Abstract
Correct delineation of the hierarchy of cardiac progenitors is a key step to understanding heart development, and will pave the way for future use of cardiac progenitors in the treatment of heart disease. Multipotent Nkx2-5 and Isl1 cardiac progenitors contribute to cardiomyocyte, smooth muscle, and endothelial lineages, which constitute the major lineages of the heart. Recently, progenitors located within the proepicardium and epicardium were reported to differentiate into cardiomyocytes, as well as smooth muscle and endothelial cells. However, the relationship of these proepicardial progenitors to the previously described Nkx2-5 and Isl1 cardiac progenitors is incompletely understood. To address this question, we performed in vivo Cre-loxP-based lineage tracing. Both Nkx2-5- and Isl1-expressing progenitors contributed to the proepicardium and expressed Wt1 and Tbx18, markers of proepicardial progenitor cells. Interestingly, Nkx2-5 knockout resulted in abnormal proepicardial development and decreased expression of Wt1, suggesting a functional role for Nkx2-5 in proepicardium formation. Taken together, these results suggest that Nkx2-5 and/or Isl1 cardiac progenitors contribute to proepicardium during heart development.
Introduction
Regenerative approaches hold great promise to improving the treatment of congenital and acquired heart disease, the leading causes of morbidity and mortality in infancy and adulthood, respectively. Realizing this promise will require detailed understanding of the diverse cardiac lineages and their hierarchies during heart development.
Recent work indicates that a common multipotent precursor differentiates into cardiomyocyte, smooth muscle, and endothelial lineages, the major differentiated cell types of the heart [1–3]. This precursor is marked by expression of the transcription factors Isl1 and Nkx2-5. Lineage tracing studies indicate that these Isl1-and Nkx2-5-expressing precursors contribute to most of the cardiomyocytes and a subset of the smooth muscle and endothelial cells of the heart [3].
Proper cardiac morphogenesis requires contribution of cells from the proepicardium (PE), an outpouching of the septum transversum that protrudes towards the heart. Proepicardial cells migrate onto the surface of the heart, forming an epithelial sheet termed the epicardium. Epicardial cells subsequently undergo epithelial to mesenchymal transition, migrate into the myocardium, and differentiate into smooth muscle and endothelial cells [4–6]. In the heart, expression of the transcription factor Wilm’s Tumor 1 (Wt1) is confined to the proepicardium and epicardium [5]. We recently performed genetic lineage tracing studies based on Wt1-driven recombinase expression, and found that that progenitors located within the PE and epicardium also differentiate into cardiomyocytes [7]. At the same time, Cai and colleagues reached the same conclusion using an independent lineage tracing system based on recombinase expression driven by Tbx18 regulatory elements [8].
The lineage relationship between epicardial progenitors and the previously described multipotent Nkx2-5+/Isl1+ precursor is incompletely understood. While Cai et al. suggested that Tbx18-marked epicardial progenitors represent a novel lineage that is complementary to and distinct from Isl1-marked cells [8], we found that Wt1-marked epicardial progenitors are descended from precursors that express Nkx2-5 and Isl1 [7]. In this work, we provide further evidence indicating that Wt1+ progenitors within the PE are derived from Nkx2-5+/Isl1+ precursors. Furthermore, we show that Nkx2-5 expression is required for normal development of the PE, suggesting that Nkx2-5 functionally regulates formation of this structure.
Materials and Methods
Mice
Nkx2-5IRESCre/+, Nkx2-5Cre/+, or Isl1Cre/+ or mice were crossed with Rosa26fsLacZ reporters [9–12] for fate map studies. We note that this Rosa26fsLacZ reporter was more robustly recombined than a similar, independently generated reporter [13]. To generate Nkx2-5 or Isl1 knock out embryos, Nkx2-5Cre mice or Isl1Cre mice were intercrossed. Noon of the day of the plug was defined as E0.5. All procedures were performed under protocols approved by the Institutional Animal Care and Use Committee of Children’s Hospital Boston.
Histology and Immunohistochemistry
Whole-mount X-gal staining for β-galactosidase (β-gal) was performed as previously described [14]. Immunostaining was performed as described [15]. Briefly, the embryos were fixed with 4% paraformaldehyde and cryosectioned at 5–8 µm. Cell membranes were permeabilized by washing with 0.1% Triton X-100 PBS. The tissues were subsequently pre-blocked in 5% donkey serum and PBS for 1.5 hour at room temperature. The primary antibodies Wt1 (1:100, Santa Cruz), Tbx18 (1:100, Santa Cruz), Nkx2-5 (1:100, Santa Cruz), β-gal (1:5000, MP biomedicals), Isl1 (1:100, Iowa Developmental Studies Hybridoma Bank) were incubated for 1.5 hours at room temperature. Bound antibodies were visualized by secondary antibodies (Alex488 or 555, Invitrogen) or by the ABC method (ABC kit, Vector Laboratories). Nuclei were stained with 4',6-Di-amidino-2-phenylindole dihydrochloride (DAPI). The immunostained tissues were imaged by confocal microscopy (FV1000, Olympus).
Results and Discussion
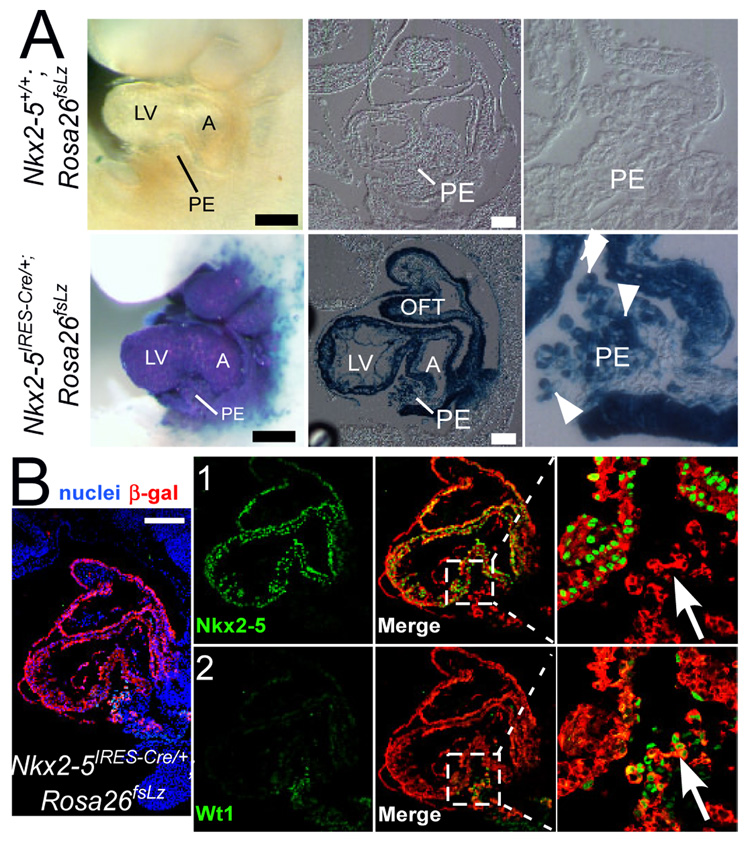
To track the descendants of Nkx2-5-expressing progenitors, we used the Cre-loxP lineage tracing approach [12]. Prior to Cre recombination, a floxed stop cassette in Rosa26fsLz prevented β-gal expression, as demonstrated by the absence of X-gal staining in Nkx2-5+/+;Rosa26fsLz embryos (Fig. 1A). In Nkx2-5IRES-Cre/+;Rosa26fsLz/+ embryos, Nkx2-5 regulatory elements drove expression of Cre recombinase, which removed the floxed stop cassette. This resulted in heritable expression of β-gal by Nkx2-5+ cells and their descendants (Fig 1A). At E9.5, Nkx2-5IRESCre/+;Rosa26fsLz embryos exhibited strong X-gal staining in the developing heart. The proepicardium (PE), residing between atrium (A) and left ventricle (LV), was robustly stained. This finding was confirmed in tissue sections, which showed that most PE cells expressed the genetic lineage β-gal (white arrowheads), indicating that cells descended from Nkx2-5+ precursors (“Nkx2-5-derived”) contribute to PE.
Fig. 1. Nkx2-5-derived cells contribute to proepicardium.
(A) Nkx2-5IRESCre/+;Rosa26fsLz and littermate control Nkx2-5+/+;Rosa26fsLz embryos, stained in whole mount with X-gal and then sagitally sectioned. Arrowheads mark the X-gal positive cells in proepicardium. (B) Co-staining of Nkx2-5 (green), Wt1 (green), and β-gal (red) on the same slide. Nuclei were stained with DAPI (blue). Arrows indicate PE cells that are β-gal and Wt1 positive, and Nkx2-5 negative. Black bar = 500 µm. White bar=100 µm. A, atrium; RV, right ventricle; LV, left ventricle; OFT, outflow tract; PE, proepicardium.
Wt1 marks a subset of PE cells that differentiate into cardiomyocyte, smooth muscle, and endothelial lineages [7, 8]. To test if Wt1+ PE cells are derived from Nkx2-5-expressing precursors, we co-stained tissue sections of Nkx2-5IRESCre/+;Rosa26fsLz embryos for Wt1 and the β-gal lineage tracer (Fig. 1B). A subset of Nkx2-5-derived PE cells (β-gal+; red) expressed Wt1 (green, Fig. 1B row 2), indicating that Wt1+ PE cells originate from Nkx2-5-expressing precursors. The Nkx2-5-derived PE cells were not actively expressing Nkx2-5 (green, Fig. 1B row 1), indicating that Nkx2-5 expression occurs transiently in PE precursors and is not maintained in PE cells.
Knock-in of Cre into the same locus by different strategies can lead to distinct recombination patterns [9, 10]. To reduce the likelihood of a false positive finding due to potentially idiosyncratic behavior of the Nkx2-5IRESCre line, we used a different, independently generated Nkx2-5 Cre knockin line, Nkx2-5Cre [10], to confirm the above findings. In Nkx2-5Cre/+;Rosa26fsLz/+ embryos, β-gal+ cells were also present within the PE, although the extent of proepicardial labeling was reduced compared to Nkx2-5IRESCre/+;Rosa26fsLz/+. These β-gal+ PE cells expressed Wt1 (Supplementary Fig. 1A) as well as Tbx18 (Supplementary Fig. 1B), another marker of the PE [8].
Collectively, these data provide strong evidence that Nkx2-5+ precursors contribute to the PE, suggesting that the PE shares a common developmental origin with the rest of the heart. The PE gives rise to much of the coronary vasculature [4, 16, 17]. Consistent with the finding that Nkx2-5+ precursors contribute to PE, our recent work indicates that Nkx2-5+ progenitors also contribute to a subset of coronary endothelial and smooth muscle cells in embryonic and adult hearts (Ma et al., submitted). While PE cells are originate from Nkx2-5+ precursors, they no longer actively express Nkx2-5, suggesting that the Nkx2-5+ progenitors are located higher in the progenitor hierarchy, before differentiation into and formation of the proepicardial/epicardial progenitors.
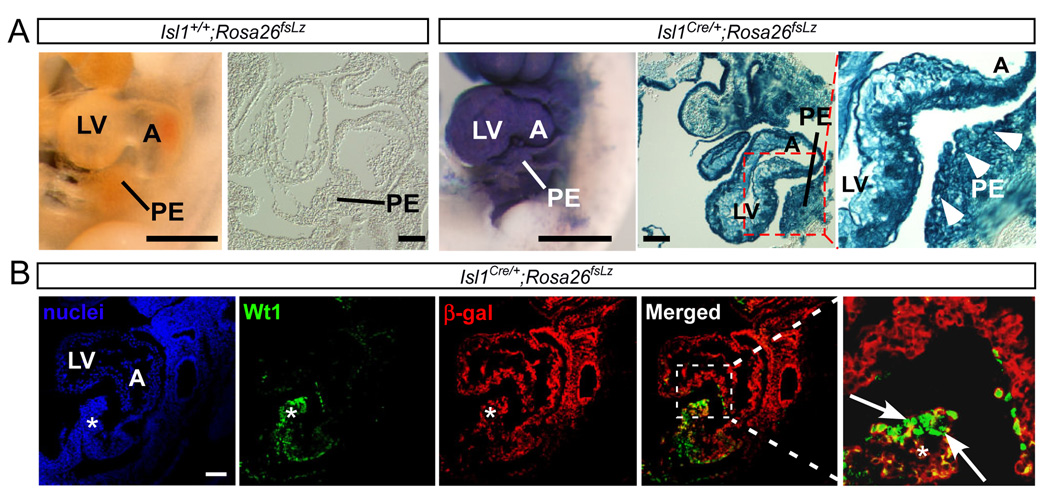
To test the hypothesis that Isl1+ progenitors contribute to the PE, we again used the Cre-loxP lineage tracing approach, now with Cre recombinase driven by Isl1 regulatory elements [11]. No β-gal+ activity was detected in negative control embryos (Isl1+/+; Rosa26fsLz) (Fig. 2A). In Isl1Cre/+;Rosa26fsLz embryos, the PE was robustly stained by X-gal (Fig. 2A). β-gal activity was also detected throughout the left ventricle (Fig. 2A). Extensive expression of β-gal within PE was confirmed in histological sections (Fig. 2A).
Fig. 2. Isl1-derived cells contribute to proepicardium.
(A) Wholemount X-gal staining of Isl1Cre/+;Rosa26fsLz and littermate control Isl1+/+;Rosa26fsLz embryos. Sagittal sections of whole mount stained embryos are shown. Arrowheads mark the X-gal positive proepicardium. (B) Immunostaining of Wt1 and β-gal on sections from Isl1Cre/+;Rosa26fsLz embryos. Asterisk indicates proepicardium. Arrows indicate cells positive for both β-gal and Wt1. Bar = 100 µm. Other abbreviations as in Fig. 1.
Next, we asked if the Isl1-derived PE cells expressed the proepicardial marker Wt1. We found that PE cells co-expressed Wt1 and the β-gal lineage tracer (Fig. 2B, arrows), indicating that Wt1+ proepicardial cells originate from Isl1-expressing precursors. Consistent with this finding, both Isl1+ cardiac progenitors and Wt1+ proepicardial/epicardial progenitors differentiate into cardiomyocytes, smooth muscle and endothelial cells in vivo and in vitro [3, 7, 18]. By immunostaining, the PE did not actively express Isl1 (data not shown), consistent with previous reports [19]. This positions the Isl1+ progenitor high in the progenitor hierarchy, before differentiation into proepicardial cells.
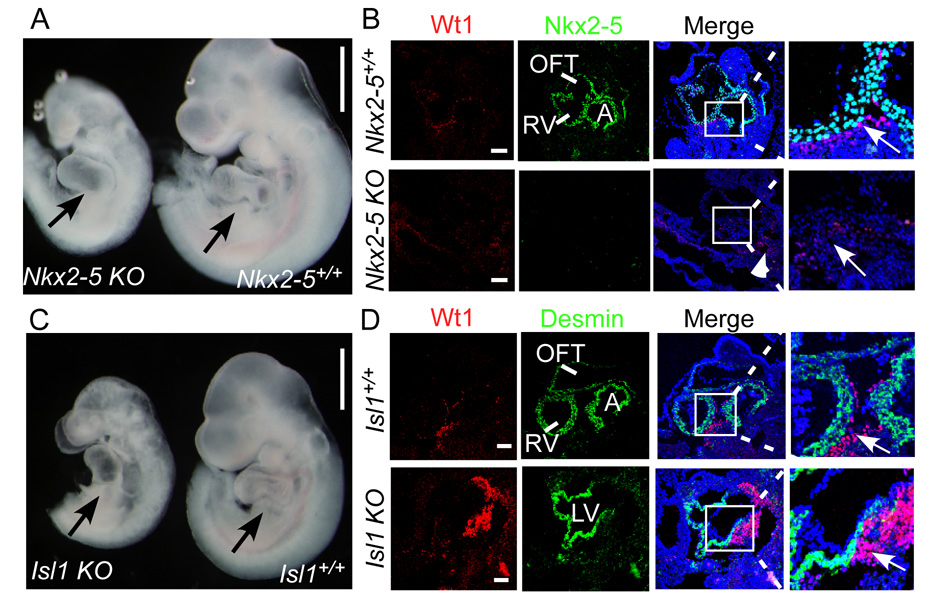
Since both Nkx2-5+ and Isl1+ cardiac progenitors contribute to PE, we asked if Nkx2-5 or Isl1 are functionally required for proepicardium development. Therefore we studied proepicardial development Nkx2-5 or Isl1 knockout embryos (Nkx2-5Cre/Cre or Isl1Cre/Cre, respectively). Nkx2-5 knockout embryos were developmentally delayed and showed severe cardiac abnormalities (Fig. 3A), consistent with previously described Nkx2-5 knockouts [20, 21]. Nkx2-5 was not detectable by immunohistochemistry (Fig. 3B), indicating that the Nkx2-5Cre allele is protein null. The PE was not clearly evident. Proepicardial Wt1 expression was markedly downregulated (Fig. 3B, arrow), while Wt1 expression in other regions of the embryo was intact (Fig. 3B, arrowheads). This suggests that Nkx2-5 is functionally required for formation of PE and expression of Wt1.
Fig. 3. Loss of Nkx2-5, but not Isl1, leads to malformation of proepicardium.
(A) Wholemount figure of Nkx2-5Cre/Cre (Nkx2-5 KO) and littermate control Nkx2-5+/+. Black arrow indicates PE in control embryo, and corresponding region of the mutant embryo. (B) Immunostaining of Wt1 (red) and Nkx2-5 (green) shows decreased Wt1 expression and no Nkx2-5 expression in mutants. Arrow indicates Wt1+ cells in PE of control embryo, and corresponding area deficient in Wt1 expression in mutant embryo. Arrowhead indicates preserved Wt1 expression outside of the heart-forming regions of the mutant embryo. (C) Wholemount figure of Isl1Cre/Cre (Isl1 KO) and littermate control Isl1+/+. Black arrow indicates PE in control and mutant embryos. (D) Immunostaining of Wt1 (red) and cardiac marker Desmin (green) shows existence of proepicardium. White arrow indicates Wt1-expressing proepicardium. A,C bar = 500 µm; B,D bar = 50 µm.
Isl1 knockout also resulted in severe growth retardation and cardiac abnormalities (Fig. 3C). Although the PE was abnormally located towards the atrial pole of the heart, it was identifiable (black arrows, Fig. 3C) and Wt1 expression was preserved (white arrows, Fig. 3D). This indicates that Isl1 is not functionally required for PE development or Wt1 expression. Furthermore, this suggests that abnormal PE development and Wt1 expression in Nkx2-5 null embryos is a specific consequence of Nkx2-5 loss of function, rather than a non-specific consequence of developmental delay or abnormal cardiac development. The mechanisms by which Nkx2-5 regulates PE development and Wt1 expression merit further investigation.
In conclusion, we found Nkx2-5- and Isl1-expressing precursors contribute to the proepicardium, marked by Wt1 and Tbx18. Nkx2-5, but not Isl1, is functionally required for PE development. These results place Wt1+ proepicardial/epicardial progenitors as an early branch of the multipotent Nkx2-5+/Isl1+ cardiac progenitor lineage.
Supplementary Material
(A) Immunostaining of Wt1 and β-gal on tissues from Nkx2-5Cre/+;Rosa26fsLz embryos. (B) Costaining of Tbx18 and β-gal on sections from Nkx2-5Cre/+;Rosa26fsLz embryos.
Acknowledgement
Nkx2-5IRESCre, Nkx2-5Cre, and Isl1Cre mice were kindly provided by Richard Harvey, Robert Schwartz, and Sylvia Evans, respectively. WTP was supported by a grant from the Harvard Stem Cell Institute. BZ was supported by a grant from the American Heart Association.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wu SM, Fujiwara Y, Cibulsky SM, Clapham DE, Lien CL, Schultheiss TM, Orkin SH. Developmental origin of a bipotential myocardial and smooth muscle cell precursor in the mammalian heart. Cell. 2006;127:1137–1150. doi: 10.1016/j.cell.2006.10.028. [DOI] [PubMed] [Google Scholar]
- 2.Kattman SJ, Huber TL, Keller GM. Multipotent flk-1+ cardiovascular progenitor cells give rise to the cardiomyocyte, endothelial, and vascular smooth muscle lineages. Dev Cell. 2006;11:723–732. doi: 10.1016/j.devcel.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 3.Moretti A, Caron L, Nakano A, Lam JT, Bernshausen A, Chen Y, Qyang Y, Bu L, Sasaki M, Martin-Puig S, Sun Y, Evans SM, Laugwitz KL, Chien KR. Multipotent embryonic isl1+ progenitor cells lead to cardiac, smooth muscle, and endothelial cell diversification. Cell. 2006;127:1151–1165. doi: 10.1016/j.cell.2006.10.029. [DOI] [PubMed] [Google Scholar]
- 4.Perez-Pomares JM, Carmona R, Gonzalez-Iriarte M, Atencia G, Wessels A, Munoz-Chapuli R. Origin of coronary endothelial cells from epicardial mesothelium in avian embryos. Int J Dev Biol. 2002;46:1005–1013. [PubMed] [Google Scholar]
- 5.Perez-Pomares JM, Phelps A, Sedmerova M, Carmona R, Gonzalez-Iriarte M, Munoz-Chapuli R, Wessels A. Experimental studies on the spatiotemporal expression of WT1 and RALDH2 in the embryonic avian heart: a model for the regulation of myocardial and valvuloseptal development by epicardially derived cells (EPDCs) Dev Biol. 2002;247:307–326. doi: 10.1006/dbio.2002.0706. [DOI] [PubMed] [Google Scholar]
- 6.Gittenberger-de Groot AC, Vrancken Peeters MP, Mentink MM, Gourdie RG, Poelmann RE. Epicardium-derived cells contribute a novel population to the myocardial wall and the atrioventricular cushions. Circ Res. 1998;82:1043–1052. doi: 10.1161/01.res.82.10.1043. [DOI] [PubMed] [Google Scholar]
- 7.Zhou B, Ma Q, Rajagopal S, Wu SM, Domian I, Rivera-Feliciano J, Jiang D, von Gise A, Ikeda S, Chien KR, Pu WT. Epicardial progenitors contribute to the cardiomyocyte lineage in the developing heart. Nature. 2008;454:109–113. doi: 10.1038/nature07060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cai CL, Martin JC, Sun Y, Cui L, Wang L, Ouyang K, Yang L, Bu L, Liang X, Zhang X, Stallcup WB, Denton CP, McCulloch A, Chen J, Evans SM. A myocardial lineage derives from Tbx18 epicardial cells. Nature. 2008 doi: 10.1038/nature06969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stanley EG, Biben C, Elefanty A, Barnett L, Koentgen F, Robb L, Harvey RP. Efficient Cre-mediated deletion in cardiac progenitor cells conferred by a 3'UTR-ires-Cre allele of the homeobox gene Nkx2-5. Int J Dev Biol. 2002;46:431–439. [PubMed] [Google Scholar]
- 10.Moses KA, DeMayo F, Braun RM, Reecy JL, Schwartz RJ. Embryonic expression of an Nkx2-5/Cre gene using ROSA26 reporter mice. Genesis. 2001;31:176–180. doi: 10.1002/gene.10022. [DOI] [PubMed] [Google Scholar]
- 11.Yang L, Cai CL, Lin L, Qyang Y, Chung C, Monteiro RM, Mummery CL, Fishman GI, Cogen A, Evans S. Isl1Cre reveals a common Bmp pathway in heart and limb development. Development. 2006;133:1575–1585. doi: 10.1242/dev.02322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- 13.Mao X, Fujiwara Y, Orkin SH. Improved reporter strain for monitoring Cre recombinase-mediated DNA excisions in mice. Proc Natl Acad Sci U S A. 1999;96:5037–5042. doi: 10.1073/pnas.96.9.5037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zeisberg EM, Ma Q, Juraszek AL, Moses K, Schwartz RJ, Izumo S, Pu WT. Morphogenesis of the right ventricle requires myocardial expression of Gata4. J Clin Invest. 2005;115:1522–1531. doi: 10.1172/JCI23769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou B, Bi YY, Han ZB, Ren H, Fang ZH, Yu XF, Poon MC, Han ZC. G-CSF-mobilized peripheral blood mononuclear cells from diabetic patients augment neovascularization in ischemic limbs but with impaired capability. J Thromb Haemost. 2006;4:993–1002. doi: 10.1111/j.1538-7836.2006.01906.x. [DOI] [PubMed] [Google Scholar]
- 16.Mikawa T, Gourdie RG. Pericardial mesoderm generates a population of coronary smooth muscle cells migrating into the heart along with ingrowth of the epicardial organ. Dev Biol. 1996;174:221–232. doi: 10.1006/dbio.1996.0068. [DOI] [PubMed] [Google Scholar]
- 17.Dettman RW, Denetclaw W, Jr, Ordahl CP, Bristow J. Common epicardial origin of coronary vascular smooth muscle, perivascular fibroblasts, and intermyocardial fibroblasts in the avian heart. Dev Biol. 1998;193:169–181. doi: 10.1006/dbio.1997.8801. [DOI] [PubMed] [Google Scholar]
- 18.Martin-Puig S, Wang Z, Chien KR. Lives of a heart cell: tracing the origins of cardiac progenitors. Cell Stem Cell. 2008;2:320–331. doi: 10.1016/j.stem.2008.03.010. [DOI] [PubMed] [Google Scholar]
- 19.Sun Y, Liang X, Najafi N, Cass M, Lin L, Cai CL, Chen J, Evans SM. Islet 1 is expressed in distinct cardiovascular lineages, including pacemaker and coronary vascular cells. Dev Biol. 2007;304:286–296. doi: 10.1016/j.ydbio.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tanaka M, Chen Z, Bartunkova S, Yamasaki N, Izumo S. The cardiac homeobox gene Csx/Nkx2.5 lies genetically upstream of multiple genes essential for heart development. Development. 1999;126:1269–1280. doi: 10.1242/dev.126.6.1269. [DOI] [PubMed] [Google Scholar]
- 21.Biben C, Harvey RP. Homeodomain factor Nkx2-5 controls left/right asymmetric expression of bHLH gene eHand during murine heart development. Genes Dev. 1997;11:1357–1369. doi: 10.1101/gad.11.11.1357. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Immunostaining of Wt1 and β-gal on tissues from Nkx2-5Cre/+;Rosa26fsLz embryos. (B) Costaining of Tbx18 and β-gal on sections from Nkx2-5Cre/+;Rosa26fsLz embryos.