integral to membrane” as distinguishing GO terms. Many of the new profiles were done in multiple replicates. Supplemental Fig. 1 shows how technical replicates (i.e., the same RNA run on independent microarrays) clustered together nearly perfectly, and biological replicates (i.e., independent biological experiments of the same tissue or conditions) also clustered together. In all cases, datasets followed the same SC vs differentiated pattern.
Fig. 3. Comparing expression profiles using GO term distributions allows clustering of stem from differentiated cells in a prospective series of additional datasets.
A) Dendrogram of multiple additional expression profiles on datasets acquired after the set depicted in Fig. 1 using the same approach. Means of previous SC and DC expression profiles are included for reference. Note: three of the new profiles are from human cells. The green branch depicts putative TAs. B) Cartesian plot as in Fig. 1 of additional expression profiles. C) Fractional representation of key distinguishing GO terms plotted vs. time following induction of differentiation in expression profiles from a population of human myoblasts. Note that whereas differentiation-enriched GO terms like “extracellular” increase as a function of differentiation and “nucleus” decreases, the non-distinguishing term “protein binding” shows no particular trend.
To further demonstrate how expression levels of a few key GO terms can identify differentiation state, we analyzed another dataset, wherein human fetal myoblasts had been induced to differentiate 33 and global gene expression assayed at 9 timepoints following induction of differentiation 34. Fig. 3C shows how the DC GO terms “membrane” and “calciumion binding” rose quickly within the first two timepoints and stayed at stable levels of representation at all subsequent time points. The differentiated term “extracellular” showed a smooth, progressive increase throughout the timecourse. SC-associated GO terms “nucleus” and “RNA binding” showed rapid sustained decrease within the first two timepoints, and the SC-associated “intracellular” decreased slowly to 36h.
Interestingly, two populations of non-DCs were outliers on the new dendrogram and the scatter plot (Fig. 3A,B). We hypothesized that those cell populations might be unusual in their high rate of cell renewal: intestinal polyps that are by definition abnormally proliferative 30 and normal crypts, containing transit amplifying cells and rare SCs, referenced to crypts where proliferation had been completely inhibited by genetic deletion of β-catenin 31. Thus, these gene expression profiles might be better biologically classified as transit amplifying cells (TAs) rather than SCs, whose capacity for self-renewal is inducible and not constant.
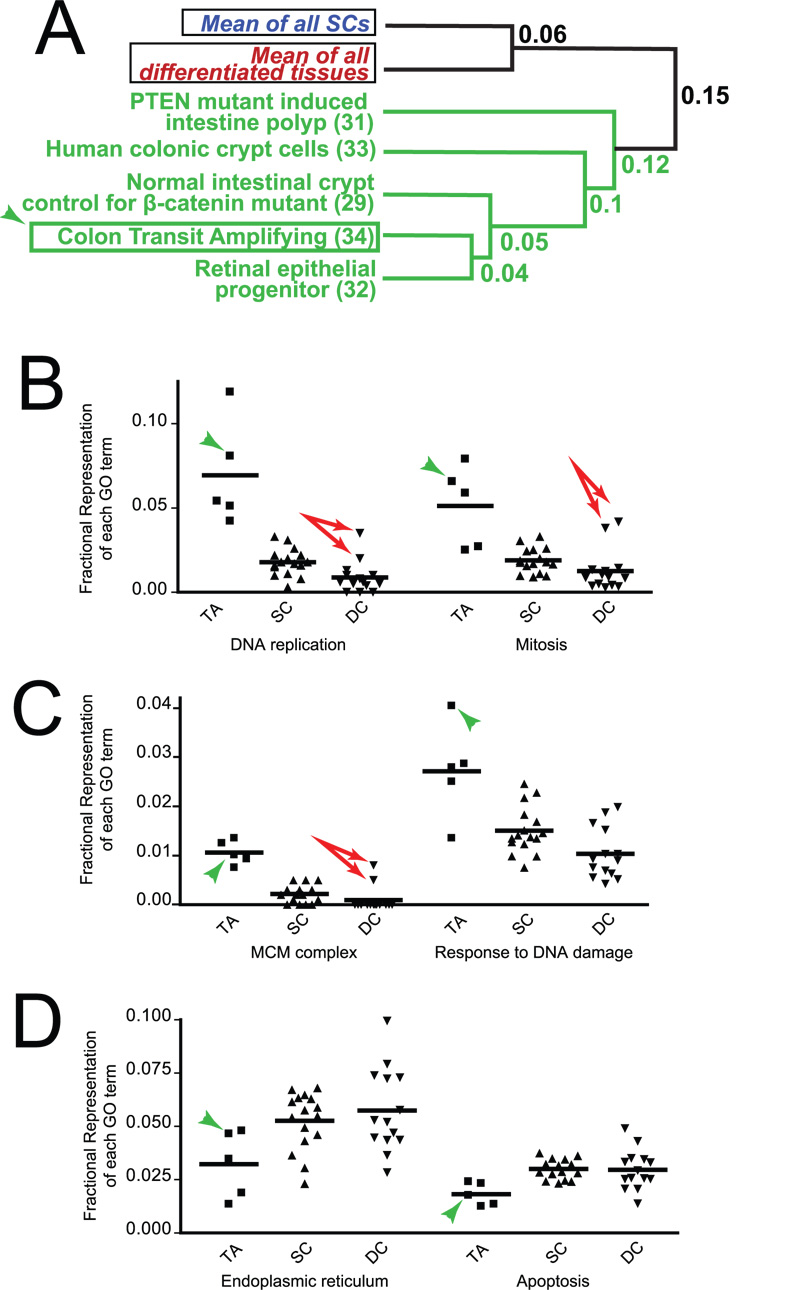
To determine whether there were patterns of gene expression associated with TAs, we acquired additional expression profiles of putative TA phenotype: 1) mouse retinal progenitor cells, which have neuroblastic properties, referenced to adult retina; 5 and 2) human colonic crypt epithelium referenced to human colonic surface epithelium 35. Both the crypt and retinal progenitor populations clustered with putative TAs and away from both SCs and DCs (Fig. 4A), whereas the corresponding differentiated populations clustered with DCs (not shown).
Fig. 4. Transit amplifying cells form a distinct cluster.
A) Putative TA expression profiles from Fig. 3A are plotted with prospectively acquired additional TA samples (all green) and with a de novo generated purified TA population (arrowheads). Means of all SC and DCs are plotted for reference. B–D) Individual expression profiles are plotted by fractional representation of GO terms that significantly distinguish TAs from SCs. Examples of B) Cell division- and C) DNA damage-related GO terms increased in TAs are plotted, as are D) examples of GO terms decreased in TAs (arrowheads=pure colonic TA population; arrows=bone marrow expression profiles, which are outliers among DC profiles in cell cycle-associated GO terms).
To directly test whether the independent cluster of expression profiles truly represented transit amplifying genes, we designed a de novo set of experiments to generate a novel expression profile of a biologically well defined transit amplifying cell population. We had previously shown that the crypts in the regions of the descending colon that surround a site of dextran sodium sulfate (DSS) induced ulceration undergo ~5-fold increase in mitotic activity. This increased proliferation is part of a temporally limited wave of expansion to attempt to repair the nearby injury 36. The result of the expansion in each crypt is that the transiently proliferating progenitor (i.e., transit amplifying) cells crowd out the other cell populations in the crypts, including the SC and the scattered differentiated goblet cells, such that 97% of the epithelial cells are TAs.
Thus, the DSS-induced crypt epithelial cells epitomize a TA population. To determine a TA gene expression profile, we laser-capture microdissected crypts from frozen sections of DSS crypts. To minimize possible contamination by inflammatory cells reacting to DSS treatment, we isolated TA cells from DSS-treated Rag−/− mice, which lack lymphocytes but show strong induction of the TA population. To control for toxic effects of DSS and to minimize contribution of SCs captured in the crypts along with the TAs, we referenced to crypt populations captured from the equivalent, ulcer-bordering regions in: 1) DSS-treated germ-free mice and 2) DSS-treated conventionally-raised mice null for the key immunomodulating gene Myd88. Germ-free and Myd88−/− mice maintain normal SC-driven proliferation, even following DSS treatment.
As expected, our prototypic TA cells clustered with the putative TA populations (Fig. 4A). To determine which GO terms were responsible for the separate TA cluster, we examined fractional representation of all GO terms associated with >1% of genes on average in the TA cell population. Supplemental table 4 shows the 22 GO terms and 8 GO terms with above threshold, statistically significantly increases and decreases (p<0.05) relative to SCs. TA-associated GO terms fell into three general categories. Cell proliferation-related genes, associated with GO terms like “DNA replication”, were uniformly increased (Fig. 4B; de novo generated mouse colonic TAs marked with arrowhead). Note how cell division related GO terms are not substantially different between SCs and DCs, as mentioned above. Interestingly, the two bone marrow expression profiles (profiles #8, 9) are clear outliers (these are the two points well above the mean in the DC population, Fig. 4B). The non-SC components of bone marrow comprise many lineage-committed but still proliferating cells, so these populations might be expected to have a TA component to their gene expression profile. DNA repair associated GO terms constituted the second category (Fig. 4C), indicating that TAs devote much of their gene expression to maintaining genomic integrity during their rapid cell division. Finally, there were miscellaneous GO terms decreased in TAs, including genes associated with the GO term “apoptosis” (Fig. 4D). Clearly, more experiments will be needed to better define how TAs differ from SCs and DCs. For example, our sample is currently heavily weighted toward the intestine, where the TA population is relatively well characterized. However, we think it unlikely that the TA cluster we define is unique to the intestine, as the expression profiles of gastrointestinal SCs (#14, 16, 17) all cluster with SCs from other tissues, and the retinal TA population clusters with the intestinal TAs.
In conclusion, our initial analysis of expression profiles of adult SCs and differentiated tissues showed that adult SCs share patterns of gene expression that distinguish them from DCs. A subsequent analysis of multiple recently published additional expression profiles and one novel unpublished expression profile from defined mouse and human progenitor and DC populations followed the same pattern and, additionally, highlighted a possible gene expression signature for TAs. Thus, we provide GO term-based metrics that can help classify future gene expression profiles of unknown cell populations based on differentiation state. Further, we show how patterns of gene expression reflect fundamental features of biology: progenitor cells preferentially express genes whose function is internal, whereas DCs express genes whose function is to communicate with the extracellular environment.
Supplementary Material
Acknowledgments
The authors thank Seth Bloom, Jessica Geahlen, Drs. Indira Mysorekar, Andrew Bredemeyer, and Jochen Lennerz for review of the manuscript.
Funding
We thank the Washington University Digestive Diseases Research Core Center Murine Models Core, supported by grant #P30 DK52574 and the Multiplexed Gene Analysis Core of the Siteman Cancer Center (supported in part by NCI grant #P30 CA91842). Grant support: JCM (K08 DK066062; R01 079798) and TSS (R03 DK55489).
Abbreviations
- SCs
Stem/Progenitor Cells
- DCs
Differentiated Cells
- TAs
Transit Amplifying Cells
- GO
Gene Ontology
References
- 1.Ivanova NB, Dimos JT, Schaniel C, Hackney JA, Moore KA, Lemischka IR. A stem cell molecular signature. Science. 2002;298:601–604. doi: 10.1126/science.1073823. [DOI] [PubMed] [Google Scholar]
- 2.Ramalho-Santos M, Yoon S, Matsuzaki Y, Mulligan RC, Melton DA. "Stemness": transcriptional profiling of embryonic and adult stem cells. Science. 2002;298:597–600. doi: 10.1126/science.1072530. [DOI] [PubMed] [Google Scholar]
- 3.Vogel G. Stem cells. 'Stemness' genes still elusive. Science. 2003;302:371. doi: 10.1126/science.302.5644.371a. [DOI] [PubMed] [Google Scholar]
- 4.Evsikov AV, Solter D. Comment on " 'Stemness': transcriptional profiling of embryonic and adult stem cells" and "a stem cell molecular signature" (II) Science. 2003;302:393. doi: 10.1126/science.1082380. [DOI] [PubMed] [Google Scholar]
- 5.Fortunel NO, Otu HH, Ng HH, et al. Comment on " 'Stemness': transcriptional profiling of embryonic and adult stem cells" and "a stem cell molecular signature" (I) Science. 2003;302:393. doi: 10.1126/science.1086384. [DOI] [PubMed] [Google Scholar]
- 6.Wong DJ, Liu H, Ridky TW, Cassarino D, Segal E, Chang HY. Module map of stem cell genes guides creation of epithelial cancer stem cells. Cell Stem Cell. 2008;2:333–344. doi: 10.1016/j.stem.2008.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ben-Porath I, Thomson MW, Carey VJ, et al. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet. 2008;40:499–507. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Phillips RL, Ernst RE, Brunk B, et al. The genetic program of hematopoietic stem cells. Science. 2000;288:1635–1640. doi: 10.1126/science.288.5471.1635. [DOI] [PubMed] [Google Scholar]
- 9.D'Amour KA, Gage FH. Genetic and functional differences between multipotent neural and pluripotent embryonic stem cells. Proc Natl Acad Sci U S A. 2003;100 Suppl 1:11866–11872. doi: 10.1073/pnas.1834200100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tumbar T, Guasch G, Greco V, et al. Defining the epithelial stem cell niche in skin. Science. 2004;303:359–363. doi: 10.1126/science.1092436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feezor RJ, Paddock HN, Baker HV, et al. Temporal patterns of gene expression in murine cutaneous burn wound healing. Physiol Genomics. 2004;16:341–348. doi: 10.1152/physiolgenomics.00101.2003. [DOI] [PubMed] [Google Scholar]
- 12.Venezia TA, Merchant AA, Ramos CA, et al. Molecular signatures of proliferation and quiescence in hematopoietic stem cells. PLoS Biol. 2004;2:e301. doi: 10.1371/journal.pbio.0020301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chateauvieux S, Ichante JL, Delorme B, et al. The Molecular Profile of Mouse Stromal Mesenchymal Stem Cells. Physiol Genomics. 2006 doi: 10.1152/physiolgenomics.00197.2006. [DOI] [PubMed] [Google Scholar]
- 14.Zipori D. The nature of stem cells: state rather than entity. Nat Rev Genet. 2004;5:873–878. doi: 10.1038/nrg1475. [DOI] [PubMed] [Google Scholar]
- 15.Eckfeldt CE, Mendenhall EM, Verfaillie CM. The molecular repertoire of the 'almighty' stem cell. Nat Rev Mol Cell Biol. 2005;6:726–737. doi: 10.1038/nrm1713. [DOI] [PubMed] [Google Scholar]
- 16.Cai J, Weiss ML, Rao MS. In search of "stemness". Exp Hematol. 2004;32:585–598. doi: 10.1016/j.exphem.2004.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mikkers H, Frisen J. Deconstructing stemness. Embo J. 2005;24:2715–2719. doi: 10.1038/sj.emboj.7600749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mills JC, Andersson N, Hong CV, Stappenbeck TS, Gordon JI. Molecular characterization of mouse gastric epithelial progenitor cells. Proc Natl Acad Sci U S A. 2002;99:14819–14824. doi: 10.1073/pnas.192574799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mills JC, Andersson N, Stappenbeck TS, Chen CC, Gordon JI. Molecular characterization of mouse gastric zymogenic cells. J Biol Chem. 2003;278:46138–46145. doi: 10.1074/jbc.M308385200. [DOI] [PubMed] [Google Scholar]
- 20.Stappenbeck TS, Mills JC, Gordon JI. Molecular features of adult mouse small intestinal epithelial progenitors. Proc Natl Acad Sci U S A. 2003;100:1004–1009. doi: 10.1073/pnas.242735899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ramsey VG, Doherty JM, Chen CC, Stappenbeck TS, Konieczny SF, Mills JC. The maturation of mucus-secreting gastric epithelial progenitors into digestive-enzyme secreting zymogenic cells requires Mist1. Development. 2007;134:211–222. doi: 10.1242/dev.02700. [DOI] [PubMed] [Google Scholar]
- 22.Huh WJ, Pan XO, Mysorekar IU, Mills JC. Location, allocation, relocation: isolating adult tissue stem cells in three dimensions. Curr Opin Biotechnol. 2006;17:511–517. doi: 10.1016/j.copbio.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 23.Doherty JM, Carmichael LK, Mills JC. GOurmet: a tool for quantitative comparison and visualization of gene expression profiles based on gene ontology (GO) distributions. BMC Bioinformatics. 2006;7:151. doi: 10.1186/1471-2105-7-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nam D, Kim SB, Kim SK, Yang S, Kim SY, Chu IS. ADGO: analysis of differentially expressed gene sets using composite GO annotation. Bioinformatics. 2006;22:2249–2253. doi: 10.1093/bioinformatics/btl378. [DOI] [PubMed] [Google Scholar]
- 26.Larsson O, Wennmalm K, Sandberg R. Comparative microarray analysis. Omics. 2006;10:381–397. doi: 10.1089/omi.2006.10.381. [DOI] [PubMed] [Google Scholar]
- 27.Irizarry RA, Warren D, Spencer F, et al. Multiple-laboratory comparison of microarray platforms. Nat Methods. 2005;2:345–350. doi: 10.1038/nmeth756. [DOI] [PubMed] [Google Scholar]
- 28.Ramos CA, Bowman TA, Boles NC, et al. Evidence for diversity in transcriptional profiles of single hematopoietic stem cells. PLoS Genet. 2006;2:e159. doi: 10.1371/journal.pgen.0020159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lugus JJ, Chung YS, Mills JC, et al. GATA2 functions at multiple steps in hemangioblast development and differentiation. Development. 2007;134:393–405. doi: 10.1242/dev.02731. [DOI] [PubMed] [Google Scholar]
- 30.He XC, Yin T, Grindley JC, et al. PTEN-deficient intestinal stem cells initiate intestinal polyposis. Nat Genet. 2007;39:189–198. doi: 10.1038/ng1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fevr T, Robine S, Louvard D, Huelsken J. Wnt/beta-catenin is essential for intestinal homeostasis and maintenance of intestinal stem cells. Mol Cell Biol. 2007;27:7551–7559. doi: 10.1128/MCB.01034-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Park IH, Zhao R, West JA, et al. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2007 doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- 33.Parise G, O'Reilly CE, Rudnicki MA. Molecular regulation of myogenic progenitor populations. Appl Physiol Nutr Metab. 2006;31:773–781. doi: 10.1139/h06-055. [DOI] [PubMed] [Google Scholar]
- 34.Perez-Iratxeta C, Palidwor G, Porter CJ, et al. Study of stem cell function using microarray experiments. FEBS Lett. 2005;579:1795–1801. doi: 10.1016/j.febslet.2005.02.020. [DOI] [PubMed] [Google Scholar]
- 35.Kosinski C, Li VS, Chan AS, et al. Gene expression patterns of human colon tops and basal crypts and BMP antagonists as intestinal stem cell niche factors. Proc Natl Acad Sci U S A. 2007;104:15418–15423. doi: 10.1073/pnas.0707210104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pull SL, Doherty JM, Mills JC, Gordon JI, Stappenbeck TS. Activated macrophages are an adaptive element of the colonic epithelial progenitor niche necessary for regenerative responses to injury. Proc Natl Acad Sci U S A. 2005;102:99–104. doi: 10.1073/pnas.0405979102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.