Abstract
MicroRNAs are a class of small RNAs that are increasingly being recognized as important regulators of gene expression. Although hundreds of microRNAs are present in the mammalian genome, genetic studies addressing their physiological roles are at an early stage. We have shown that mice deficient for bic/microRNA-155 are immunodeficient and display increased lung airway remodeling. We demonstrate a requirement of bic/microRNA-155 for the function of B and T lymphocytes and dendritic cells. Transcriptome analysis of bic/microRNA-155–deficient CD4+ T cells identified a wide spectrum of microRNA-155–regulated genes, including cytokines, chemokines, and transcription factors. Our work suggests that bic/microRNA-155 plays a key role in the homeostasis and function of the immune system.
MicroRNAs (miRNAs) posttranscriptionally regulate gene expression by forming imperfect base pairing with sequences in the 3′ untranslated region (3′ UTR) of genes to prevent protein accumulation by repressing translation or by inducing mRNA degradation (1, 2). More than 500 miRNAs have been identified in mammals, although their functions are only now being elucidated (3). In the immune system, the enzyme responsible for regulatory RNA biogenesis, Dicer, is required for T cell function, which suggests regulatory roles for miRNAs in lymphocytes (4, 5). One miRNA, miR-155 (6), maps within, and is processed from, an exon of the noncoding RNA known as bic (7, 8), its primary miRNA precursor (9). bic/miR-155 shows greatly increased expression in activated B and T cells (9–11), as well as in activated macrophages and dendritic cells (DCs) (12, 13). Overexpression of bic/miR-155 has been reported in B cell lymphomas and solid tumors (14), and transgenic miR-155 mice have also been shown to develop B cell malignancies in vivo (15), indicating that the locus may also be linked to cancer.
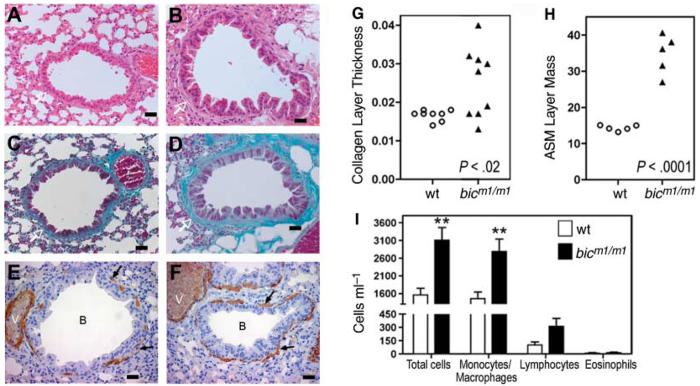
To define the in vivo role of bic/miR-155 (bic), we generated mutant alleles in embryonic stem cells (16) to obtain bic-deficient (bicm1/m1 and bicm2/m2) mice (fig. S1, A and B). bic-deficient mice were viable and fertile but developed lung pathology with age. At 320 to 350 days, 56% (5 out of 9) of bicm1/m1 mice displayed significant remodeling of lung airways, with increased bronchiolar subepithelial collagen deposition and increased cell mass of sub-bronchiolar myofibroblasts (Fig. 1, B, D, and F), relative to age-matched control mice (n = 8) (Fig. 1, A, C, and E). A statistically significant increase in the ratio of collagen thickness/bronchiolar diameter and smooth muscle cell area/bronchiolar diameter could be measured in bic-deficient mice, compared with wild-type controls (Fig. 1, G and H). Increased airway remodeling in aged bicm1/m1 mice was accompanied by a significant increase in the numbers of leukocytes in bronchoalveolar lavage fluids (BAL) (Fig. 1, I) but not the lung interstitium. These changes are reminiscent of the lung fibrosis that often complicates systemic autoimmune processes with lung involvement (17, 18). We also noted that many bicm1/m1 mice developed enteric inflammation, a trait we have not investigated further. Thus, the phenotype we observed suggested that bic/miR-155 may participate or play a role in regulating the homeostasis of the immune system.
Fig. 1.
Mice deficient for bic/miR-155 show increased lung airway remodeling (A to F) Histological examination of sections of lung bronchioles from control wild-type (A, C, and E) and bicm1/m1 mice (B, D, and F). Scale bar, 100 μm. (A and B) Haematoxylin and eosin stain; (C and D) Masson Trichrome stain; (E and F) Immunohistochemical staining for smooth muscle actin. Collagen layer (white arrows), lung myofibroblasts (black arrows), bronchioles (B), and blood vessels (V) are indicated. (G) Quantitation of peribronchiolar collagen thickness or (H) airways smooth muscle cell (ASM) mass in bicm1/m1 mice compared with that of wild-type mice. (G) P < 0.02 or (H) P < 0.0001, in comparison with wild-type group, Student's two-tailed t test. Open circles, control mice; filled triangles, bicm1/m1 mice. Notably, bicm1/m1 mice with increased collagen layer thickness also had increased ASM mass. (I) Total and differential cell counts in BAL from the indicated mice. Data are the mean + SE from seven bic-deficient mice and six control mice. **P < 0.01 in comparison with wild-type group, Student's two-tailed t test.
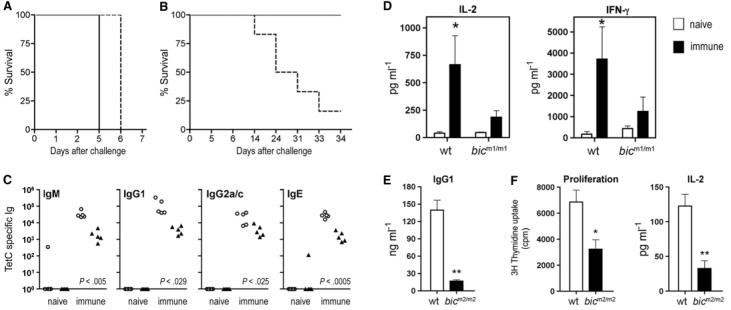
The pathology observed in bic-deficient mice prompted us to examine the requirement of bic/miR-155 in immunity. Although no gross defect in myeloid or lymphoid development in bic-deficient mice was observed (tables S1 and S2), protective immunity did appear to be impaired. Thus, after intravenous immunization with the live attenuated form of the enteric pathogen Salmonella typhimurium (aroA mutant strain), mice were assessed for their ability to resist oral challenge with virulent S. typhimurium bacteria (19, 20). Both unvaccinated bicm2/m2 and wild-type control mice (5 out of 5; n = 5) died within 7 days after infection (Fig. 2A). However, unlike their wild-type counterparts, bicm2/m2 mice were less readily protected by aroA vaccination, and the majority of mice (5 out of 6; n = 6) succumbed to challenge with the virulent strain by 33 days after infection (Fig. 2B). Thus, immunized bic-deficient mice, unlike wild-type mice, could not be protected by immunization to this pathogen.
Fig. 2.
Defective adaptive immunity by bic-deficient mice. (A) Survival curve for mice (n = 5 in each group) infected orally with 1 × 108 colony-forming units (CFU)–virulent S. typhimurium strain SL344. As expected for mice of this genetic background, all failed to survive challenge. (B) Survival of mice (n = 6 in each group) infected intravenously with 1 × 104 CFU of S. typhimurium aroA strain followed by oral challenge with S. typhimurium SL344 6 weeks after prime. In contrast with control mice, bicm2/m2 mice demonstrate reduced survival after challenge. (A and B) Line, control C57BL/6J (wild-type) mice; dashed line, N5 C57BL/6J backcross bicm2/m2 mice. (C) TetC-specific Ig levels from control mice (open circles) or bic-deficient mice (filled triangles) immunized with TetC at days 1 and 21 and analyzed 13 days after secondary immunization. P values denote significant differences; Student's two-tailed t test. (D) Production of IL-2 and IFN-γ by splenocytes isolated from wild-type or bicm1/m1-naïve mice (open bars) or immunized with TetC as in (C) (closed bars) and cultured for 48 hours in the presence of TetC. Data are the mean ± SE from four mice. *P < 0.05 versus naïve mice; Student's two-tailed t test. (E) Reduced IgG1 production by bicm2/m2 B cells cultured in the presence of LPS and IL-4 for 4 days. Data are the mean + SE from 3 mice. **P < 0.01 versus wild-type; Student's two-tailed t test. (F) Significantly reduced proliferation and IL-2 production by ovalbumin T cell receptor transgenic (OT-II) cells cultured with LPS-matured, bone marrow–derived, bic-deficient DCs in the presence of cognate (2.5 μM) ovalbumin protein. Cell proliferation was determined by [3H]-thymidine incorporation at 72 hours. IL-2 was measured from supernatants by enzyme-linked immunosorbent assay (ELISA) at 48 hours. Data are the mean + SE from five mice of each genotype. *P < 0.05 versus wild-type; Student's two-tailed t test.
Protective immunity requires the function of T and B lymphocytes. Therefore, we next examined the in vivo B and T cell responses of bic-deficient mice immunized with the T-dependent antigen, tetanus toxin fragment C protein (TetC). Immunized bicm1/m1 mice produced significantly reduced amounts of immunoglobulin M (IgM) and switched antigen-specific antibodies (Fig. 2C), indicative of impaired B cell responses. For examination of T cell function, splenocytes from mice immunized with TetC were restimulated in vitro, and the levels of interleukin (IL)–2 and interferon (IFN)–γ cytokines were measured. As expected, splenocytes from wild-type mice immunized with TetC produced significantly increased levels of IL-2 and IFN-γ relative to naive mice (Fig. 2D). In contrast, bicm1/m1 immunized mice failed to produce significant levels of these cytokines (Fig. 2D). Thus, B and T cell responses were diminished in bic-deficient mice, possibly contributing to their impaired enteric immunity.
To understand the nature of defective immune responses in vivo, we explored the possibility of an intrinsic requirement for bic/miR-155 in B cells and T cells. Dendritic cell (DC) function was also tested, because these cells act as professional antigen presenting cells (APCs) with the ability to influence T cell activation and differentiation. Production of IgG1 by lipopolysaccharide (LPS)– and IL-4–stimulated bicm2/m2 B cells was significantly reduced (Fig. 2E), although this defect did not appear to correspond with abnormal proliferation (fig. S2). After encountering antigen, DCs increase their immunostimulatory capacity (21) through a process that is mimicked in vitro by stimulation with LPS. After treatment with LPS bicm2/m2, bone marrow–derived DCs expressed levels of major histocompatibility complex–II and costimulatory molecules similar to those seen on identically treated matured wild-type DCs (fig. S3, A and B), which indicates that bic/miR-155 is not required for maturation. Nevertheless, bicm2/m2 DCs failed to efficiently activate T cells, consistent with defective antigen presentation or costimulatory function (Fig. 2F). Collectively, these results suggest that the effects of bic/miR-155 may operate in part on T cells through its influence on DC function.
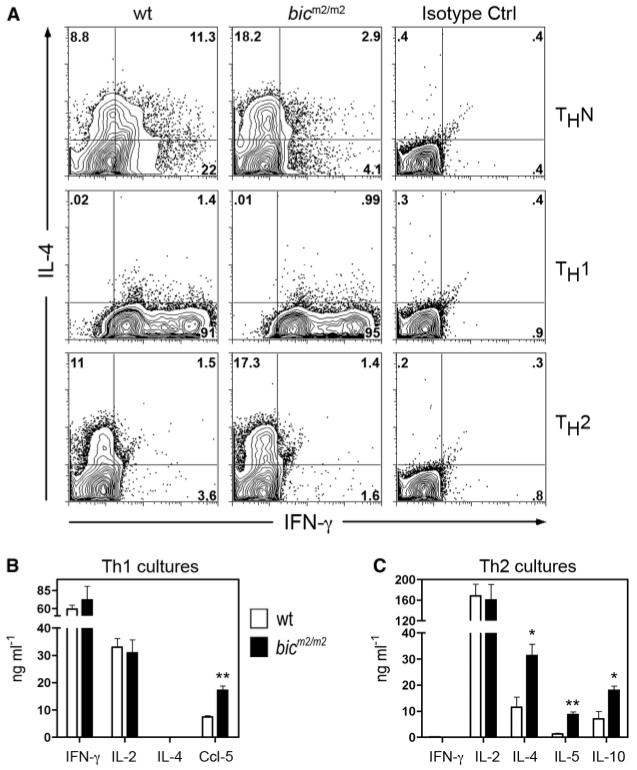
To establish whether there is also an intrinsic requirement for bic/miR-155 in T cell function, the response of receptor-stimulated naïve bicm2/m2 CD4+ T cells was tested. Despite normal proliferation, uncommitted bicm2/m2 CD4+ cells showed a significant reduction of the T helper (Th)–1 cytokine, IFN-γ, after stimulation with antibodies to CD3 and CD28 (fig. S4). A reduction by a factor of 5 in the number of IFN-γ–producing cells was also observed after restimulation of bicm2/m2 CD4+ T cells cultured under conditions designed not to polarize Th responses (Fig. 3A); and was accompanied by a doubling in the number of IL-4 single-producing cells (Fig. 3A). In light of the expression of bic/miR-155 in both Th1 and Th2 cell lineages (fig. S5, A and B), we next examined the phenotype of bicm2/m2 CD4+ T cells after culture in conditions that promote Th1 or Th2 cell differentiation. The levels of IFN-γ, as well as the number of bicm2/m2 Th1 cells secreting cytokine, were similar to controls, which indicates that bic/miR-155 is not required for Th1 differentiation (Fig. 3, A and B). However, phenotypic alterations were observed as bicm2/m2 Th1 cells produced elevated levels of CCL-5 (Fig. 3B and table S3). By contrast, increased commitment to the Th2 pathway was evident in bicm2/m2 Th2 cell cultures as higher numbers of IL-4–producing cells were observed (Fig. 3A). In support of this result, enhanced levels of the Th2 cytokines IL-4, IL-5, and IL-10 were generated by bicm2/m2 cells after culture in Th2 polarizing conditions (Fig. 3C). Taken together, these data demonstrate that bic-deficient CD4+ T cells are intrinsically biased toward Th2 differentiation. Moreover, Th1 cells may have altered function despite normal production of IFN-γ.
Fig. 3.
Increased Th2 polarization and amplified Th2 cytokine production by bic-deficient CD4+ T cells. CD4+CD62L+ cells of indicated genotypes were cultured under (A, middle panel, and B) Th1 conditions, (A, lower panel, and C) Th2 in vitro differentiation conditions, or (A, upper panel) nonpolarizing (ThN) conditions and restimulated with immobilized antibody to CD3 (10 μg/ml) and soluble 2 μg/ml antibody to CD28 on day 6. (A) Intracellular cytometric analysis for IFN-γ and IL-4 production (16). The panel shows a representative result of three mice of each genotype analyzed in the same experiment. Data are representative of two independent experiments (n = 3 per genotype). Numbers in each quadrant are percentages of cells of indicated phenotype. (B and C) Cytokine levels were assayed by ELISA 21 hours after restimulation of cells cultured under (B) Th1 or (C) Th2 polarizing conditions. Data are the mean + SE from three individual mice. *P < 0.05 or **P < 0.01 versus wild-type; Student's two-tailed t test.
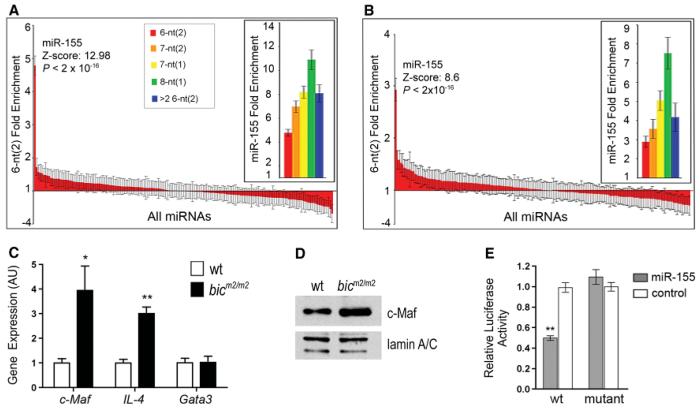
To understand how bic/miR-155 regulates Th2 commitment and to gain a more global insight into the extent of deregulation in Th1 cells, we analyzed gene expression in bicm2/m2 Th1 or Th2 cells using microarray analysis. In addition, because the 5′ region of miRNAs (referred to as the “seed” region) is believed to be crucial for target mRNA recognition (1, 2), we searched the 3′ UTRs of significantly up-regulated genes in microarrays for the presence of seed matches specific for miR-155. In bic-deficient Th1 cells, we identified 46 of 53 up-regulated transcripts as potential miR-155 targets (table S3 and fig. S6). In bic-deficient Th2 cells, 53 out of 99 up-regulated transcripts were predicted targets (table S4 and fig. S7). To confirm these genes as likely targets of miR-155, we then searched the 3′ UTRs for seed matches specific for all of the known mouse miRNAs in the miRbase public database (3). miR-155 seed sequences were significantly overrepresented over all other tested mouse miRNAs, indicating a significant probability that these genes are direct targets of miR-155 (Fig. 4, A and B). This computational data strongly suggests that miR-155 represses a wide assortment of genes in CD4+ T cells and lends support for the hypothesis that miRNA targets are generally abundant in mammals (22).
Fig. 4.
miR-155 pattern sequences are enriched in the Th1 and Th2 cell up-regulated genes, and c-Maf is a bona fide target of miR-155. (A and B) Fold enrichment of 5′ miRNA pattern sequences of the indicated types contained in the 3′ UTRs of the (A) Th1 or (B) Th2 cDNA microarray significantly up-regulated gene sets. The standard deviation, Z score, and P value were calculated by sampling 1000 random sets of 53 (for Th1 set) or 99 (for Th2 set) genes from the mouse genome (16). Data are fold enrichment ± SD. (C) Quantitative PCR analysis for Gata3, c-Maf, and IL-4 transcript levels from Th2 cells re-stimulated with antibodies to CD3 and CD28. Data are the mean + SE from three mice. *P < 0.05 versus wild-type; Student's two-tailed t test. (D) c-MAF protein levels were assessed by Western blot of nuclear extracts of Th2 cells isolated from the indicated genotypes. Expression of lamin A/C was used as loading control. (E) miR-155–dependent repression of c-Maf reporter in vitro. A luciferase (Rluc) reporter was used to validate c-Maf as a direct target of miR-155. Wild-type (wt) or mutant plasmids (mut) were contransfected with the indicated duplex miRNA for miR-155 (open bars) or control Cel-miR-64 (filled bars) into HeLa S3 cells. Data are mean ± SE from three experiments. **P < 0.0001 in comparison with wild-type plasmid treated with nonspecific RNA duplex, Cel-miR-64; Student's two-tailed t test.
A wide spectrum of miR-155 target genes with diverse molecular roles, such as T cell costimulation (e.g., Tnfsf9), chemotaxis (e.g., Ccl-5), and signaling (e.g., Ikbke), were identified. Among these, we noted that the transcription factor c-Maf contains phylogenetically conserved miR-155 seed matches in the 3′ UTR (fig. S8). c-Maf is a potent transactivator of the IL-4 promoter, and ectopically expressed c-Maf is sufficient to cause increased IL-4, IL-5 and IL-10 production by Th2 cells (23–25). In concordance with the microarray results, a significant induction of c-Maf mRNA was detected in bicm2/m2 Th2 cells, and the levels of c-Maf protein were correspondingly increased (Fig. 4, C and D). By contrast, levels of Gata3 transcript, which does not contain a miR-155 seed, were not elevated (Fig. 4C). Increased expression of c-Maf may thus contribute, at least in part, to the increased Th2 cytokine production phenotype observed in bicm2/m2 Th2 cells. To further confirm whether c-Maf is a direct target of miR-155, we cloned its 3′ UTR into a luciferase reporter plasmid. The wild-type c-Maf reporter exhibited significant miR-155–dependent repression relative to the reporter with a mutant seed sequence, which indicates that this is a direct target for miR-155 (Fig. 4E). We conclude from these experiments that bic/miR-155 modulates levels of c-Maf in CD4+ T cells and this is likely to contribute to the attenuation of Th2 cell responses in vivo.
Our data demonstrate that mice carrying a null mutation in the bic/miR-155 gene display altered immune responses. Thus, along with an increase in airways remodeling suggestive of altered homeostasis, we observed that bic/miR-155 regulates the function of both lymphocytes and DCs, leading to an overall diminution of immune responses. The identification of multiple novel potential targets of miR-155 supports the view that bic/miR-155 is a core regulator of gene expression in multiple cell types, with a “targetome” optimized to modulate the immune response. Interestingly, bic-deficient mice share some of the cellular features observed in CD4-Cre/DicerFL mice, including defects in CD4+ T cell cytokine production and immune homeostasis (3, 4). It will now be important to define the patho-physiology of bic-deficient lymphocytes and further test the role of miR-155–dependent repression of c-Maf on immune responses in vivo. The strength of the bic/miR-155 mutant phenotype more generally suggests critical roles for miRNAs in vivo, with potentially severe loss-of-function phenotypes directly relevant to human disease. In this regard, it is intriguing that the human BIC/miR-155 gene maps to an asthma, pollen sensitivity, and atopic dermatitis susceptibility region on chromosome 21q21 (26–28). Given the severe phenotypes noted in these mice, BIC/miR-155 should be investigated as a potential immune disease locus in humans.
Acknowledgments
The authors acknowledge D. Corry, F. Colucci, and members of the Bradley laboratory for critical reading of the manuscript. We also thank Beverley Haynes for histology work. A.R. was supported by a Ruth Kirschstein Fellowship. E.V. was supported by a Babraham Institute Career Progression Fellowship. M.T. was supported by the Medical Research Council and Biotechnology and Biological Sciences Research Council. A.B., G.D., A.E. were supported by the Wellcome Trust and Sanger Institute grant number 79643.
Footnotes
Supporting Online Material
www.sciencemag.org/cgi/content/full/316/5824/608/DC1 Materials and Methods Figs. S1 to S8 Tables S1 to S4 References
References and Notes
- 1.Bartel DP. Cell. 2004;116:281. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Engels BM, Hutvagner G. Oncogene. 2006;25:6163. doi: 10.1038/sj.onc.1209909. [DOI] [PubMed] [Google Scholar]
- 3.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. Nucleic Acids Res. 2006;34:D140. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Muljo SA, et al. J. Exp. Med. 2005;202:261. doi: 10.1084/jem.20050678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cobb BS, et al. J. Exp. Med. 2006;203:2519. doi: 10.1084/jem.20061692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lagos-Quintana M, et al. Curr. Biol. 2002;12:735. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- 7.Tam W, Ben-Yehuda D, Hayward WS. Mol. Cell. Biol. 1997;17:1490. doi: 10.1128/mcb.17.3.1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tam W. Gene. 2001;274:157. doi: 10.1016/s0378-1119(01)00612-6. [DOI] [PubMed] [Google Scholar]
- 9.Eis PS, et al. Proc. Natl. Acad. Sci. U.S.A. 2005;102:3627. doi: 10.1073/pnas.0500613102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haasch D, et al. Cell. Immunol. 2002;217:78. doi: 10.1016/s0008-8749(02)00506-3. [DOI] [PubMed] [Google Scholar]
- 11.van den Berg A, et al. Genes Chrom. Cancer. 2003;37:20. doi: 10.1002/gcc.10186. [DOI] [PubMed] [Google Scholar]
- 12.Taganov KD, Boldin MP, Chang KJ, Baltimore D. Proc. Natl. Acad. Sci. U.S.A. 2006;103:12481. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stetson DB, Medzhitov R. Immunity. 2006;24:93. doi: 10.1016/j.immuni.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 14.Calin GA, Croce CM. Nat. Rev. Cancer. 2006;6:857. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 15.Costinean S, et al. Proc. Natl. Acad. Sci. U.S.A. 2006;103:7024. doi: 10.1073/pnas.0602266103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Materials and methods are available as supporting material on Science Online.
- 17.Lai CK, Wallace WD, Fishbein MC. Semin. Respir. Crit. Care Med. 2006;27:613. doi: 10.1055/s-2006-957333. [DOI] [PubMed] [Google Scholar]
- 18.Jindal SK, Agarwal R. Curr. Opin. Pulm. Med. 2005;11:438. doi: 10.1097/01.mcp.0000170522.71497.61. [DOI] [PubMed] [Google Scholar]
- 19.Mastroeni P, Chabalgoity JA, Dunstan SJ, Maskell DJ, Dougan G. Vet. J. 2001;161:132. doi: 10.1053/tvjl.2000.0502. [DOI] [PubMed] [Google Scholar]
- 20.Clare S, et al. Infect. Immun. 2003;71:5881. doi: 10.1128/IAI.71.10.5881-5891.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Banchereau J, Steinman RM. Nature. 1998;392:245. doi: 10.1038/32588. [DOI] [PubMed] [Google Scholar]
- 22.Lim LP, et al. Nature. 2005;433:769. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 23.Ho IC, Lo D, Glimcher LH. J. Exp. Med. 1998;188:1859. doi: 10.1084/jem.188.10.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ho IC, Hodge MR, Rooney JW, Glimcher LH. Cell. 1996;85:973. doi: 10.1016/s0092-8674(00)81299-4. [DOI] [PubMed] [Google Scholar]
- 25.Hwang ES, White IA, Ho IC. Proc. Natl. Acad. Sci. U.S.A. 2002;99:13026. doi: 10.1073/pnas.202474499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ober C, et al. Hum. Mol. Genet. 1998;7:1393. doi: 10.1093/hmg/7.9.1393. [DOI] [PubMed] [Google Scholar]
- 27.Blumenthal MN, et al. J. Allergy Clin. Immunol. 2006;117:79. doi: 10.1016/j.jaci.2005.09.038. [DOI] [PubMed] [Google Scholar]
- 28.Bu LM, et al. Allergy. 2006;61:617. doi: 10.1111/j.1398-9995.2006.01072.x. [DOI] [PubMed] [Google Scholar]