Abstract
Degeneration of the dopaminergic neurons of the substantia nigra is a hallmark of Parkinson’s disease. To facilitate the study of the differentiation and maintenance of this population of dopaminergic neurons both in vivo and in vitro, we generated a knock-in reporter line in which the yellow fluorescent protein (YFP) replaced the first exon and the first intron of the tyrosine hydroxylase (TH) gene in one allele by homologous recombination. Expression of YFP under the direct control of the entire endogenous 5′ upstream region of the TH gene was predicted to closely match expression of TH from the wild type allele, thus marking functional dopaminergic neurons. We found that YFP was expressed in dopaminergic neurons differentiated in vitro from the knock-in mouse embryonic stem cell line and in dopaminergic brain regions in knock-in mice. Surprisingly, however, YFP expression did not overlap completely with TH expression, and the degree of overlap varied in different TH-expressing brain regions. Thus, the reporter gene did not identify functional TH-expressing cells with complete accuracy. A DNaseI hypersensitivity assay revealed a cluster of hypersensitivity sites in the first intron of the TH gene, which was deleted by insertion of the reporter gene, suggesting that this region may contain cis-acting regulatory sequences. Our results suggest that the first intron of the rodent TH gene may be important for accurate expression of TH.
Keywords: homologous recombination, mouse embryonic stem cells, in vitro differentiation, DNaseI hypersensitivity, cis-acting regulatory elements, intron sequence
Parkinson’s disease (PD) is a debilitating neurological disorder that is characterized by the degeneration of the midbrain dopaminergic neurons of the substantia nigra and the subsequent depletion of dopamine in the striatum (Martin, 1999). A better understanding of the differentiation and maintenance of this clinically important population of neurons could lead to therapeutic advances to protect these cells from pathological changes and enable surviving or transplanted cells to maximally replace the function of degenerated neurons (Bjorklund and Lindvall, 2000; Isacson, 2003).
Mouse embryonic stem cells (mES) can be induced in vitro to differentiate specifically into neurons that express markers of a midbrain dopaminergic phenotype (Kawasaki et al., 2000; Lee et al., 2000; Chung et al., 2002, 2005; Kim et al., 2002; Barberi et al., 2003; Andersson et al., 2006). This production of midbrain dopaminergic neurons in culture provides a useful in vitro system for studying the differentiation of dopaminergic neurons and for generating cells for transplantation (Bjorklund and Lindvall, 2000; Hynes and Rosenthal, 2000; Isacson, 2003). Transplantation of these differentiated cells in rodent models of PD has been shown to lead to behavioral recovery (Kim et al., 2002; Barberi et al., 2003). Even at their most efficient, however, these in vitro methods produce only a subset of cells in culture that are dopaminergic neurons, which can then be identified by staining for appropriate markers. A reporter system using a vital fluorescent marker to readily and accurately identify dopaminergic neurons in vitro and in vivo would facilitate further study of the molecular mechanisms that control the specification of the midbrain dopaminergic cell fate. This would lead to more effective production of dopaminergic tissue for transplantation as well as better methods to ensure the proper function and maintenance of transplanted neurons.
Most existing dopaminergic reporter systems have used transgenic approaches to drive reporter gene expression in mice in vivo using varying lengths of the upstream 5′ tyrosine hydroxylase (TH) promoter. TH is the first and rate-limiting enzyme in the synthesis of dopamine, and is a well-established marker for dopaminergic neurons. Extensive study of the regulation of the rodent TH gene has focused on the upstream 5′ flanking region, which has been shown to contain multiple important regulatory elements (Harrington et al., 1987; Cambi et al., 1989; Fader and Lewis, 1990; Gizang-Ginsberg and Ziff, 1990; Fung et al., 1992; Icard-Liepkalns et al., 1992; Kilbourne et al., 1992; Yoon and Chikaraishi, 1992, 1994; Kim et al., 1993a,b, 2001; Dawson et al., 1994; Wong et al., 1994; Best et al., 1995; Lazaroff et al., 1995; Patankar et al., 1997; Tinti et al., 1997; Ghee et al., 1998; Yang et al., 1998; Sakurada et al., 1999; Cazorla et al., 2000; Iwawaki et al., 2000). In transgenic mice, TH sequences from 2.5 kb up to 11.0 kb of the upstream 5′ TH region have been shown to be sufficient to drive reporter gene expression in catecholaminergic areas, including the midbrain (Banerjee et al., 1992; Sasaoka et al., 1992; Min et al., 1994; Morgan et al., 1996; Liu et al., 1997; Trocme et al., 1998; Schimmel et al., 1999; Sawamoto et al., 2001; Matsushita et al., 2002; Kessler et al., 2003). However, even in the most accurate of these transgenic reporter mice there is incomplete overlap of TH and reporter gene expression, ectopic expression of the reporter gene, and/or developmental differences between TH and reporter gene expression.
The lack of accurate expression overlap could be due to the presence of regulatory elements at a greater distance from the flanking 5′ region than can be incorporated in a transgenic insertion. Distal cis-acting regulatory domains can be located at distances of up to hundreds of kilobases upstream or downstream from the flanking promoter region (Bulger and Groudine, 1999; Carter et al., 2002; Spitz et al., 2003; Sabo et al., 2004; Pipkin and Lichtenheld, 2006). In addition, necessary and appropriate transcriptional regulation could be affected by the integration site of the transgene. To avoid these possible limitations, we chose instead to pursue a knock-in reporter system approach to insert yellow fluorescent protein (YFP) in the endogenous locus of the TH gene on one allele through homologous recombination in mES cells. This would place the reporter gene under the direct control of the entire endogenous upstream region. We hypothesized that, as a result, expression of YFP would closely match expression of TH from the wild type allele in the same cell. Thus, this TH–YFP reporter system would provide a readily detectable marker for functional dopaminergic neurons and could facilitate the development of therapeutic applications using mouse models of PD. Here we report the analysis of a mES cell line and mouse in which YFP has been inserted in the endogenous locus of the TH gene. Our surprising results comparing the expression patterns of TH and YFP led us to analyze regions of the rodent TH gene that had not been previously shown to contain sequences important for regulating TH expression.
EXPERIMENTAL PROCEDURES
TH targeting vector
A targeting vector was designed to replace exon 1, intron 1, and the first 15 bp of exon 2 of the mouse TH gene with the gene for YFP and a phosphoglycerate kinase-neo gene cassette (Neo) flanked by loxP sites for positive selection through antibiotic resistance. The vector was designed to place the YFP gene in frame with the endogenous ATG start codon at +6. The targeting vector was composed of 5.2 kb of 5′ homology sequence and 0.8 kb of 3′ homology sequence, encompassing the remaining sequence of exon 2 and part of intron 2. The targeting vector also contained the gene sequence for diphtheria toxin A (DTA) for negative selection (Yagi et al., 1990) (Fig. 1A).
Fig. 1.

Generation of THYFP/+ knock-in mES cells and mice. (A) Schematic of targeting strategy used to insert the gene for YFP into the TH gene locus by homologous recombination. Southern blot digestion sites and PCR primers for genotyping are indicated. (B) Southern blot analysis of genomic DNA after digestion with SpeI confirmed homologous recombination and in vitro Neo excision. (C) PCR of mouse tail DNA confirmed heterozygous TH/YFP genotype and excision of Neo.
Generation of TH knock-in mES cell line
R1 mouse blastocyst-derived mES cells (Feng et al., 1999) were transfected by electroporation and grown on irradiated mouse embryonic fibroblasts (MEFs) with positive selection using 300 μg/ml G418 (Promega, Fitchburg, WI, USA) and negative selection using DTA production from non-homologous insertion. Surviving clones were screened by PCR to identify homologous recombination, and one positive clone was identified out of 391 screened. Homologous recombination in this positive clone was confirmed by Southern blot of genomic DNA after digestion with the enzyme SpeI (Fig. 1B). This positive clone was expanded and cells were transiently transfected by electroporation with a vector expressing Cre recombinase. Cells were screened by PCR to identify clones with successful excision of the Neo gene, which was confirmed by Southern blot of genomic DNA after digestion with the enzyme SpeI (Fig. 1B), and then expanded to establish a line of THYFP/+ mES cells.
In vitro differentiation of mES cells
The mouse blastocyst-derived mES cell lines wild-type R1 and THYFP/+ were propagated on mitomycin-treated (10 μg/ml media; Sigma-Aldrich, St. Louis, MO, USA) MEFs (Stemcell Technologies, Vancouver, BC, Canada) in Dulbecco’s modified minimal essential medium (DMEM; Invitrogen, Carlsbad, CA, USA) supplemented with 2 mM l-glutamine (Invitrogen), 1 mM β-mercaptoethanol, 1× non-essential amino acids (NEAA; Invitrogen), 1× nucleosides (Specialty Media, Chemicon, Temecula, CA, USA), 15% fetal bovine serum (FBS; Sigma-Aldrich), 100 U/ml penicillin, 100 μg/ml streptomycin (Invitrogen) and 2000 U/ml human recombinant leukemia inhibitory factor (LIF; R&D Systems, Minneapolis, MN, USA).
mES cell were purified from MEFs by attachment to gelatin-coated dishes for 30 min. Purified mES cells were subsequently differentiated on PA6 cells (Riken, Kobe, Japan) (Kawasaki et al., 2000). PA6 cells were propagated in alpha minimum essential media (α-MEM; Gibco Invitrogen), 10% FBS (Sigma) and 2 mM l-glutamine. For differentiation on PA6, mES cells were seeded at a density of 500 cells/cm2 onto contact-inhibited PA6 in differentiation media consisting of Glasgow Minimum Essential Medium (G-MEM; Invitrogen), 10% Knockout Serum Replacement (KSR; Invitrogen), 1× NEAA, 1 mM sodium pyruvate, 100 U/ml penicillin, 100 μg/ml streptomycin (Invitrogen) and 0.1 mM β-mercaptoethanol. Medium was changed as needed, and days 8–14 of differentiation were continued in N2 medium consisting of G-MEM, 1× N2-supplement (Invitrogen), 0.1 mM NEAA, 1 mM sodium pyruvate and 0.1 mM β-mercaptoethanol.
Immunocytochemistry of differentiated mES cells
In vitro differentiated mES cells were fixed at day 9, 12 or 14 of differentiation in 4% formaldehyde (Electron Microscopy Sciences, Ft. Washington, PA, USA) for 30 min and rinsed with phosphate-buffered saline (PBS). The coverslips were subsequently incubated with blocking buffer (PBS, 10% normal donkey serum (NDS) and 0.1% Triton X-100) for 1 h. Coverslips were then incubated overnight at 4 °C with primary antibodies diluted in PBS, 10% NDS/normal goat serum (NGS), 0.1% Triton X-100: Rabbit anti-GFP at 1:2000 (a gift of Dr. Don Lo, Duke University, Durham, NC, USA), Sheep anti-TH at 1:300 (Pel-Freeze, Rogers, AR, USA), Rabbit anti-class III β-tubulin (TuJ1) at 1:500 (Covance, Princeton, NJ, USA), and anti-NeuN at 1:200 (Chemicon). The coverslips were subsequently incubated in fluorescent-labeled secondary antibodies at 1:500 (Jackson Immunoresearch, West Grove, PA, USA) in PBS and 10% NDS for 1 h at room temperature. After rinsing for 3×10 min in PBS, Hoechst 33342 (4 μg/ml) was used for counterstaining. The coverslips were mounted onto slides in Gel/Mount (Biomedia Corporation, Foster City, CA, USA). Confocal analysis was performed using a Zeiss LSM510/Meta Station (Thornwood, NY, USA). For identification of signal colocalization within a cell, optical thickness was kept to a minimum, and orthogonal reconstructions were obtained. Stereology was performed using Stereo Investigator image-capture equipment and software (MicroBrightField, Willinston, VT, USA) and a Zeiss Axioplan I fluorescent microscope. The extent of overlap between TH and YFP was determined by counting cells in 40× magnification, selected by random sampling within a colony, using Stereo Investigator. In total, 30 colonies, randomly selected for counting in the Hoechst counterstaining and distributed over three different coverslips, were analyzed.
Generation of TH knock-in mice
To generate THYFP/+ knock-in mice, blastocysts were injected with mES cells derived from the original positive clone (prior to excision of the Neo gene in culture). Four high percentage male chimeras were produced. These males were crossed to C57BL/6J females. The resulting F1 generation was screened by PCR to identify heterozygous THYFP/+ male mice. These were then crossed to females expressing Cre recombinase under the β-actin promoter to excise the Neo resistance gene in the germline. These matings produced a line of heterozygous THYFP/+ knock-in mice, as confirmed by PCR analysis of genomic DNA (Fig. 1C).
Immunohistochemistry of brain sections
Mice were perfused transcardially with 4% paraformaldehyde in PBS. Brains were removed, frozen, and cryosectioned at 20 μm. Sections were mounted on slides for immunostaining. Sections were blocked in 2% bovine serum albumin (BSA)/5% NGS/1% Triton in PBS for one hour to overnight and then incubated for 3–4 days with primary antibodies diluted in blocking solution: rabbit anti-GFP at 1:2000 (a gift of Dr. Don Lo, Duke University) and mouse anti-TH at 1:2000 (DiaSorin, Inc., Stillwater, MN, USA). Slides were washed five times with PBS and incubated for 4–6 h with secondary antibodies diluted in blocking solution: FITC-conjugated goat anti-rabbit at 1:400 (Jackson ImmunoResearch) and Cy3 goat anti-mouse IgG1 at 1:2000 (Jackson ImmunoResearch). After washing five times with PBS, slides were mounted under coverslips in 90% glycerol with 0.1% p-phenylenediamine.
DNaseI hypersensitivity assay
PC12 or C6 glioma cells were grown to 90% confluence as a monolayer and collected after washing with PBS supplemented with 2.5 mM EDTA. Nuclei were prepared by treating the cells with 0.5 ml of ice-cold cell lysing buffer for 3 min at 4 °C as described previously (Ishiguro et al., 1993). After centrifugation at 500× for 10 min, nuclear pellets were washed again in the same buffer, then twice in digestion buffer and resuspended in digestion buffer at a concentration of 25 A260/ml. Different concentrations of DNaseI (0, 25, 50, 100 units/ml) were added to the above resuspension and incubated for 5 min at 20 °C. The reaction was stopped by adding SDS to 1%, EDTA to 50 mM and proteinase K to 200 μg/ml at 37 °C for 1 h. Purified genomic DNA from DNaseI-treated nuclei were digested with EcoRI, separated by 1% agarose gel electrophoresis and transferred to a nylon membrane using the capillary transfer method and were further subjected to Southern blot analysis. Rat cDNA fragments were gel-purified, labeled with 32P by random priming and used as a probe, including a 1137 bp EcoRI-PstI fragment spanning exons 2–11, a 177 bp BamHI-EcoRI fragment spanning exons 8–11, and a 1200 bp EcoRI-KpnI fragment (Grima et al., 1985; Brown et al., 1987; Harrington et al., 1987). After hybridization and washing of the membrane, it was exposed for 2–4 days using XAR film and intensifying screen at −70 °C.
RESULTS
Generation of TH–YFP mES cell line
In order to generate a reporter system to label dopaminergic neurons we used a knock-in approach to insert cDNA encoding YFP into the TH locus on one allele through homologous recombination in mES cells, thus placing YFP under the direct control of the entire endogenous 5′ upstream region of the mouse TH gene. Because these cells contain one wild-type allele expressing TH and one knock-in allele expressing YFP, we predicted that in this system YFP and TH would be expressed in a nearly identical pattern, thus accurately marking functional, dopamine-expressing neurons.
The targeting vector was designed to replace sequences from exon 1, intron 1 and the first 15 bp of exon 2 of the mouse TH gene with the YFP coding sequence (Fig. 1A). mES cells were transfected by electroporation, and one clone was identified as positive for homologous recombination by PCR screening. Homologous recombination was then confirmed by Southern blot (Fig. 1B). To remove the Neo gene, mES cells were transiently transfected with Cre recombinase. Excision of the Neo gene was confirmed by Southern blot (Fig. 1B). This clone was used to generate a line of heterozygous THYFP/+ mES cells, and all subsequent in vitro experiments were conducted using the Neo-excised mES cell line.
YFP expression in dopaminergic cells generated by in vitro differentiation of mES cells
A primary reason for generating this reporter system was to provide a line of mES cells in which dopaminergic neurons could be accurately and readily identified by a fluorescent reporter. Using this reporter mES cell line for established in vitro dopaminergic differentiation techniques would facilitate the study of the specification of dopaminergic neurons and the isolation of a pure, labeled population of dopaminergic neurons for transplantation experiments by methods such as fluorescence activated cell sorting (FACS).
Different methods have been developed for the in vitro dopaminergic differentiation of mES cells, including the use of defined medium and timed application of growth factors (Lee et al., 2000) and the growth of ES cells on two bone-marrow-derived stromal cell lines: PA6 without signaling molecules (Kawasaki et al., 2000) or MS5 with the addition of signaling molecules (Barberi et al., 2003). To test whether our knock-in THYFP/+ mES cells could be reliably differentiated into fluorescently labeled dopaminergic neurons, we differentiated our THYFP/+ mES cell line on PA6 cells for 9, 12 or 14 days. After 9 days of differentiation, some parts of the cultures had a high overlap between YFP and TH (Fig. 2A), with some colonies containing as many as 80% of TH+ neurons that also expressed YFP. However, other regions were almost completely YFP+/TH− (Fig. 2B, left) or YFP−/TH+ (Fig. 2B, right), with as few as 20% of TH+ neurons also expressing YFP. Overall, 36% of TH-expressing cells also expressed YFP. Only 29% of cells expressed both YFP and TH, while 54% expressed TH alone and 17% expressed YFP alone (Fig. 2G). Further analysis of the THYFP/+ mES cell line after differentiation on PA6 for 12 days or 14 days showed similar results to 9 days of differentiation (data not shown).
Fig. 2.
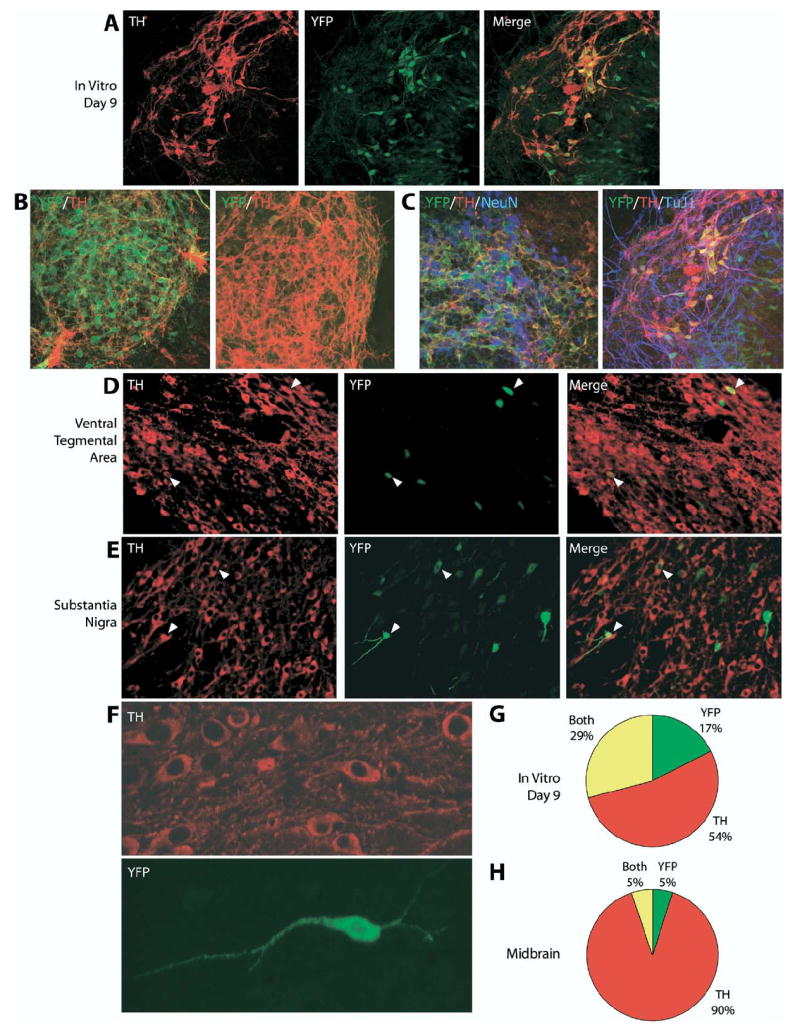
YFP reporter expression in differentiated THYFP/+ knock-in mES cells and adult THYFP/+ knock-in mouse midbrain. (A, B) THYFP/+ mES cells were differentiated in vitro for 9 days on PA6 cells into dopaminergic neurons and co-stained with antibodies to TH (red) and YFP (green). (C) THYFP/+ mES cells were differentiated for 9 days and co-stained for TH (red), YFP (green) and the nuclear neuronal marker NeuN (blue) or for TH (red), YFP (green) and the neuronal marker TuJ1 (blue). Triple overlap in expression appears purple. Cryosections of THYFP/+ heterozygous adult brain were co-stained with antibodies to TH (red) and YFP (green) in the ventral tegmental area (D) and substantia nigra (E). Examples of cells with overlapping expression are indicated by arrowheads. A higher magnification confocal stack image of neurons in the substantia nigra is seen in (F). (G) Quantification of differentiated TH and YFP expressing cells in vitro. Of the total cells (n=1508), 54% expressed TH alone (n=804), 17% expressed YFP alone (n=262), and 29% expressed both TH and YFP (n=442). (H) Quantification of TH and YFP expressing cells in the mouse midbrain. Of the total cells (n=426), 90% expressed TH alone (n=382), 5% expressed YFP alone (n=21), and 5% expressed both TH and YFP (n=23).
These results were surprising given that the YFP gene was placed in the locus of the TH gene. Most of the cells expressing YFP either alone or with TH also expressed the neuronal markers NeuN (Fig. 2C, left) and TuJ1 (Fig. 2C, right). This confirmed the successful differentiation of this THYFP/+ mES cell line into a neuronal phenotype, suggesting that the YFP reporter gene insertion and expression did not affect the in vitro neuronal differentiation process. Instead, the partial overlap between YFP and TH in the differentiated THYFP/+ mES cells may be due to differences in the stability of YFP and TH and/or to variability in TH promoter regulation under the cell culture conditions.
YFP expression in vivo in TH-expressing cell groups
A reporter mouse that accurately labels dopaminergic neurons would allow for in vivo studies of the developing and adult dopaminergic systems. To test whether the knock-in THYFP/+ mES cells could reliably differentiate into fluorescently labeled dopaminergic neurons in vivo, we generated a reporter mouse line by injecting blastocysts with mES cells derived from the original homologous recombinant clone. Four high percentage male chimeras were crossed to C57BL/6J females. The resulting F1 generation was screened by PCR analysis of genomic DNA to identify heterozygous THYFP/+ male mice (Fig. 1C). These were then crossed to females expressing Cre recombinase under the β-actin promoter to excise the Neo gene, as confirmed by PCR analysis (Fig. 1C). All subsequent in vivo experiments were conducted using the Neo-excised THYFP/+ mouse line. Although mice with a homozygous null TH deletion die before birth, heterozygous mice are viable and appear normal (Kobayashi et al., 1995; Zhou et al., 1995; Rios et al., 1999). Consistent with this, in our mouse line homozygous knock-in mice did not survive to birth and heterozygous THYFP/+ reporter mice were indistinguishable from wild type mice.
We examined the brains of adult THYFP/+ knock-in mice to assess whether YFP was expressed in catecholaminergic cell groups in vivo and whether neurons expressing TH also expressed YFP. The regional specificity of YFP expression was generally accurate, with expression in each of the major catecholaminergic cell groups in the brain and no detectable YFP expression in areas of the brain where TH is not normally expressed. Because the primary goal of this reporter system was to label dopaminergic neurons, our analysis focused on dopaminergic cell groups in the THYFP/+ mouse brain.
We found that YFP expression was present in the dopaminergic midbrain regions: the ventral tegmental area (Fig. 2D) and substantia nigra (Fig. 2E, F). A few cells in these areas expressed both YFP and TH (Fig, 2D, E arrowheads), and cell morphology in these cells was clearly visualized with the reporter gene (Fig. 2F). However, only 6% of TH cells also expressed YFP. In addition, a few cells expressed YFP that did not express TH. Only 5% of cells expressed both YFP and TH, while 90% expressed TH alone and 5% expressed YFP alone (Fig. 2H). Thus, the fidelity of expression in vivo in the midbrain was even lower than in the mES cells differentiated in vitro.
To determine whether the lack of overlap between YFP and TH expression occurred in other dopaminergic areas of the brain in THYFP/+ mice, we examined other dopaminergic cell groups. TH is expressed in dopaminergic peri-glomerular cells in the olfactory bulb. YFP expression was seen in these cells in THYFP/+ mice. As in the midbrain, most TH-expressing cells did not express YFP (Fig. 3A). However, the overlap of YFP with TH expression was higher in the olfactory bulb than in the midbrain (Fig. 3A arrowheads), with 38% of TH cells expressing YFP. Compared with the midbrain, more cells expressed YFP and, as in the midbrain, some cells expressed YFP but did not express TH. In the olfactory bulb, 22% of cells expressed both YFP and TH, while 37% expressed TH alone and 41% expressed YFP alone (Fig. 3C). Thus, the pattern of relative TH and YFP expression is distinct from that seen in the ventral midbrain region, with more overall YFP expression and more cells expressing YFP alone.
Fig. 3.

YFP and TH expression in other dopaminergic and non-dopaminergic TH-expressing cell groups. Cryosections of THYFP/+ heterozygous adult brains were co-stained with antibodies to TH (red) and YFP (green) in the olfactory bulb (A) and hypothalamus (B). Examples of cells with overlapping expression are indicated by arrowheads. (C) Quantification of TH and YFP expressing cells in the olfactory bulb. Of the total cells (n=174), 37% expressed TH alone (n=65), 41% expressed YFP alone (n=70), and 22% expressed both TH and YFP (n=39). (D) Quantification of TH and YFP expressing cells in the dopaminergic cell groups in the hypothalamus. Of the total cells (n=274), 35% expressed TH alone (n=97), 36% expressed YFP alone (n=98), and 29% expressed both TH and YFP (n=79). Cryosections of THYFP/+ heterozygous adult brain were co-stained with antibodies to TH (red) and YFP (green) in the locus ceruleus (E) and C1 cell group (F). Examples of cells with overlapping expression are indicated by arrowheads.
TH is also expressed in dopaminergic cell groups in the hypothalamus. YFP expression was seen in the hypothalamus, but as in the midbrain and the olfactory bulb, many TH-expressing cells did not express YFP (Fig. 3B). However, the overlap of YFP expression with TH in the hypothalamus was again higher than in the midbrain (Fig. 3B arrowheads). In the dopaminergic cell groups of the hypothalamus, 45% of TH cells co-expressed YFP. Similar to the olfactory bulb, there were more cells expressing YFP than in the midbrain, and some cells expressed YFP that did not express TH. In the hypothalamus, 29% of cells expressed both YFP and TH, while 35% expressed TH alone and 36% expressed YFP alone (Fig. 3D).
TH is also expressed in other, non-dopaminergic catecholaminergic cells that synthesize norepinephrine and epinephrine from dopamine. To compare the pattern of YFP expression in non-dopaminergic TH-expressing regions of the brain, we examined several catecholaminergic cell groups in the locus ceruleus and the brainstem. YFP expression followed a similar pattern to that seen in dopaminergic areas of the brain, with some cells expressing both TH and YFP, some cells expressing only TH and some cells expressing only YFP, as shown here in the norepinephrinergic locus ceruleus cell group (Fig. 3E) and the epinephrinergic C1 cell group in the brainstem (Fig. 3F).
In summary, the expression of the YFP reporter gene in the THYFP/+ knock-in mice was specific to TH-expressing regions of the brain, with no detectable YFP expression in ectopic brain regions. Within TH-expressing regions of the brain, some cells did express both TH and YFP, but most TH-expressing cells did not express YFP and some YFP-expressing cells did not express TH. This result was similar to that seen after in vitro differentiation of the THYFP/+ mES cells, suggesting that the in vitro result was not solely due to an effect of the cell culture conditions. The absence of YFP expression in some TH-expressing cells could be due to differences in stability between the endogenous TH mRNA or protein and the reporter YFP mRNA or protein; however, the high levels of YFP expression in some cells indicates that the protein can in fact be highly and stably expressed.
Potential regulatory role for deleted TH gene sequence
The knock-in reporter system described here was designed to maximize the accurate labeling of dopaminergic neurons by placing a reporter gene in the TH locus under the direct control of the entire endogenous 5′ upstream region of the TH gene on one allele while maintaining the intact wild type TH gene on the other allele. Cells expressing TH were predicted to express TH from the intact allele and the YFP reporter from the knock-in allele in a nearly identical manner. Surprisingly, however, the expression pattern of TH and YFP differed substantially and the relative expression pattern varied by brain region. One explanation for the difference is that the TH gene on one allele and the knock-in YFP gene on the other allele are actually subject to differential transcriptional regulation. In the targeting strategy used, the 2 kb of TH sequence downstream of the transcription initiation site (exon 1, intron 1, and 15 bp of exon 2) were deleted by replacement with the YFP gene. Although previous studies of TH gene regulation point to the importance of the 5′ region, it is possible that the deleted sequences contain additional, previously unidentified regulatory elements that are important for specific expression.
As an initial test of this possibility, we assayed the TH gene for DNaseI hypersensitivity sites (DHSs) in regions including the sequence that was deleted in our reporter system. The presence of DHSs serves as an indication of the relative accessibility of regions of intact chromatin and is a hallmark of transcriptional regulatory elements (Gross and Garrard, 1988; Crawford et al., 2004; Sabo et al., 2004). We isolated nuclei from TH-positive PC12 and TH-negative C6 glioma cells that had been treated with increasing amounts of DNaseI (0, 25, 50, 100 units/ml). Isolated chromosomal DNA was digested with EcoRI and subjected to Southern blot analysis using a rat TH cDNA probe that spans exons 2–11 (Fig. 4B).
Fig. 4.

DNaseI hypersensitivity assay of the TH gene in C6 glioma cells and PC12 cells. (A) Southern blot of chromosomal DNA from TH-negative C6 glioma cells and TH-positive PC12 cells treated with increasing concentrations of DNaseI. DNA was digested with EcoRI and probed using an EcoRI-PstI rat cDNA fragment that spans from nucleotides 117–1137 bp (exons 2–11) within the TH coding region. (B) Schematic of hypersensitivity sites identified in the TH gene. Sites HS3-6 fall within the region deleted in the reporter system. (C) Schematic map of the first exon, first intron and second exon of the mouse, rat and human TH genes.
Southern blot analysis revealed several DHSs in the intact chromatin of the TH-positive PC12 cells (Fig. 4A, B). This included three prominent DHSs (HS3–HS5) and one weaker potential DHS (HS6) within the first intron of the rat TH gene in PC12 cells, in which the TH gene is actively transcribed, but not in the TH-negative C6 glioma cells. Almost identical results were obtained in a separate experiment using a shorter probe that spans exons 8–11 (unpublished observation). These cell-specific DHSs may represent active, tissue-specific regulatory elements important for TH gene expression in PC12 cells. A prominent PC12 cell-specific DHS (HS2) also appeared in the 5′ core promoter region, where the TATA box, the cyclic AMP response element (CRE) and the transcription initiation site are located, consistent with the idea that the sequence motifs critical for basal TH transcription (Kim et al., 1993a; Lazaroff et al., 1995) are kept open in the TH-expressing PC12 cells but remain as a closed structure in the TH-negative C6 cells.
This analysis also revealed a possible DHS (HS1) with modest signal in the further 5′ upstream region at approximately −6 kb. This DHS appeared in both PC12 and C6 glioma cells, indicating that this sequence may not be directly involved in cell-specificity. The upstream region between −5.6 kb and −9.0 kb exerts a strong enhancer activity both in PC12 and C6 glioma cells (Yang et al., 1998), and it is possible that this DHS may represent the sequence domain responsible for this enhancer activity. We also further analyzed the 3′ side of the TH gene, ranging from exon 13 to +15 kb, by a similar DHS mapping analysis. No DHSs with evident signals were detected in this region (data not shown).
DISCUSSION
The initial goal of this work was to generate a reporter system that would accurately label dopamine-expressing neurons and could be used for both in vitro and in vivo studies of dopaminergic differentiation and maintenance as well as for transplantation studies in mouse models of PD. Previous reporter systems using transgenic approaches to express reporter genes from the 5′ flanking sequences of the TH gene have not replicated the endogenous expression pattern of TH with complete accuracy (Banerjee et al., 1992; Sasaoka et al., 1992; Min et al., 1994; Morgan et al., 1996; Liu et al., 1997; Trocme et al., 1998; Schimmel et al., 1999; Sawamoto et al., 2001; Matsushita et al., 2002; Kessler et al., 2003). This could be because of variability in transcriptional regulation due to transgene integration site or because accurate expression may require regulatory sequences at a greater distance from the 5′ flanking region than can be incorporated in a transgenic approach.
We chose instead to use a knock-in approach to place a YFP reporter gene in the locus of the TH gene on one allele while maintaining the endogenous TH gene on the other. By expressing the reporter gene under the endogenous control of the entire 5′ upstream sequence of the TH gene we predicted that neurons expressing TH would simultaneously express YFP, thus closely reflecting normal physiological expression of TH. In our THYFP/+ knock-in mouse we found that the YFP reporter gene was specifically expressed in TH-expressing regions of the mouse brain. YFP was also expressed in dopaminergic neurons differentiated in vitro from our THYFP/+ knock-in mES cell line. Surprisingly, however, many TH-positive cells both in vivo and in vitro did not express YFP and some cells expressing YFP did not express TH. Thus, the reporter gene did not accurately identify all functional TH-expressing cells. In addition, different dopaminergic brain regions had different relative TH/YFP expression patterns. The differences in expression pattern suggest that the transcription of the TH gene on one allele and the knock-in YFP gene on the other allele may actually be regulated differently, and that this regulation differs depending on the brain region.
In our THYFP/+ knock-in targeting strategy the first 2 kb of the mouse TH genomic sequence, including the first exon, the first intron and 15 bp from the second exon, were replaced by homologous recombination with the YFP reporter gene. Intron sequences have been shown to contain regulatory elements (Galvagni and Oliviero, 2000; Karadsheh and Delpire, 2001; Hermann and Heckert, 2005; Lee and Friedman, 2005; Meng et al., 2005), and the difference in this sequence could explain the differences in expression pattern between the YFP and TH alleles in our knock-in system. Indeed, we identified TH-expressing cell-specific DHSs in the first intronic sequence of the rat TH gene, which is highly homologous to the mouse sequence. Because DHSs are a hallmark of cis-acting regulatory elements, including promoters, enhancers, silencers, insulators and locus control regions (Gross and Garrard, 1988; Crawford et al., 2004; Sabo et al., 2004), this suggests that this region of the TH gene may contain regulatory elements that might be important for controlling TH expression. Although the function of these hypersensitivity sites remains to be elucidated, their presence provides preliminary support for the hypothesis that the loss of this sequence may have affected the endogenous expression pattern of the allele carrying the inserted YFP gene. The cells within each brain region that were induced to express only from the knock-in YFP allele, only from the wild type TH allele, or from both alleles could represent distinct subsets of cells in which transcription from the endogenous TH locus is regulated differently. In addition, tissue specific differences in the role of the deleted TH sequence could explain the different relative TH and YFP expression patterns we observed in different dopaminergic brain regions.
The discrepancies in previously reported transgenic reporter systems, which did not include intron and exon sequences, may also be due in part to the absence of these intronic regulatory elements. A recent knock-in mouse placed Cre-recombinase at the 3′ untranslated region of the TH gene using an internal ribosomal entry sequence (Lindeberg et al., 2004). The Cre-recombinase gene was inserted without disrupting the TH intron and exon sequences and co-expressed with TH with greater accuracy in the dopaminergic cell groups in the midbrain than our THYFP/+ knock-in reporter gene. This provides additional support for the potential importance of the first intronic sequence of the rodent TH gene in regulating specific TH expression.
Interestingly, the first intron of the human TH gene contains a tetranucleotide repeat that has been shown to have a regulatory function in TH expression and is a binding site for transcriptional regulators (Meloni et al., 1998; Albanese et al., 2001; Lenartowski and Goc, 2002). However, the overall sequence identity in the first intron is only about 50% both between human and rat and between human and mouse. Some regions within the first intron sequence have a higher degree of homology while other regions have little shared sequence, and the tetranucleotide repeat in the human TH gene is not conserved in mouse and rat. Therefore, to our knowledge this study represents the first evidence for a potential regulatory role of the first intronic sequence of the rodent TH gene. Although the regulation of TH expression appears to be generally preserved among species, there are in fact only a few regions of high homology between the human and rodent promoter sequences (Gandelman et al., 1990; Kessler et al., 2003; Kim et al., 2003), and differences in TH gene regulation between human and rodent have been demonstrated (Romano et al., 2005; Jin et al., 2006). Differences in regulatory elements between the rodent first intron sequence and the human first intron sequence may also contribute to species-specific regulation.
In hindsight, it is clear that deletion of any of the endogenous TH gene sequence was a limitation to our knock-in reporter strategy. Our results suggest that, in general, knock-in reporter strategies should preserve as much of the endogenous gene sequence as possible in order to maintain all regulatory elements and thus most accurately reflect the endogenous expression pattern. However, the unanticipated consequences of deleting some of the TH gene sequence in this study led to interesting and potentially important results suggesting that, in addition to the well-characterized 5′ upstream regulatory region of the TH gene, there may be other, previously unidentified regulatory elements in the first intron sequence that are important for accurate gene expression in rodents. Further study will be needed to elucidate the functional characteristics of the putative regulatory elements revealed through our DNaseI hypersensitivity assay. This will expand the understanding of the regulation of TH expression and should improve the precision with which reporter systems can be generated and subsequently used to study molecular determinants of dopaminergic differentiation in vitro, to isolate dopaminergic cells for transplantation, and to monitor dopaminergic cells in vivo during development and after transplantation of cells derived from reporter mice or mES cell cultures.
Acknowledgments
This research was supported by an Academic Research Initiation grant from the North Carolina Biotechnology Center and a research grant from the Whitehall Foundation (G.F.), Udall Parkinson’s Disease Center of Excellence grant P50 NS39793 (O.I., K.S.K.), National Institutes of Health grant NS22675 (D.M.C.), the Duke University Medical Center Medical Scientist Training Program (B.B.K.) and a fellowship from the Swedish Brain Foundation (E.H.). We thank Dr. Rashmi Chandra, Marybeth Groelle and Jimmy Gross for technical assistance and Dr. Fan Wang and Dr. Mike Ehlers for helpful comments on this work. We are grateful to the members of the Feng laboratory for support and critical reading of this manuscript.
Abbreviations
- DHS
DNaseI hypersensitivity site
- DTA
diptheria toxin A
- EDTA
ethylenediaminetetraacetic acid
- FBS
fetal bovine serum
- G-MEM
Glasgow Minimum Essential Medium
- MEF
mouse embryonic fibroblast
- mES cells
mouse embryonic stem cells
- NDS
normal donkey serum
- NEAA
non-essential amino acid
- NGS
normal goat serum
- PBS
phosphate-buffered saline
- PD
Parkinson’s disease
- TH
tyrosine hydroxylase
- TuJ1
class III β-tubulin
- YFP
yellow fluorescent protein
References
- Albanese V, Biguet NF, Kiefer H, Bayard E, Mallet J, Meloni R. Quantitative effects on gene silencing by allelic variation at a tetranucleotide microsatellite. Hum Mol Genet. 2001;10:1785–1792. doi: 10.1093/hmg/10.17.1785. [DOI] [PubMed] [Google Scholar]
- Andersson E, Tryggvason U, Deng Q, Friling S, Alekseenko Z, Robert B, Perlmann T, Ericson J. Identification of intrinsic determinants of midbrain dopamine neurons. Cell. 2006;124:393–405. doi: 10.1016/j.cell.2005.10.037. [DOI] [PubMed] [Google Scholar]
- Banerjee SA, Hoppe P, Brilliant M, Chikaraishi DM. 5′ Flanking sequences of the rat tyrosine hydroxylase gene target accurate tissue-specific, developmental, and transsynaptic expression in transgenic mice. J Neurosci. 1992;12:4460–4467. doi: 10.1523/JNEUROSCI.12-11-04460.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barberi T, Klivenyi P, Calingasan NY, Lee H, Kawamata H, Loonam K, Perrier AL, Bruses J, Rubio ME, Topf N, Tabar V, Harrison NL, Beal MF, Moore MA, Studer L. Neural subtype specification of fertilization and nuclear transfer embryonic stem cells and application in parkinsonian mice. Nat Biotechnol. 2003;21:1200–1207. doi: 10.1038/nbt870. [DOI] [PubMed] [Google Scholar]
- Best JA, Chen Y, Piech KM, Tank AW. The response of the tyrosine hydroxylase gene to cyclic AMP is mediated by two cyclic AMP-response elements. J Neurochem. 1995;65:1934–1943. doi: 10.1046/j.1471-4159.1995.65051934.x. [DOI] [PubMed] [Google Scholar]
- Bjorklund A, Lindvall O. Cell replacement therapies for central nervous system disorders. Nat Neurosci. 2000;3:537–544. doi: 10.1038/75705. [DOI] [PubMed] [Google Scholar]
- Brown ER, Coker GT, 3rd, O’Malley KL. Organization and evolution of the rat tyrosine hydroxylase gene. Biochemistry. 1987;26:5208–5212. doi: 10.1021/bi00390a046. [DOI] [PubMed] [Google Scholar]
- Bulger M, Groudine M. Looping versus linking: toward a model for long-distance gene activation. Genes Dev. 1999;13:2465–2477. doi: 10.1101/gad.13.19.2465. [DOI] [PubMed] [Google Scholar]
- Cambi F, Fung B, Chikaraishi D. 5′ Flanking DNA sequences direct cell-specific expression of rat tyrosine hydroxylase. J Neurochem. 1989;53:1656–1659. doi: 10.1111/j.1471-4159.1989.tb08567.x. [DOI] [PubMed] [Google Scholar]
- Carter D, Chakalova L, Osborne CS, Dai YF, Fraser P. Long-range chromatin regulatory interactions in vivo. Nat Genet. 2002;32:623–626. doi: 10.1038/ng1051. [DOI] [PubMed] [Google Scholar]
- Cazorla P, Smidt MP, O’Malley KL, Burbach JP. A response element for the homeodomain transcription factor Ptx3 in the tyrosine hydroxylase gene promoter. J Neurochem. 2000;74:1829–1837. doi: 10.1046/j.1471-4159.2000.0741829.x. [DOI] [PubMed] [Google Scholar]
- Chung S, Hedlund E, Hwang M, Kim DW, Shin BS, Hwang DY, Jung Kang U, Isacson O, Kim KS. The homeodomain transcription factor Pitx3 facilitates differentiation of mouse embryonic stem cells into AHD2-expressing dopaminergic neurons. Mol Cell Neurosci. 2005;28:241–252. doi: 10.1016/j.mcn.2004.09.008. [DOI] [PubMed] [Google Scholar]
- Chung S, Sonntag KC, Andersson T, Bjorklund LM, Park JJ, Kim DW, Kang UJ, Isacson O, Kim KS. Genetic engineering of mouse embryonic stem cells by Nurr1 enhances differentiation and maturation into dopaminergic neurons. Eur J Neurosci. 2002;16:1829–1838. doi: 10.1046/j.1460-9568.2002.02255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford GE, Holt IE, Mullikin JC, Tai D, Blakesley R, Bouffard G, Young A, Masiello C, Green ED, Wolfsberg TG, Collins FS. Identifying gene regulatory elements by genome-wide recovery of DNase hypersensitive sites. Proc Natl Acad Sci U S A. 2004;101:992–997. doi: 10.1073/pnas.0307540100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson SJ, Yoon SO, Chikaraishi DM, Lillycrop KA, Latchman DS. The Oct-2 transcription factor represses tyrosine hydroxylase expression via a heptamer TAATGARAT-like motif in the gene promoter. Nucleic Acids Res. 1994;22:1023–1028. doi: 10.1093/nar/22.6.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fader D, Lewis EJ. Interaction of cyclic AMP and cell-cell contact in the control of tyrosine hydroxylase RNA. Brain Res Mol Brain Res. 1990;8:25–29. doi: 10.1016/0169-328x(90)90005-x. [DOI] [PubMed] [Google Scholar]
- Feng G, Krejci E, Molgo J, Cunningham JM, Massoulie J, Sanes JR. Genetic analysis of collagen Q: roles in acetylcholinesterase and butyrylcholinesterase assembly and in synaptic structure and function. J Cell Biol. 1999;144:1349–1360. doi: 10.1083/jcb.144.6.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung BP, Yoon SO, Chikaraishi DM. Sequences that direct rat tyrosine hydroxylase gene expression. J Neurochem. 1992;58:2044–2052. doi: 10.1111/j.1471-4159.1992.tb10945.x. [DOI] [PubMed] [Google Scholar]
- Galvagni F, Oliviero S. Utrophin transcription is activated by an intronic enhancer. J Biol Chem. 2000;275:3168–3172. doi: 10.1074/jbc.275.5.3168. [DOI] [PubMed] [Google Scholar]
- Gandelman KY, Coker GT, 3rd, Moffat M, O’Malley KL. Species and regional differences in the expression of cell-type specific elements at the human and rat tyrosine hydroxylase gene loci. J Neurochem. 1990;55:2149–2152. doi: 10.1111/j.1471-4159.1990.tb05811.x. [DOI] [PubMed] [Google Scholar]
- Ghee M, Baker H, Miller JC, Ziff EB. AP-1, CREB and CBP transcription factors differentially regulate the tyrosine hydroxylase gene. Brain Res Mol Brain Res. 1998;55:101–114. doi: 10.1016/s0169-328x(97)00370-7. [DOI] [PubMed] [Google Scholar]
- Gizang-Ginsberg E, Ziff EB. Nerve growth factor regulates tyrosine hydroxylase gene transcription through a nucleoprotein complex that contains c-Fos. Genes Dev. 1990;4:477–491. doi: 10.1101/gad.4.4.477. [DOI] [PubMed] [Google Scholar]
- Grima B, Lamouroux A, Blanot F, Biguet NF, Mallet J. Complete coding sequence of rat tyrosine hydroxylase mRNA. Proc Natl Acad Sci U S A. 1985;82:617–621. doi: 10.1073/pnas.82.2.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross DS, Garrard WT. Nuclease hypersensitive sites in chromatin. Annu Rev Biochem. 1988;57:159–197. doi: 10.1146/annurev.bi.57.070188.001111. [DOI] [PubMed] [Google Scholar]
- Harrington CA, Lewis EJ, Krzemien D, Chikaraishi DM. Identification and cell type specificity of the tyrosine hydroxylase gene promoter. Nucleic Acids Res. 1987;15:2363–2384. doi: 10.1093/nar/15.5.2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann BP, Heckert LL. Silencing of Fshr occurs through a conserved, hypersensitive site in the first intron. Mol Endocrinol. 2005;19:2112–2131. doi: 10.1210/me.2004-0244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynes M, Rosenthal A. Embryonic stem cells go dopaminergic. Neuron. 2000;28:11–14. doi: 10.1016/s0896-6273(00)00079-9. [DOI] [PubMed] [Google Scholar]
- Icard-Liepkalns C, Biguet NF, Vyas S, Robert JJ, Sassone-Corsi P, Mallet J. AP-1 complex and c-fos transcription are involved in TPA provoked and trans-synaptic inductions of the tyrosine hydroxylase gene: insights into long-term regulatory mechanisms. J Neurosci Res. 1992;32:290–298. doi: 10.1002/jnr.490320219. [DOI] [PubMed] [Google Scholar]
- Isacson O. The production and use of cells as therapeutic agents in neurodegenerative diseases. Lancet Neurol. 2003;2:417–424. doi: 10.1016/s1474-4422(03)00437-x. [DOI] [PubMed] [Google Scholar]
- Ishiguro H, Kim KT, Joh TH, Kim KS. Neuron-specific expression of the human dopamine beta-hydroxylase gene requires both the cAMP-response element and a silencer region. J Biol Chem. 1993;268:17987–17994. [PubMed] [Google Scholar]
- Iwawaki T, Kohno K, Kobayashi K. Identification of a potential nurr1 response element that activates the tyrosine hydroxylase gene promoter in cultured cells. Biochem Biophys Res Commun. 2000;274:590–595. doi: 10.1006/bbrc.2000.3204. [DOI] [PubMed] [Google Scholar]
- Jin H, Romano G, Marshall C, Donaldson AE, Suon S, Iacovitti L. Tyrosine hydroxylase gene regulation in human neuronal progenitor cells does not depend on Nurr1 as in the murine and rat systems. J Cell Physiol. 2006;207:49–57. doi: 10.1002/jcp.20534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karadsheh MF, Delpire E. Neuronal restrictive silencing element is found in the KCC2 gene: molecular basis for KCC2-specific expression in neurons. J Neurophysiol. 2001;85:995–997. doi: 10.1152/jn.2001.85.2.995. [DOI] [PubMed] [Google Scholar]
- Kawasaki H, Mizuseki K, Nishikawa S, Kaneko S, Kuwana Y, Nakanishi S, Nishikawa SI, Sasai Y. Induction of midbrain dopaminergic neurons from ES cells by stromal cell-derived inducing activity. Neuron. 2000;28:31–40. doi: 10.1016/s0896-6273(00)00083-0. [DOI] [PubMed] [Google Scholar]
- Kessler MA, Yang M, Gollomp KL, Jin H, Iacovitti L. The human tyrosine hydroxylase gene promoter. Brain Res Mol Brain Res. 2003;112:8–23. doi: 10.1016/s0169-328x(02)00694-0. [DOI] [PubMed] [Google Scholar]
- Kilbourne EJ, Nankova BB, Lewis EJ, McMahon A, Osaka H, Sabban DB, Sabban EL. Regulated expression of the tyrosine hydroxylase gene by membrane depolarization. Identification of the responsive element and possible second messengers. J Biol Chem. 1992;267:7563–7569. [PubMed] [Google Scholar]
- Kim HS, Hong SJ, LeDoux MS, Kim KS. Regulation of the tyrosine hydroxylase and dopamine beta-hydroxylase genes by the transcription factor AP-2. J Neurochem. 2001;76:280–294. doi: 10.1046/j.1471-4159.2001.00044.x. [DOI] [PubMed] [Google Scholar]
- Kim JH, Auerbach JM, Rodriguez-Gomez JA, Velasco I, Gavin D, Lumelsky N, Lee SH, Nguyen J, Sanchez-Pernaute R, Bankiewicz K, McKay R. Dopamine neurons derived from embryonic stem cells function in an animal model of Parkinson’s disease. Nature. 2002;418:50–56. doi: 10.1038/nature00900. [DOI] [PubMed] [Google Scholar]
- Kim KS, Lee MK, Carroll J, Joh TH. Both the basal and inducible transcription of the tyrosine hydroxylase gene are dependent upon a cAMP response element. J Biol Chem. 1993a;268:15689–15695. [PubMed] [Google Scholar]
- Kim KS, Park DH, Wessel TC, Song B, Wagner JA, Joh TH. A dual role for the cAMP-dependent protein kinase in tyrosine hydroxylase gene expression. Proc Natl Acad Sci U S A. 1993b;90:3471–3475. doi: 10.1073/pnas.90.8.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TE, Park MJ, Choi EJ, Lee HS, Lee SH, Yoon SH, Oh CK, Lee BJ, Kim SU, Lee YS, Lee MA. Cloning and cell type-specific regulation of the human tyrosine hydroxylase gene promoter. Biochem Biophys Res Commun. 2003;312:1123–1131. doi: 10.1016/j.bbrc.2003.11.029. [DOI] [PubMed] [Google Scholar]
- Kobayashi K, Morita S, Sawada H, Mizuguchi T, Yamada K, Nagatsu I, Hata T, Watanabe Y, Fujita K, Nagatsu T. Targeted disruption of the tyrosine hydroxylase locus results in severe catecholamine depletion and perinatal lethality in mice. J Biol Chem. 1995;270:27235–27243. doi: 10.1074/jbc.270.45.27235. [DOI] [PubMed] [Google Scholar]
- Lazaroff M, Patankar S, Yoon SO, Chikaraishi DM. The cyclic AMP response element directs tyrosine hydroxylase expression in catecholaminergic central and peripheral nervous system cell lines from transgenic mice. J Biol Chem. 1995;270:21579–21589. doi: 10.1074/jbc.270.37.21579. [DOI] [PubMed] [Google Scholar]
- Lee SH, Lumelsky N, Studer L, Auerbach JM, McKay RD. Efficient generation of midbrain and hindbrain neurons from mouse embryonic stem cells. Nat Biotechnol. 2000;18:675–679. doi: 10.1038/76536. [DOI] [PubMed] [Google Scholar]
- Lee TK, Friedman JM. Analysis of NF1 transcriptional regulatory elements. Am J Med Genet A. 2005;137:130–135. doi: 10.1002/ajmg.a.30699. [DOI] [PubMed] [Google Scholar]
- Lenartowski R, Goc A. Tissue-specific association of the human tyrosine hydroxylase gene with the nuclear matrix. Neurosci Lett. 2002;330:151–154. doi: 10.1016/s0304-3940(02)00746-2. [DOI] [PubMed] [Google Scholar]
- Lindeberg J, Usoskin D, Bengtsson H, Gustafsson A, Kylberg A, Soderstrom S, Ebendal T. Transgenic expression of Cre recombinase from the tyrosine hydroxylase locus. Genesis. 2004;40:67–73. doi: 10.1002/gene.20065. [DOI] [PubMed] [Google Scholar]
- Liu J, Merlie JP, Todd RD, O’Malley KL. Identification of cell type-specific promoter elements associated with the rat tyrosine hydroxylase gene using transgenic founder analysis. Brain Res Mol Brain Res. 1997;50:33–42. doi: 10.1016/s0169-328x(97)00163-0. [DOI] [PubMed] [Google Scholar]
- Martin JB. Molecular basis of the neurodegenerative disorders. N Engl J Med. 1999;340:1970–1980. doi: 10.1056/NEJM199906243402507. [DOI] [PubMed] [Google Scholar]
- Matsushita N, Okada H, Yasoshima Y, Takahashi K, Kiuchi K, Kobayashi K. Dynamics of tyrosine hydroxylase promoter activity during midbrain dopaminergic neuron development. J Neurochem. 2002;82:295–304. doi: 10.1046/j.1471-4159.2002.00972.x. [DOI] [PubMed] [Google Scholar]
- Meloni R, Albanese V, Ravassard P, Treilhou F, Mallet J. A tetranucleotide polymorphic microsatellite, located in the first intron of the tyrosine hydroxylase gene, acts as a transcription regulatory element in vitro. Hum Mol Genet. 1998;7:423–428. doi: 10.1093/hmg/7.3.423. [DOI] [PubMed] [Google Scholar]
- Meng F, Zolova O, Kokorina NA, Dobretsova A, Wight PA. Characterization of an intronic enhancer that regulates myelin proteolipid protein (Plp) gene expression in oligodendrocytes. J Neurosci Res. 2005;82:346–356. doi: 10.1002/jnr.20640. [DOI] [PubMed] [Google Scholar]
- Min N, Joh TH, Kim KS, Peng C, Son JH. 5′ Upstream DNA sequence of the rat tyrosine hydroxylase gene directs high-level and tissue-specific expression to catecholaminergic neurons in the central nervous system of transgenic mice. Brain Res Mol Brain Res. 1994;27:281–289. doi: 10.1016/0169-328x(94)90011-6. [DOI] [PubMed] [Google Scholar]
- Morgan WW, Walter CA, Windle JJ, Sharp ZD. 3.6 kb Of the 5′ flanking DNA activates the mouse tyrosine hydroxylase gene promoter without catecholaminergic-specific expression. J Neurochem. 1996;66:20–25. doi: 10.1046/j.1471-4159.1996.66010020.x. [DOI] [PubMed] [Google Scholar]
- Patankar S, Lazaroff M, Yoon SO, Chikaraishi DM. A novel basal promoter element is required for expression of the rat tyrosine hydroxylase gene. J Neurosci. 1997;17:4076–4086. doi: 10.1523/JNEUROSCI.17-11-04076.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pipkin ME, Lichtenheld MG. A reliable method to display authentic DNase I hypersensitive sites at long-ranges in single-copy genes from large genomes. Nucleic Acids Res. 2006;34:e34. doi: 10.1093/nar/gkl006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rios M, Habecker B, Sasaoka T, Eisenhofer G, Tian H, Landis S, Chikaraishi D, Roffler-Tarlov S. Catecholamine synthesis is mediated by tyrosinase in the absence of tyrosine hydroxylase. J Neurosci. 1999;19:3519–3526. doi: 10.1523/JNEUROSCI.19-09-03519.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano G, Suon S, Jin H, Donaldson AE, Iacovitti L. Characterization of five evolutionary conserved regions of the human tyrosine hydroxylase (TH) promoter: implications for the engineering of a human TH minimal promoter assembled in a self-inactivating lentiviral vector system. J Cell Physiol. 2005;204:666–677. doi: 10.1002/jcp.20319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabo PJ, Humbert R, Hawrylycz M, Wallace JC, Dorschner MO, McArthur M, Stamatoyannopoulos JA. Genome-wide identification of DNaseI hypersensitive sites using active chromatin sequence libraries. Proc Natl Acad Sci U S A. 2004;101:4537–4542. doi: 10.1073/pnas.0400678101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurada K, Ohshima-Sakurada M, Palmer TD, Gage FH. Nurr1, an orphan nuclear receptor, is a transcriptional activator of endogenous tyrosine hydroxylase in neural progenitor cells derived from the adult brain. Development. 1999;126:4017–4026. doi: 10.1242/dev.126.18.4017. [DOI] [PubMed] [Google Scholar]
- Sasaoka T, Kobayashi K, Nagatsu I, Takahashi R, Kimura M, Yokoyama M, Nomura T, Katsuki M, Nagatsu T. Analysis of the human tyrosine hydroxylase promoter-chloramphenicol acetyl-transferase chimeric gene expression in transgenic mice. Brain Res Mol Brain Res. 1992;16:274–286. doi: 10.1016/0169-328x(92)90236-5. [DOI] [PubMed] [Google Scholar]
- Sawamoto K, Nakao N, Kobayashi K, Matsushita N, Takahashi H, Kakishita K, Yamamoto A, Yoshizaki T, Terashima T, Murakami F, Itakura T, Okano H. Visualization, direct isolation, and transplantation of midbrain dopaminergic neurons. Proc Natl Acad Sci U S A. 2001;98:6423–6428. doi: 10.1073/pnas.111152398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimmel JJ, Crews L, Roffler-Tarlov S, Chikaraishi DM. 4.5 kb Of the rat tyrosine hydroxylase 5′ flanking sequence directs tissue specific expression during development and contains consensus sites for multiple transcription factors. Brain Res Mol Brain Res. 1999;74:1–14. doi: 10.1016/s0169-328x(99)00234-x. [DOI] [PubMed] [Google Scholar]
- Spitz F, Gonzalez F, Duboule D. A global control region defines a chromosomal regulatory landscape containing the HoxD cluster. Cell. 2003;113:405–417. doi: 10.1016/s0092-8674(03)00310-6. [DOI] [PubMed] [Google Scholar]
- Tinti C, Yang C, Seo H, Conti B, Kim C, Joh TH, Kim KS. Structure/function relationship of the cAMP response element in tyrosine hydroxylase gene transcription. J Biol Chem. 1997;272:19158–19164. doi: 10.1074/jbc.272.31.19158. [DOI] [PubMed] [Google Scholar]
- Trocme C, Sarkis C, Hermel JM, Duchateau R, Harrison S, Simonneau M, Al-Shawi R, Mallet J. CRE and TRE sequences of the rat tyrosine hydroxylase promoter are required for TH basal expression in adult mice but not in the embryo. Eur J Neurosci. 1998;10:508–521. doi: 10.1046/j.1460-9568.1998.00059.x. [DOI] [PubMed] [Google Scholar]
- Wong SC, Moffat MA, O’Malley KL. Sequences distal to the AP1/E box motif are involved in the cell type-specific expression of the rat tyrosine hydroxylase gene. J Neurochem. 1994;62:1691–1697. doi: 10.1046/j.1471-4159.1994.62051691.x. [DOI] [PubMed] [Google Scholar]
- Yagi T, Ikawa Y, Yoshida K, Shigetani Y, Takeda N, Mabuchi I, Yamamoto T, Aizawa S. Homologous recombination at c-fyn locus of mouse embryonic stem cells with use of diphtheria toxin A-fragment gene in negative selection. Proc Natl Acad Sci U S A. 1990;87:9918–9922. doi: 10.1073/pnas.87.24.9918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Kim HS, Seo H, Kim KS. Identification and characterization of potential cis-regulatory elements governing transcriptional activation of the rat tyrosine hydroxylase gene. J Neurochem. 1998;71:1358–1368. doi: 10.1046/j.1471-4159.1998.71041358.x. [DOI] [PubMed] [Google Scholar]
- Yoon SO, Chikaraishi DM. Tissue-specific transcription of the rat tyrosine hydroxylase gene requires synergy between an AP-1 motif and an overlapping E box-containing dyad. Neuron. 1992;9:55–67. doi: 10.1016/0896-6273(92)90220-8. [DOI] [PubMed] [Google Scholar]
- Yoon SO, Chikaraishi DM. Isolation of two E-box binding factors that interact with the rat tyrosine hydroxylase enhancer. J Biol Chem. 1994;269:18453–18462. [PubMed] [Google Scholar]
- Zhou QY, Quaife CJ, Palmiter RD. Targeted disruption of the tyrosine hydroxylase gene reveals that catecholamines are required for mouse fetal development. Nature. 1995;374:640–643. doi: 10.1038/374640a0. [DOI] [PubMed] [Google Scholar]


