Abstract
Spatial normalization and segmentation of infant brain MRI data based on adult or pediatric reference data may not be appropriate due to the developmental differences between the infant input data and the reference data. In this study we have constructed infant templates and a priori brain tissue probability maps based on the MR brain image data from 76 infants ranging in age from 9 to 15 months. We employed two processing strategies to construct the infant template and a priori data: one processed with and one without using a priori data in the segmentation step. Using the templates we constructed, comparisons between the adult templates and the new infant templates are presented. Tissue distribution differences are apparent between the infant and adult template, particularly in the gray matter (GM) maps. The infant a priori information classifies brain tissue as GM with higher probability than adult data, at the cost of white matter (WM), which presents with lower probability when compared to adult data. The differences are more pronounced in the frontal regions and in the cingulate gyrus. Similar differences are also observed when the infant data is compared to a pediatric (age 5 to 18) template. The two-pass segmentation approach taken here for infant T1W brain images has provided high-quality tissue probability maps for GM, WM, and CSF, in infant brain images. These templates may be used as prior probability distributions for segmentation and normalization; a key to improving the accuracy of these procedures in special populations.
Introduction
Spatial normalization of individual imaging data to a common reference frame allows us to make statistical inferences across and between groups of individuals. This is usually accomplished by transforming individual image data to a standardized stereotaxic space. Typically, registration and warping of an individual brain into a stereotaxic space is done using a template created from the average of a large number of brain images (Guimond et al. 2000). Talairach and Tournoux described the original method for transforming brain images into a common reference frame based on detailed anatomical measurements in a single individual (Talairach and Tournoux 1988). More recently, the coordinate frame created at the Montreal Neurological Institute (MNI) was constructed from 152 healthy adults between the ages of 18 and 44 years (Evans et al., 1993). However, the use of this template for spatial normalization in special populations such as children, infants and neonates, has been questioned by several authors (Muzik et al., 2000; Burgund et al., 2002; Wilke et al., 2003a; Hoeksma et al., 2005; Machilsen et al, 2007; Murgsova et al., 2007). To this end Wilke et al (2003b) created a pediatric template (CCHMC) using brain images from a large number of children between the ages of 5 and 18 (Wilke et al, 2003b). They demonstrated the misclassification that can result from using the adult template and prior probabilities for spatial normalization of pediatric data. There have been some attempts to construct an infant template; however, these are based on a very limited number of infants (Dehaene-Lambertz et al., 2002; Srinivasan et al, 2007; Kazemi et al., 2007).
Similarly, in morphological studies based on segmentation of the human brain into different tissue classes, most approaches use a priori reference data to improve classification of brain tissue so that classification is not only based upon the input voxel intensity information alone. The recently-proposed unified segmentation framework offered within the SPM5 environment, integrates tissue classification with bias correction and image registration, and it estimates the associated parameters by minimizing the resulting objective function (Ashburner and Friston, 2005). However, if the prior data is taken from an adult reference population, it may not be an appropriate template for data from special populations, such as that from infants.
In this study we set out to construct an infant brain template and associated probability maps for GM, WM and CSF that can be used for registration and as a priori information in segmenting new infant brain images. We constructed an infant brain template and a priori data using high quality T1-weighted 3 Tesla magnetic resonance (MR) images from 76 infants whose age ranged from 9 to 15 months. We utilized the unified segmentation procedure implemented in Statistical Parameter Mapping (SPM5) software (Wellcome Department, University College, London, UK), but modified it to include a Hidden Markov Random Field (HMRF) model as an additional spatial constraint (Cuadra et al.; 2005). We also modified the default segmentation procedure provided in SPM5 in order to omit the use of the adult a priori data from the tissue probability estimates. The new segmentation algorithm is based on a Gaussian Mixture Model, but in contrast to the SPM5 segmentation (default) algorithm the final tissue probabilities are estimated without tissue priors (Gaser et al.; 2007). In an effort to further reduce the impact of the adult prior data, we employed a two-pass approach. In the first pass the adult a priori data are used. Then in the second pass, the infant probability data generated by the first pass approach are used.
After constructing infant brain image templates and tissue probability maps using the new methods, we quantified the magnitude of differences between the newly constructed infant population data and MNI adult reference data or CCHMC pediatric reference data. Quantitative comparisons are summarized in Table 2 for convenience.
Table 2.
Summary of the different comparisons made. Except for comparison 4, all comparisons are based on second pass images. Bold letters indicate image reference to Figure 1.
| Comparison | Image 1 | Image 2 | Differences |
|---|---|---|---|
| 1 | New Infant GM/WM/CSF constructed under the new and default strategies. (Initial registration was with adult reference data). (i) & (g) |
Adult prior GM/WM/CSF (a) |
Figure 3 |
| 2 | New Infant GM/WM/CSF constructed under the new and default strategies. (Initial registration was with pediatric reference data). (j) & (h) |
Pediatric prior GM/WM/CSF (b) |
Figure 4 |
| 3 | New Infant GM/WM/CSF constructed under the new strategy (i) |
New Infant GM/WM/CSF constructed under the default strategy (g) |
Figure 5 |
| 4 | First pass infant GM/WM/CSF constructed under both strategies (e) & (c) |
Second pass infant GM/WM/CSF constructed under both strategies (i) & (g) |
Figure 6 |
Materials and Methods
Subjects
Our subjects were drawn from two different, currently ongoing protocols. The first protocol is testing fMRI of auditory language stimulation in hearing impaired infants at or near 12 months of age. Infants with normal hearing and hearing impairment were recruited to compare fMRI results with auditory and speech stimulation between the two groups. This protocol is designed to test whether fMRI of auditory-language stimulation in a hearing-impaired infant can provide specific, clinically relevant details about the child’s auditory perception and processing ability. Specifically infants between the ages of 9 and 15 months (12 ± 3) were included in the protocol, and currently data for 28 (9 male and 19 female) hearing-impaired and normal hearing control subjects is available for processing
In order to increase the number of subjects for the template construction we include additional infants from a second protocol. This protocol includes infants referred for clinical brain imaging with MRI for clinical diagnosis of various indications. From this protocol, forty nine infants (22 male and 27 female; age range 9–15 months) were found to have normal brain anatomy, confirmed by the attending neuroradiologist at the time of scan. They were thus included in the current study.
Overall, MR images from 77 infants (46 female and 31 male) aged between 9 and 15 months, were acquired from sedated infants being scanned for clinical indications as described above. Imaging data from one 9 month old female infant was excluded, as image artifacts interfered with the segmentation procedure. The remaining 76 MR images were visually confirmed to be free of artifacts and were thus used for all subsequent analyses. The age distribution of the resulting 76 infants (45 female and 31 male) is presented in Table 1. Infants in both protocols were sedated using either chloral hydrate 75 mg/kg or Nembutal 5 mg/kg. Institutional Review Board approval was obtained for this study which involves retrospective data analysis.
Table 1.
Age and gender distribution of the study population
| Gender | Total | ||
|---|---|---|---|
| Age (months) | Female | Male | |
| 9 | 6 | 8 | 14 |
| 10 | 5 | 2 | 7 |
| 11 | 7 | 5 | 12 |
| 12 | 7 | 7 | 14 |
| 13 | 7 | 3 | 10 |
| 14 | 10 | 2 | 12 |
| 15 | 3 | 4 | 7 |
| Total | 45 | 31 | 76 |
Data acquisition and preparation
Infants were imaged with a clinical 3 Tesla clinical MRI Scanner (Siemens Trio, Siemens Medizintechnik, Erlangen, Germany). High resolution T1-weighted, 3D brain images were acquired using the Magnetization Prepared Rapid Acquisition Gradient Echo (MPRAGE) method (Mugler and Brookeman, 1990). The imaging parameters used were TR=2000 ms, TE=2.93 ms for n=68 subjects, and TR=1900 ms, TE=4.13 ms for n=8 subjects; flip angle=12°, FOV 15×20×19.2 cm and matrix [208–512] × [256–512] × [128–192] resolution. All data was imported from DICOM format into ANALYZE format for analysis using the SPM5 DICOM import function. All processing was done using SPM5 or stand-alone scripts running in MATLAB. Instead of manually orienting the data using the display option in SPM5, an automated center of mass approach was used.
Processing: Template and a priori data construction
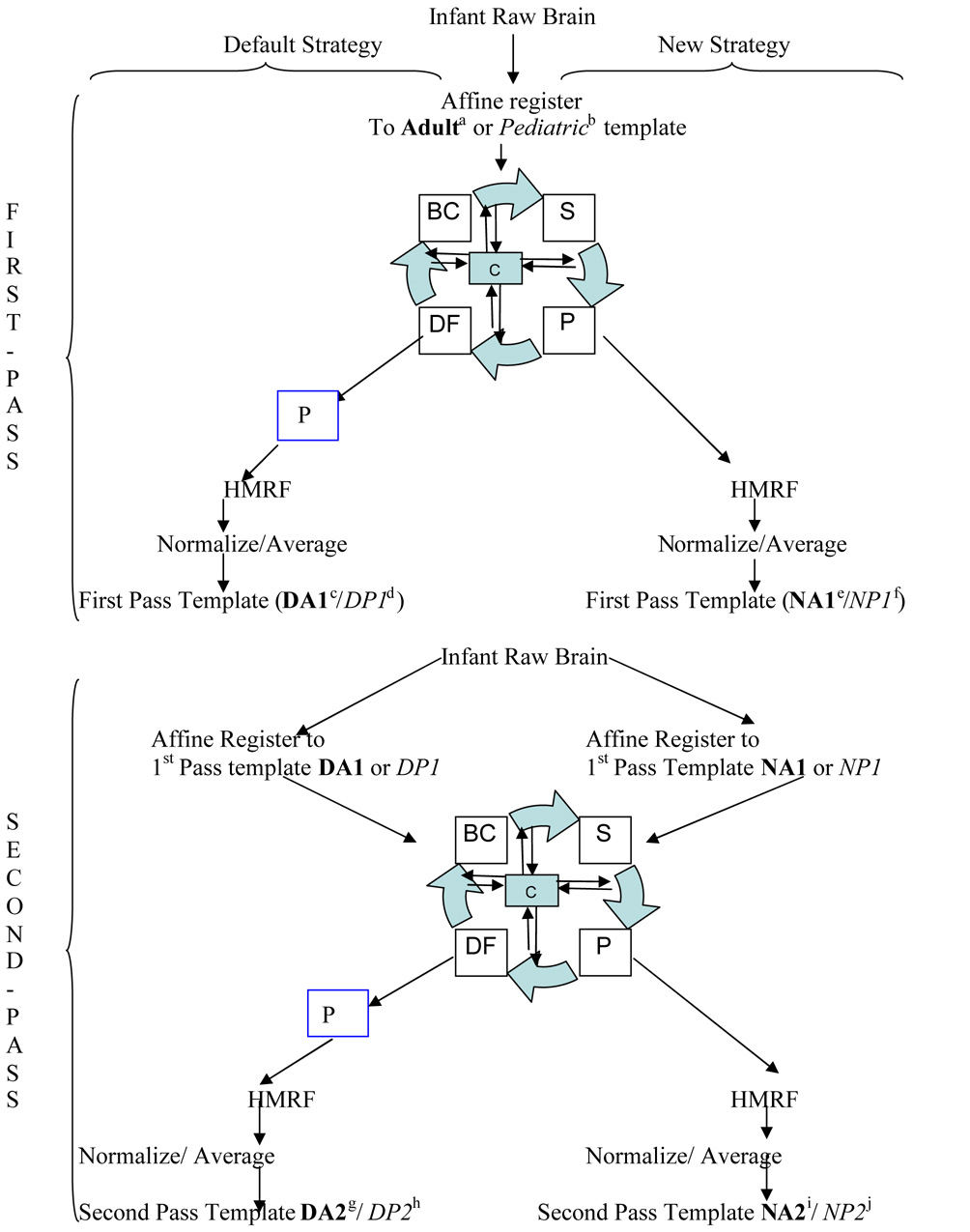
Our objective was to construct an infant template and a priori data that can be used for improved spatial normalization and the morphological study of infant data. In order to achieve this we employed two different processing strategies. The first strategy uses the standard unified segmentation approach that utilizes adult a priori data for segmentation. This is the default strategy in SPM5, though we modified it to incorporate the HMRF model to additionally introduce spatial constraint. This modification to the algorithm helps in determining the probability of a given voxel to belong to a tissue class which is achieved by calculating the MRF energy for a given voxel, based on its proximity to the surrounding voxels. The unified segmentation approach in SPM5 finds a Maximum a Posteriori (MAP) solution by repeatedly alternating among classification, bias correction and registration steps in a unified generative model involving a mixture of Gaussians, bias correction component and warping (non-linear registration) component (Ashburner and Friston, 2005). This process is indicated by the segmentation engine diagramed in Figure 1. This strategy will be referred to as the default strategy for the rest of the paper. The second processing strategy, basing segmentation solely on current voxel intensity and not using prior information, has only recently been implemented as a custom function within the unified segmentation approach in SPM5. The segmentation is based on a Gaussian mixture model and the final tissue probabilities are estimated without tissue priors. We additionally applied the HMRF model to introduce spatial constraints (Cuadra et al., 2005). This strategy will be referred to as the new strategy for the rest of the paper. A flow chart outlining both strategies is diagrammed in Figure 1.
Figure 1.
Overview of infant template construction. Initially all images are registered to the adult /pediatric template. After registration a segmentation estimation procedure is used to estimate the parameters by iteratively going through segmentation (S), bias correction (BC), deformation (DF) and priors (P) until convergence criteria (C) are met. After this step two strategies are used for calculation of the tissue probability maps. The first (default) strategy includes adult prior probabilities for distribution of GM, WM and CSF based on adult / pediatric data indicated by the inclusion of the blue box (P). The second (new) strategy does not use prior probability distributions in the calculation of the tissue probability maps; instead it only uses the intensity of the T1 images. Then a Hidden Markov Random Field (HMRF) process is applied to the resulting image, normalized and averaged to produce the first pass template. Second pass templates are obtained in similar fashion except the first pass template is used for registration and normalization.
Key: DA1/DP1=First pass adult/pediatric template via default strategy;
DA2/DP2=Second pass adult/pediatric template via default strategy;
NA1/NP1=First pass adult/ pediatric template via new strategy;
NA2/NP2=Second pass adult/pediatric template via new strategy. Bold font indicates adult data while italic font indicates pediatric data.
Comparisons: 1. (g-a) and (i-a); 2. (h-b) and (j-b); 3. (g-i); 4. (g-c), (i-e) and (h-d), (j-f); using number conventions from table 2.
Each strategy was deployed using two different approaches, where first we established a first pass template and a priori data based on the adult or pediatric reference data. These templates are then used in a second iteration as reference data to yield a second pass template and a priori data. To clarify the two strategies combined with two approaches employed and the comparisons resulting from these combinations, a flow chart is provided in Figure 1.
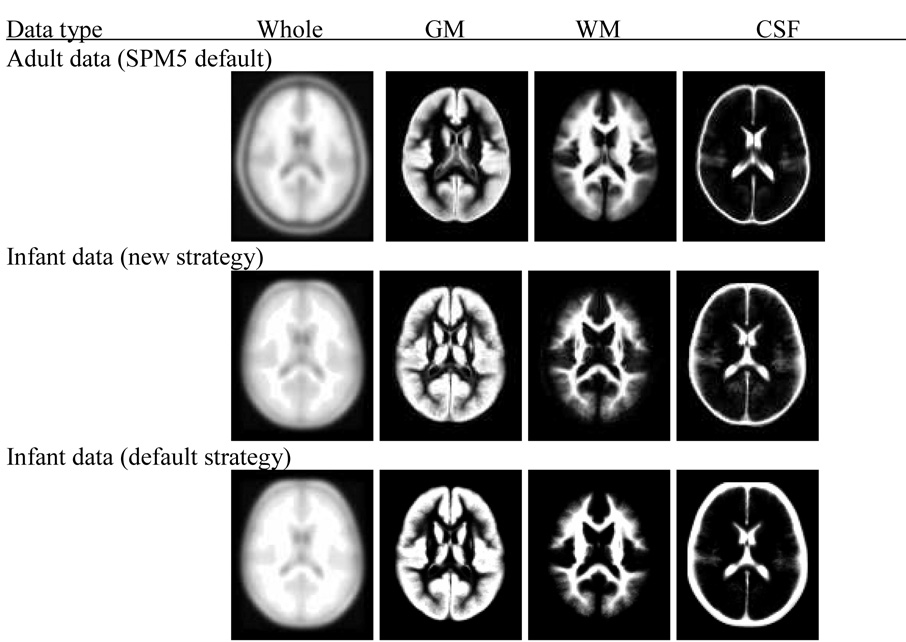
For all processing strategies and approaches, a light cleaning procedure within the SPM5 segmentation procedure was used in order to remove residual non-brain tissue from the segmented images. All normalized data are written out with the same resolution and dimension (157×189×156 voxels at 1×1×1 mm resolution) and, then averaged to create a priori probability maps of GM, WM and CSF. Similarly, we applied the normalization parameters obtained from the segmentation step to write out each individual normalized T1-weighted image and calculated the average of all normalized T1-images (Figure 2).
Figure 2.
Display of adult reference data and infant templates including GM, WM and CSF probability maps. The top panel is for the adult data (SPM 5 default), the middle panel is for infant data processed using the new strategy and the bottom panel is for infant data processed using the default strategy as outlined in Figure 1. The displayed infant data is based on second pass output.
Assessment of differences
Comparison with adult and pediatric reference data
The first round of comparison (comparison 1, Table2) involves data obtained from the two processing strategies against the default SPM5 adult a priori data. The adult data is based on the scans of 152 young, healthy subjects from Montreal Neurological Institute (MNI-152). The second round of comparison (comparison 2, Table2) uses data obtained from the two processing strategies against the pediatric a priori data. The pediatric data is based on 200 healthy children between the ages of 5 and 18 years, and is provided by Cincinnati Children’s Hospital Medical Center (CCHMC2-200)), available at www.irc.cchmc.org/software/pedbrain.php. An outline of the comparison strategies is presented in Table 2.
Comparison between processing strategies and approaches
In addition to these comparisons we also assessed the impact on the infant template and the corresponding infant a priori data images resulting from the two processing strategies (comparison 3, Table 2). Using this comparison we are able to evaluate the impact of the use of adult prior data in the resulting images. We also compared the images obtained from the two approaches used for each strategy. Specifically the difference between first pass (use of adult data) and second pass (use of infant data) data are compared (comparison 4, Table2).
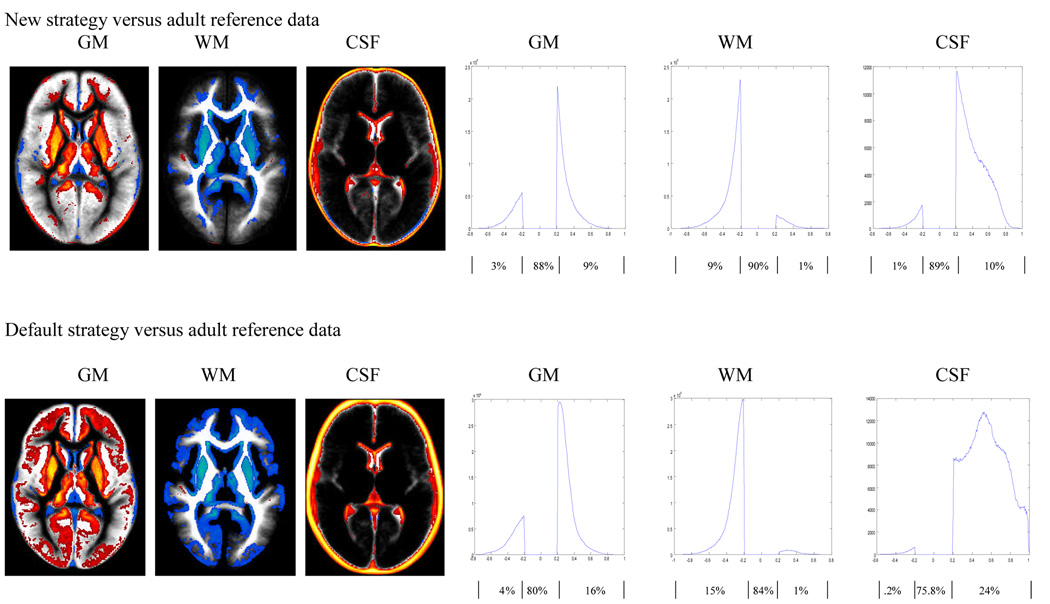
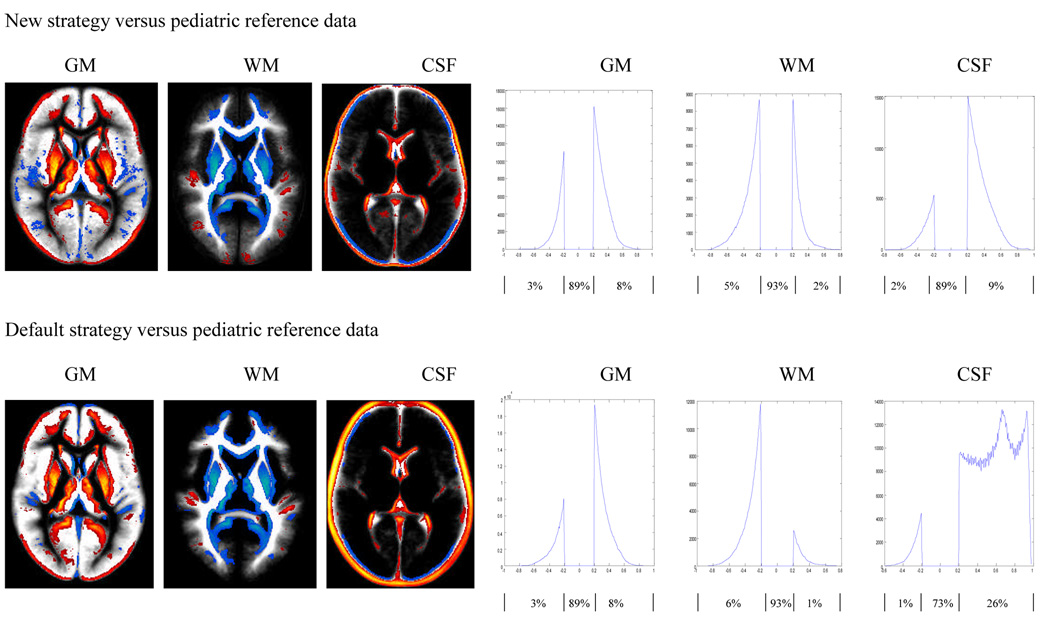
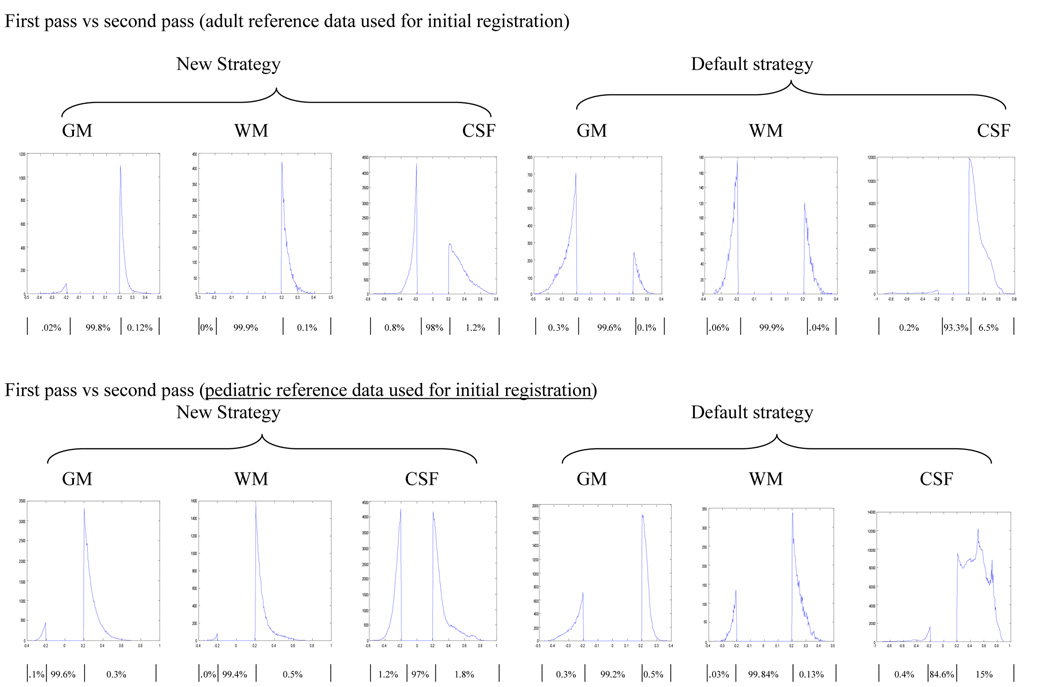
We used the SPM image calculation function to calculate the differences between pairs of images. We displayed the difference between two images using MRIcro with color coding, where red and yellow indicated higher tissue concentration in the infant data (i.e. infant>adult, or infant>pediatric) and blue indicated lower tissue concentration in the infant data (infant<adult, or infant<pediatric). Only tissue probability differences of at least 20% are displayed in the color overlays (Figure 3 and 4). All results are in neurological orientation. We also display these differences using a histogram, illustrating the distribution of these differences. The histograms are obtained by retaining voxels from the difference image that are greater than 0.2 and less that −0.2. Only tissue probability differences of 20% or more (i.e. differences in tissue probabilities that are >0.2 or <−0.2) are plotted in the histograms in Figure 3–Figure 6. The percentage of voxels classified consistently (within the 20% limit) or inconsistently between the strategies and approaches are also indicated in the figures.
Figure 3.
Comparison of adult a priori data and infant GM, WM and CSF distributions constructed using the new (top panel) and default (bottom panel) strategies as outlined in Figure 1. Differences are displayed as an overlay on the infant data to show spatial location (left panels, where red and yellow indicates infant probability is greater than adult probability while blue is for the reverse effect), and as a histogram to show their distribution (right panels). Results shown are for differences of at least 20% in tissue probability and are based on the second pass output. For the histograms, the x-axis represents the magnitude of differences and the y-axis the corresponding number of voxels. The flat line between −0.2 and 0.2 indicates the 20% threshold used, while the curves on the right and left indicate where the infant tissue probability is greater or less than the adult data respectively. Percentages of voxels in each segment of the histogram (i.e. <−0.2, no difference, >0.2) are listed below the horizontal axis.
Figure 4.
Comparison of pediatric a priori data versus infant GM, WM and CSF distributions constructed using new (top panel) and default (bottom panel) strategies as outlined in Figure 1. Differences are displayed as an overlay on the infant data to show spatial location (left panels, where red and yellow indicates infant probability is greater than adult probability while blue is for the reverse effect), and as a histogram to show their distribution (right panels). Results shown are for differences of at least 20% in tissue probability and are based on the second pass output. For the histograms, the x-axis represents the magnitude of differences and the y-axis the corresponding number of voxels. The flat line between −0.2 and 0.2 indicates the 20% threshold used, while the curves on the right and left indicate where the infant tissue probability is greater or less than the pediatric data respectively. Percentages of voxels in each segment of the histogram (i.e. <−0.2, no difference, >0.2) are listed below the horizontal axis.
Figure 6.
Comparison of first pass and second pass probability maps for images prepared using the new (top panel) and default (bottom panel) strategies. The histograms show the distribution of these differences for each tissue class and strategy. Results shown are for differences of at least 20% in tissue probability. The x-axis represents the magnitude of differences and the y-axis the corresponding number of voxels The flat line between −0.2 and 0.2 indicates the 20% threshold used, while the curves on the right and left indicate where the infant tissue probability obtained using the second pass approach is greater or less than the infant data obtained from the first pass approach respectively. Percentages of voxels in each segment of the histogram (i.e. <−0.2, no difference, >0.2) are listed below the horizontal axis
Results
All processing strategies, including the use of adult a priori data as a reference template for registration and segmentation, produce high quality images as displayed in Figure 2. However there are significant differences in tissue classifications between the newly created infant and the adult (Figure 3) as well the infant and pediatric (Figure 4) GM, WM and CSF tissue probability maps in both strategies employed here. The degree of discordance seems to be higher when the default strategy is used (via the default SPM5 segmentation routine) to generate the resulting infant data. In general the results show that in the infant reference data, brain tissues tend to be classified as GM with higher probability compared to adult reference data (Figure 3) and to a lesser extent with pediatric reference data (Figure 4), as indicated by the red and yellow colors.
However this seems to occur at the expense of WM, which is presented with higher probability in both the adult and pediatric reference data as indicated by the blue color. With infant data generated using the new strategy versus adult reference data, GM is classified with greater probability at the superior frontal gyrus, medial frontal gyrus, anterior cingulate, putamen, and thalamus as shown in Figure 3 (top). In addition to these areas, comparison of infant data obtained using the default strategy versus the adult reference data results in additional pixels in the inferior frontal gyrus, middle occipital gyrus and superior frontal gyrus classified as GM with a higher probability: Figure 3 (bottom).
We also used the proportion of voxels that showed a difference of 20% or more in classification probability, to additionally quantify the graphically depicted data shown in Figure 3 and Figure 4 (left panel). These proportions are calculated by counting the number of voxels that are below −0.2 or above 0.2 (20% threshold rule) and dividing it by the total number of voxel in the tissue under consideration (4.6 million pixels). Each histogram shown in Figure 3 to Figure 6 is divided into three distinct areas. The left and right “bumps” represent the proportion of voxels that are below −0.2 and above 0.2 respectively when looking at the distribution of the difference of the two images considered. The middle section between (⩲0.2) corresponds to the proportion of voxels that are classified consistently by both methods, according to the 20% threshold rule used in this paper. The proportion of voxels in each segment of the histogram is indicated below the horizontal axis of Figure 3–Figure 6. The sum of the proportion in the three areas should add to 1.
Additionally, if one strategy or approach causes voxels to misclassify as one tissue class (for example GM) then we expect an equivalent number of voxels to be lost from the rest of the tissue classes combined (for example WM + CSF). However in our case since we used a 20% threshold rule the numbers may not always balance out because some voxels may also be shifted in and out of the 20% threshold group. For example in Figure 3 (right top panel) the proportion of voxels that are misclassified as high for GM is 9%, while the proportion of voxels that are misclassified as low for the combined WM and CSF is 10% (9% + 1%). Although close the proportions are not exactly the same, this is because 1% of the voxels classified as high for GM shifts into the concordant classification group.
When we compare the newly created infant GM, WM and CSF using the new strategy with the corresponding adult reference data: Figure 3 (top) the proportion of voxels that showed a difference of 20% or more were 12%, 10% and 11% for GM, WM and CSF respectively. The magnitude of these proportions increases to 20%, 16%, and 24.2% when the infant data is constructed using the default strategy: Figure 3 (bottom). This indicates that the use of adult a priori data in the segmentation step, as is the case for the default strategy, increases the number of discordant (beyond 20% difference in probability) voxels. Among the voxels that show a difference of 20% or more, the proportion of voxels that the newly constructed infant data classifies with higher probability, were 75%, 10% and 91% for GM, WM and CSF respectively when the data is obtained using the new strategy. These proportions were 80%, 6.2% and 99% when the comparison infant data was created with the default strategy. These distributions are displayed on the right panel of Figure 3. This shows that the newly created infant data classifies GM and CSF with higher probability but at the expense of WM which was classified with lower probability.
Similar patterns were observed when we compare the newly created infant data with the corresponding pediatric reference data. When comparing the infant data created using the new strategy with the pediatric reference data, the proportions of voxels that show a difference of 20% or more in classification probability were 11%, 7% and 11% for GM, WM and CSF respectively: Figure 4 (top). These proportions were 11%, 7% and 27% when the infant data was constructed using the default strategy: Figure 4 (bottom). Unlike the comparison of the infant data with the adult reference data, where the use of the default strategy increased the proportion of voxels that shows a difference of 20 % or more; here the proportions are very similar (except for CSF) for both strategies used. In addition the proportion of voxels classified with high probability as GM, WM, and CSF in the second pass infant data that is constructed using the new strategy are 72%, 28.6% and 81.8% respectively. These proportions were 72.7%, 14.3% and 96% when the infant data was constructed using the default strategy. These distributions are displayed in the right panel of Figure 4.
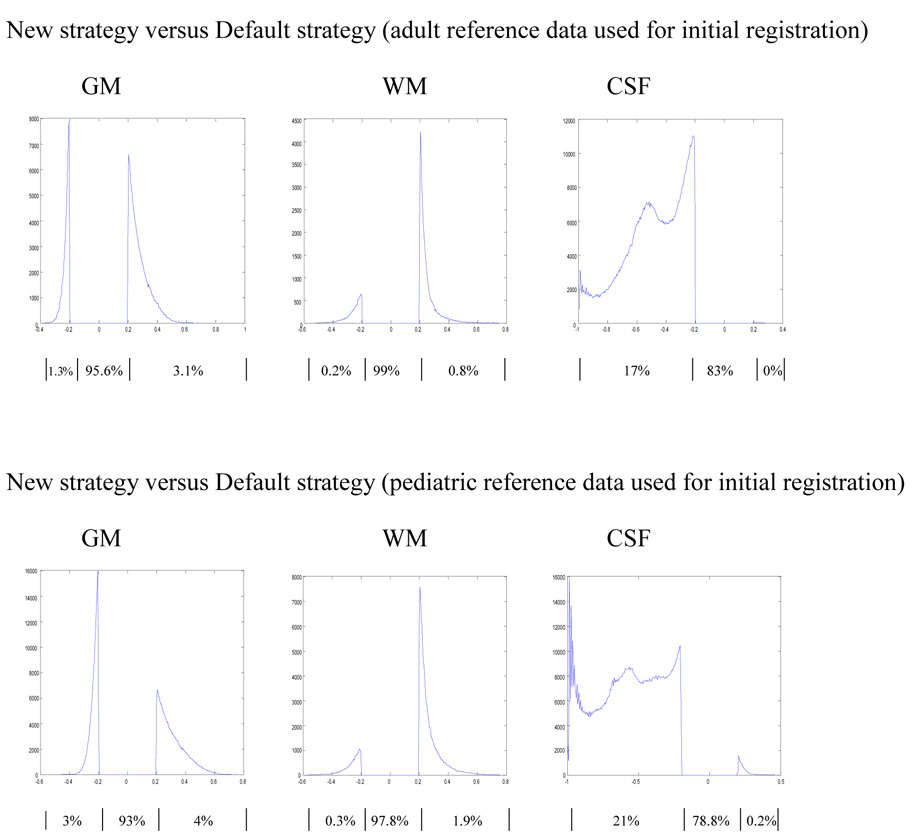
The comparisons between the images obtained using the new and the default strategy resulted in similar images with only moderate differences as defined here. The proportion of voxels that show a difference of 20% or more between the images resulted from the two strategies where adult reference data was used for initial registration (Figure 1: DA2 and NA2), were 4.4%, 1% and 17% for GM, WM and CSF respectively (Figure 5: top). Similar but consistently greater differences were observed when comparing the newly created infant data using the two strategies where the initial registration was based on pediatric reference data (Figure1: DP2 AND NP2). In this case the proportions of voxels that differ by more than 20% were 7%, 2.2% and 21.2% for GM, WM and CSF respectively (Figure 5: bottom).
Figure 5.
Comparison of GM, WM and CSF probability distributions in second pass infant data obtained with the new and default strategy. In the top panel adult and in the bottom pediatric reference data were used for initial registration. The histograms indicate the distribution of the differences for each tissue class. Results shown are for differences of at least 20% in tissue probability. For the histograms, the x-axis represents the magnitude of differences and the y-axis the corresponding number of voxels. The flat line between −0.2 and 0.2 indicates the 20% threshold used, while the curves on the right and left indicate where the infant tissue probability obtained using the new strategy is greater or less than the infant data obtained using the default strategy respectively. Percentages of voxels in each segment of the histogram (i.e. <−0.2, no difference, >0.2) are listed below the horizontal axis.
The two pass approach we employed to minimize the impact of the adult prior data results in similar tissue classification in the final infant templates, with very little difference between the images obtained using the two approaches as shown in Figure 6. The proportion of voxels that show a difference of 20% or more were less than 1% for both GM and WM regardless of the strategy used to generate the infant data when the initial registration were with adult data. However 2% of voxels showed a difference of 20% or more for CSF probability, when the infant data is constructed using the new strategy and 6.7% of voxels when using the default strategy. (Figure 6: top) Similar patterns but with slightly higher percentages of discordant voxels were observed when the first pass and second pass approaches were compared for images generated using the pediatric reference data for initial registration. Regardless of the strategy used to generate the infant images, the proportions of voxels that exhibit 20% or greater differences were less than 1% for both GM and WM. However for CSF these proportions were 3% when infant data is generated under the new strategy and 15.4% under the default strategy (Figure 6: bottom).
Discussion
In this article, we present a procedure to create an infant template and a priori probability maps of GM, WM and CSF. We used a novel approach that does not require the use of prior data for segmenting brain images in an effort to reduce the impact of inappropriate adult or pediatric prior data. We showed that the use of the default adult or pediatric template to segment the infant data will result in misclassifications of infant tissue, as shown in Figure 3 and Figure 4. This is consistent with the findings reported when comparing the use of adult a priori data for segmenting pediatric data (Wilke et al. 2003b) and is likely due to the different shape and size of developing infant brains as well as different GM/WM ratios in the infant brain relative to children and adults.
The observed difference between the newly created infant data and the pediatric reference data (Figure1: h-b and j-b) is less severe compared to the difference between the infant and adult reference data (Figure 1: g-a and i-a) as indicated by the proportion of discordant voxels, particularly when the default strategy was used to generate the infant data (Figure 1: h-b and g-a). However, the appearance of the observed discordance between the infant and pediatric data mimicked the difference observed between the infant and adult data suggesting that the pediatric data is closer to the adult data in tissue distribution than to the infant data (Figure 3 and 4). This is apparent when we recognize that the proportion of voxels classified with high probability in the second pass infant data are very similar whether adult or pediatric reference data were used for initial registration (Figure 1 : g-a and h-b: i-a and j-b). This is depicted in the right panel of Figure 3 and Figure 4.
The indirect comparison between the two processing strategies through their comparison with the adult reference data as shown in Figure 3, suggests that the use of the default strategy results in a higher number of discordant voxel classifications than the new strategy. This difference is more precisely depicted in the direct comparison of images obtained from the two strategies as displayed on Figure 5. This suggests that the new strategy with less discordant voxels may provide more consistent classification for infant brain image data.
However, regardless of the strategy employed to obtain the infant template, our results demonstrate the importance of using appropriately constructed infant templates for future normalization and segmentation of infant images, since the alternative of using adult or pediatric templates will result in misclassification of tissue class as shown in Figure 3 and Figure 4.
The comparison between images produced using first-pass (using adult data) and second-pass (using the resulting infant data from first pass procedure) images shows very little difference (Figure 6). This is likely due to the fact that the priors are warped to the input data during the first pass, so that the influence of the priors is diminished in final images. In addition the use of affine and non-linear transformation in both approaches (first and second pass) coupled with the introduction of the HMRF model, to apply spatial constraints from neighboring voxels, might further reduce the impact of the adult prior data during the first pass segmentation step.
Limitations
The MR data is obtained from infants sedated by either chloral hydrate or Nembutal. The impact of the sedation, if any, on the resulting MR images is unknown. Although unlikely, it is possible that the sedation might have an impact on T1 image intensity distribution and the resulting template and probability maps through either a direct effect or indirectly due to alterations in cerebral perfusion induced by the drugs.
The purpose of spatial normalization is to be able to compare brain activations or structural changes across individuals, which will always entail deforming the contributing subjects to a common spatial reference frame. Comparing the activation patterns from children and adults and finding the evolution that takes place in childhood is only possible if a common reference frame is used for the subjects being compared. Considering the advantages that such a common spatial frame offers over myriads of study-specific custom reference frames, we opted to use the most commonly-used reference frame for our study here (MNI-152). While this choice is arguable, we still believe that the advantages outweigh the disadvantages at this point.
The original study that motivated construction of the template and probability maps using the new methods reported here focused on infants at nominally 12 months of age (± 3 mo.) as described above in the Methods: Subjects section. So for our purposes, the age span of infants selected for template construction matches our population exactly. However, even this extremely narrow age range might be too wide to preserve fine scale changes over this 6 month period of rapid brain development (Huttenlocher 1979). Also, the applicability of the specific template and probability maps we offer may be limited to other studies of 1 year olds. The methods described here provide potential users with a completely general formula to construct their own prior-free, templates as needed. In this context, our specific data set is just an example of the power of the new method. For readers who may find it helpful to use our template directly we have made it available on our website at http://www.irc.cchmc.org/software.
Conclusion
In the absence of a gold standard, a direct comparison between the proposed new procedure and the default procedure may not establish which procedure is more accurate. However the comparison presented here of the new and default procedure with a priori reference data from adults or children, suggests that the new procedure is superior for use in segmentation of infant brain images as it results in less severe misclassification when compared to the default procedure using adult a priori probability maps. The new infant template and a priori data obtained from the new procedure is therefore recommended for use as a reference data for spatial normalization and segmentation of infant brain MRI data.
Acknowledgements
This work was partially supported by the National Institute of Deafness and Communication Disorders (RO1-DC07186)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ashburner J, Friston KJ. Unified segmentation. NeuroImage. 2005;26:839–851. doi: 10.1016/j.neuroimage.2005.02.018. [DOI] [PubMed] [Google Scholar]
- Burgund ED, Kang HC, Kelly JE, Buckner RL, Snyder AZ, Petersen SE, Schlaggar BL. The feasibility of a common stereotactic space for children and adults in fMRI studies of development. NeuroImage. 2002;17:184–200. doi: 10.1006/nimg.2002.1174. [DOI] [PubMed] [Google Scholar]
- Cuadra MB, Cammoun L, Butz T, Cuisenaire O, Thiran JP. Comparison and validation of tissue modelization and statistical classification methods in T1-weighted MR brain images. IEEE Trans Med Imaging. 2005;24:1548–1565. doi: 10.1109/TMI.2005.857652. [DOI] [PubMed] [Google Scholar]
- Dehaene-Lambertz G, Dehaene S, Hertz-Pannier L. Functional neuroimaging of speech perception in infants. Science. 2002;298:2013–2015. doi: 10.1126/science.1077066. [DOI] [PubMed] [Google Scholar]
- Evans AC, Collins DL, Mills SR, Brown ED, Kelly RL, Peters TM. 3D statistical neuroanatomical models from 305 MRI volumes. Proc. IEEE-Nucl Sci Symp Med Imaging Conf. 1993:1813–1817. [Google Scholar]
- Gaser C, Altaye M, Wilke M, Holland SK. Unified segmentation without tissue priors. NeuroImage. 2007;36 Suppl. 1:S68. [Google Scholar]
- Guimond A, Meunier J, Thirion JP. Average brain models: a convergence study. Comput Vis Image Underst. 2000;77(2):192–210. [Google Scholar]
- Hoeksma MR, Kenemans JL, Kemner C, Van Engeland H. Variability in spatial normalization of pediatric and adult brain images. Clin Neurophysiol. 2005;116:1188–1194. doi: 10.1016/j.clinph.2004.12.021. [DOI] [PubMed] [Google Scholar]
- Huttenlocher P. Synaptic density in human frontal cortex - developmental changes and effects of age. Brain Res. 1979;163:195–205. doi: 10.1016/0006-8993(79)90349-4. [DOI] [PubMed] [Google Scholar]
- Kazemi K, Moghaddam HA, Grebe R, Gondry-Jouet C, Wallois F. A neonatal atlas template for spatial normalization of whole-brain magnetic resonance images of newborns: preliminary results. NeuroImage. 2007;37:463–473. doi: 10.1016/j.neuroimage.2007.05.004. [DOI] [PubMed] [Google Scholar]
- Machilsen B, D'Agostino E, Maes F, Vandermeulen D, Hahn HK, Lagae L, Stiers P. Linear normalization of MR brain images in pediatric patients with periventricular leukomalacia. NeuroImage. 2007;35:686–697. doi: 10.1016/j.neuroimage.2006.12.037. [DOI] [PubMed] [Google Scholar]
- Mugler JP, Brookeman JR. Three-dimensional magnetization –prepared rapid gradient-echo imaging (3D MP RAGE) Magn Reson Med. 1990;15:152–157. doi: 10.1002/mrm.1910150117. [DOI] [PubMed] [Google Scholar]
- Murgasova M, Dyet L, Edwards D, Rutherford M, Hajnal JV, Rueckert D. Segmentation of brain MRI in young children. Academic Radiology. 2007;14:1350–1366. doi: 10.1016/j.acra.2007.07.020. [DOI] [PubMed] [Google Scholar]
- Muzik O, Chugani DC, Juhász C, Shen C, Chugani HT. Statistical parametric mapping: assessment of application in children. NeuroImage. 2000;12:538–549. doi: 10.1006/nimg.2000.0651. [DOI] [PubMed] [Google Scholar]
- Srinivasan L, Dutta R, Counsell SJ, Allsop JM, Boradman JP, Rutherford MA, Edwards AD. Quantification of deep gray matter in preterm infants at term-equivalent age using manual volumetry of 3-Tesla magnetic resonance images. Pediatrics. 2007;119:759–765. doi: 10.1542/peds.2006-2508. [DOI] [PubMed] [Google Scholar]
- Talairach J, Tournoux P. Co-planar Stereotaxic Atlas of the Human Brain. Thieme, Stuttgart, Germany: 1988. [Google Scholar]
- Wilke M, Holland SK. Variability of gray and white matter during normal development: a voxel-based MRI analysis. Neuroreport. 2003a;14:1887–1890. doi: 10.1097/01.wnr.0000090951.15465.c8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilke M, Schmithorst VJ, Holland SK. Normative pediatric brain data for spatial normalization and segmentation differs from standard adult data. Magn Reson Med. 2003b;50:749–757. doi: 10.1002/mrm.10606. [DOI] [PubMed] [Google Scholar]
- Wilke M, Holland SK, Altaye M, Gaser C. Template-O-Matic: A toolbox for creating customized pediatric templates. NeuroImage. 2008 doi: 10.1016/j.neuroimage.2008.02.056. (in press) [DOI] [PubMed] [Google Scholar]