Abstract
Background
Aberrant activation of the signal transducer and activator of transcription (STAT)3 occurs in many human tumors. Moreover, studies utilizing genetic and pharmacological approaches to modulate constitutive STAT3 activity have provided compelling evidence for the critical role of aberrant STAT3 activity in malignant transformation and tumor progression, and thereby validated STAT3 as a novel cancer drug target.
Objective
This review is intended to be a full coverage of the efforts to develop direct STAT3 inhibitors and will provide a discussion on the inhibitory modalities developed to date.
Methods
Review of the literature focused on the modalities and mechanisms that directly target and inhibit the STAT protein or its functions.
Results/conclusion
While a variety of STAT3 inhibitors have been identified that induce antitumor cell effects in vitro and in vivo, the landscape remains murky. With a few exceptions, most of the STAT3 inhibitors reported to date have not undergone an in vivo efficacy, pharmacology or toxicity testing. Also, there is no evidence, per the published literature of an impending clinical development for the few agents that were reported to exhibit in vivo efficacy. Overall, there is the need for a reassessment of the ongoing strategies to target STAT3 intended not only for refinement, but also for incorporating some new technologies to strengthen our efforts and ensure the success – sooner, rather than later – of identifying suitable anti-STAT3 agents for development into clinically useful anticancer therapeutics.
Keywords: antitumor therapeutics, cancer, molecular targeted therapy, small-molecule inhibitors, STAT3
1. Introduction
Signal transducer and activator of transcription (STAT) proteins were originally discovered as a family of latent cytoplasmic transcription factors that mediate normal cellular responses to cytokines, growth factors, and other polypeptide ligands [1,2]. The activation of STATs is an important event for the mediation of cytokine and growth factor-induced cellular and biological processes, including proliferation, differentiation, survival, development and inflammation (Figure 1). Seven members of the STAT family of proteins have been identified in mammalians: STAT1, 2, 3, 4, 5a, 5b, and 6. All of the family members share six distinct structural domains, including the N-terminal, coiled-coil, DNA-binding, Src homology 2 (SH2), and the transactivation domains, and contain a critical tyrosine (Tyr) residue at the C-terminus (Tyr705 for STAT3), which is phosphorylated during activation (Figure 2). STAT activation by phosphorylation is mediated by growth factor receptor tyrosine kinases, and the cytoplasmic kinases, such as cytokine receptor-associated Janus kinases (JAKs), and Src family kinases. Phosphorylation induces STAT:STAT dimer formation between two monomers via a reciprocal phosphoTyr (pTyr)-SH2 domain interactions [1,2], although pre-existing complexes between inactive, STAT monomers have also been detected [3,4]. From the cytoplasm, STATs accumulate in the nucleus where they mediate gene transcription by binding to specific DNA response elements [1]. Nuclear accumulation of STATs as monomers or dimers remains to be fully defined and may involve several mechanisms [4], including active shuttling between the cytoplasm and nucleus [5].
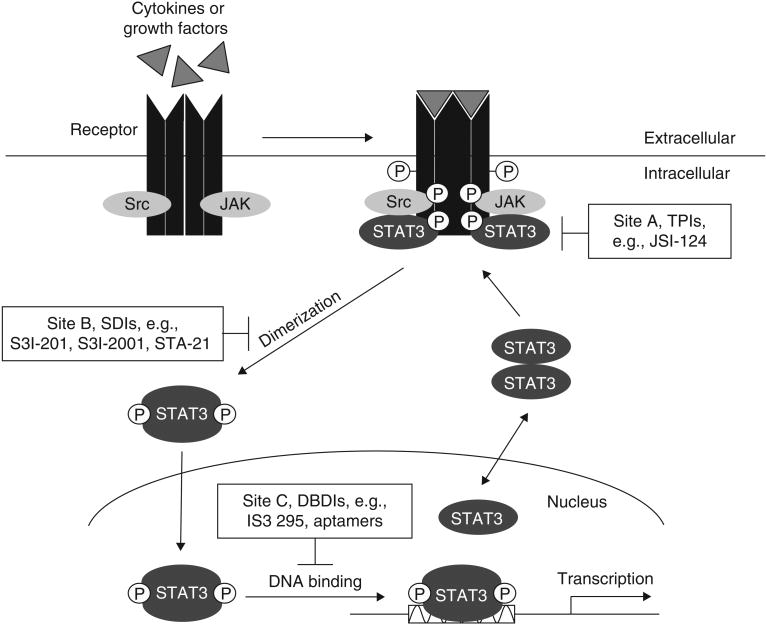
Figure 1. STAT3 signaling pathway and the specific targeting sites for inhibitors.
Polypeptides binding to their cognate receptors activates the tyrosine (Tyr) kinase (TK) activities of the receptors, JAKs, or Src. Latent STAT3 is recruited to the activated receptor for phosphorylation (site A) by TKs on critical Tyr residue, leading to STAT3:STAT3 dimerization (site B) and activation. STAT3 accumulates in the nucleus, where it binds to specific DNA response elements (site C) in the promoters of target genes, leading to gene transcription. Currently identified STAT3 inhibitors can block STAT3 biological activity and function via interrogating at specific sites A, B, C, and D (shown in Figure 1 and Figure 2).
DBDIs: DNA binding domain inhibitors; JAKs: Janus kinases; P: Phosphate; SDIs: SH2 domain inhibitors or dimerizaton inhibitors; Src: Src family kinases; STAT: Signal transducer and activator of transcription; TPIs: Tyr phosphorylation inhibitors.
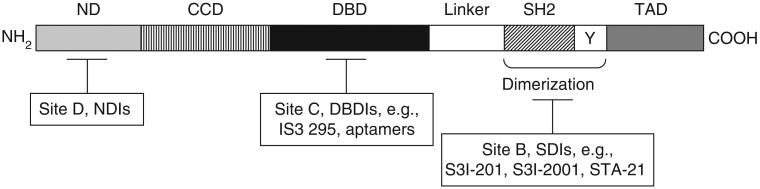
Figure 2. Schematic representation of the structural domains of the STAT3 protein.
STAT3 protein is modular in structure. The N-terminus contains a domain (ND) that mediates STAT dimer–dimer interactions in tetramer formation that is essential for stabilizing the binding of the dimers to DNA. The coiled-coil domain (CCD) links the ND to the DNA-binding domain (DBD) and participates in the interactions with other proteins. The DBD makes physical contact with STAT3-response elements in the promoters of target genes and is linked to the Src homology 2 (SH2) domain by the linker domain (Linker). The SH2 domain is important for dimerization between two STAT3 monomers. Upon the phosphorylation of the critical Tyr (Y) residue in the transcriptional activation domain (TAD), the SH2 domain of one monomer and the pTyr residue of another engage in reciprocal pTyr-SH2 domain interaction. The transactivation domain mediates the transcriptional activation of target genes and may contain a serine residue essential for maximal transcriptional activity in the case of some STAT proteins, including STAT3. Sites B, C, and D indicate the specific sites where currently identified STAT3 inhibitors interact with the STAT3 protein.
DBDIs: DNA binding domain inhibitors; NDIs: N-terminal domain inhibitors; SDIs: SH2 domain inhibitors or dimerizaton inhibitors.
In contrast to the transient nature of STAT activation in normal cells, many human solid and hematological tumors harbor constitutive STAT3 activity. Evidence strongly implicates aberrantly active STAT3 in tumor formation. While the mechanisms by which constitutively active STAT3 induces tumor formation continue to be elucidated, a large body of evidence supports the dysregulation of growth and survival, the promotion of angiogenesis, and the suppression of host's immune surveillance of tumor [2,6-9]. Moreover, aberrant STAT3 promotes invasion and metastasis, thereby contributing to tumor progression [2,6-9]. It is now established that persistently active STAT3 functions as a master regulator of molecular and biological events that promote tumorigenesis [2,9,10]. Thus, the inhibition of aberrant STAT3 activity by genetic or pharmacological approaches induced growth arrest and apoptosis of tumor cells in vitro, as well as tumor regression in vivo [9,11-14]. Indeed, numerous studies have validated STAT3 as a cancer drug target, thereby providing the rationale to pursue the protein for the discovery and development of novel anticancer agents.
2. Targeting of the STAT3 signaling pathway
Although in principle targeting the STAT3 signaling pathway to undermine the malignant phenotype could be approached in many different ways, only a few of those strategies have been explored. Figures 1 and 2 show the more explored strategies, which are: i) the direct targeting of the STAT3 protein by way of the SH2 domain inhibitors or dimerization inhibitors (SDIs, site B), the DNA binding domain inhibitors (DBDIs, site C), and the N-terminal domain inhibitors (NDIs, site D); and ii) the indirect targeting of the upstream components of the STAT3 pathway (site A, tyrosine phosphorylation inhibitors, TPIs). Within the scope of this review, the discussion will mainly focus on the modalities that directly target and inhibit STAT3.
2.1 Modalities that directly inhibit the STAT3 protein
Since the original report of the STAT3 inhibitor development in 2001 [15], several others followed suit, a reflection of the high significance of STAT3 as a target for anticancer drug discovery. Table 1 lists the names and properties of agents reported to date that target STAT3. The present modalities that directly target STAT3 could be grouped into four main strategies, described below.
Table 1. Known STAT3 inhibitors identified to date, their modes of action, cellular effects and models of tumors tested.
| Inhibitor | Target site | Mode of inhibition of STAT3 function | Cellular effects | Cell line tested | Tumor model | Ref. |
|---|---|---|---|---|---|---|
| PY*LKTK | STAT3 SH2 | Dimerization | ↓ Malignant cell growth & transformation | NIH3T3/vSrc | n/d | [15,17] |
| Y*LPQTV | STAT3 SH2 | Dimerization | n/d | n/d | n/d | [21,22] |
| SS 610 | STAT3 SH2 | Dimerization | ↓ Malignant cell growth & transformation, ↑ apoptosis | NIH3T3/vSrc, MDA-MB-231, MDA-MB-435 | n/d | [20] |
| S3I-M2001 | STAT3 SH2 | Dimerization | ↓ Malignant cell growth, ↑ apoptosis, ↓ migration | NIH3T3/vSrc, MDA-MB-231, MDA-MB-435, Panc-1 | MDA-MB-231 | [12] |
| STA-21 | STAT3 SH2 | Dimerization | ↑ Apoptosis | MDA-MB-231, MDA-MB-435, MDA-MB-468 | n/d | [25,26] |
| S3I-201 | STAT3 SH2 | Dimerization | ↓ Cell growth, ↑ apoptosis | NIH3T3/vSrc, MDA-MB-231, MDA-MB-435 | MDA-MB-231 | [11] |
| Stattic | STAT3 SH2 | Phosphorylation | ↑ Apoptosis | MDA-MB-231, MDA-MB-435, HepG2 | n/d | [28] |
| Catechol-containing compounds | STAT3 SH2 | DNA-binding | n/d | n/d | n/d | [29] |
| IS3 295 | STAT3 DBD | DNA binding | ↓ Cell growth, ↑ apoptosis | NIH3T3/vSrc, MDA-MB-468, MDA-MB-435 | n/d | [30] |
| CPA-1, CPA-7 | STAT3 DBD | DNA binding | ↓ Cell growth, ↑ apoptosis | NIH3T3/vSrc, MDA-MB-231, MDA-MB-435, CT26 | CT26 | [31] |
| Galiellalactone | STAT3 DBD | DNA binding | ↑ Apoptosis | HepG2, DU145, PC3 | DU145 | [32,33] |
| Peptide aptamers | STAT3 DBD | DNA binding | ↓ Cell growth, ↑ apoptosis | B16, U266, glioblastoma | n/d | [24,34] |
| Decoy ODN | STAT3 DBD | Compete against endogenous DNA cis-element | ↓ Cell growth, ↑ apoptosis | HNSCC, A549, DU145, HepG2, PLC/PRF/5 | A549, SCCHN | [42-47] |
| G-quartet ODN | STAT3 SH2 | Phosphorylation | ↓ Cell growth, ↑ apoptosis | MDA-MB-468, PC3, HepG2, SCCHN | MDA-MB-468, PC3, A549 | [13,14,51] |
| Peptides | STAT3 ND | Transcriptional activity | ↓ Cell growth, ↑ apoptosis | MCF-7, MDA-MB-231, MDA-MB-435 | n/d | [53] |
| JSI-124 & derivatives | JAK? | Phosphorylation | ↓ Cell growth, ↑ apoptosis, ↓ invasiveness | MDA-MB-468, A549, HCT8/S11, Human lymphoma cells | A549, MDA-MB-435, NIH3T3/vSrc | [54-57] |
| Withacnistin | JAK? | Phosphorylation | ↓ Proliferation, ↑ apoptosis | A549, MDA-MB-435, NIH 3T3/vSrc | MDA-MB-435 | [59] |
↑: Increase; ↓: Decrease; DBD: DNA binding domain; JAKs: Janus kinases; n/d: Not determined; ND: N-terminal domain; ODN: Oligonucleotide; Y*: pTyr.
2.1.1 Strategy I: Inhibitors targeting the STAT3 SH2 domain
2.1.1.1 SH2 domain binding peptides and peptidomimetics or dimerization disruptors
The SH2 domain of STAT is a critical module to facilitate the binding to specific pTyr motifs of receptors for the activation of the protein, as well as to mediate the dimerization between two STAT monomers via the reciprocal interactions between the SH2 domain of one monomer and the pTyr motif of the other. STAT3 dimerization via the pTyr-SH2 domain interaction thus represents an important molecular event for STAT3 functioning. Exploiting this interaction to disrupt STAT3 dimerization (Figure 1, site B) therefore appears to represent an excellent conceptual framework to selectively block the aberrant activity of the protein, and hence its functions in cancer cells. In general, the targeting of SH2 domain to inhibit protein functions as a means for drug development has remained a daunting task [16] and therefore been largely unexplored. In spite of the associated challenges, one of the earliest work on developing STAT3 inhibitors explored the pTyr interaction with the STAT3 SH2 domain [15]. In that study, structure-based design led to the identification of a phosphopeptide inhibitor derived from the STAT3 SH2 domain-binding peptide sequence, PY*LKTK (where Y * represents pTyr). The initial evaluation showed the PY*LKTK to selectively block STAT3 DNA-binding activity in vitro, albeit at a high micromolar concentration (IC50 of 235 μM), probably a reflection of the difficulty of disrupting the large protein surface area. Mechanistically, PY*LKTK disrupted STAT3:STAT3 dimers in vitro. Its selectivity for STAT3 over STAT5 was high, as it lacked any significant effect on STAT5 DNA-binding activity but was only twofold for STAT3 over STAT1, in part because of the close identity in this interaction region for those two STAT proteins. Nonetheless, this earlier work provided a validation of the concept to disrupt STAT3:STAT3 dimerization and activity using a small-molecule peptide.
Two key challenges facing a peptidic approach to drug discovery are membrane permeability and stability. With reference to PY*LKTK, the requirement for the phosphorylation on Tyr for the inhibitory activity presented one more level of challenge to making the phosphopeptide a useful tool for cellular applications. Low membrane permeability was overcome by the covalent attachment of a membrane-translocating sequence (mts) of hydrophobic amino acids (AAVLLPVLLAAP) to the C-terminus of the peptide. Cellular studies showed the PY*LKTK-mts suppressed STAT3-mediated gene transcription and malignant transformation, and induced apoptosis in NIH3T3/v-Src fibroblasts that harbor aberrant STAT3 [15], albeit at 1 mm concentration [15,17], underscoring the potential difficulty of converting this approach into an effective therapeutic modality. However, this work provided the initial proof of principle supporting peptide-mediated approach to block aberrant STAT3 activity and thereby induce antitumor cell effects. Subsequent successful applications of the phosphopeptide as a STAT3 inhibitor were demonstrated, including the usage in the study of the role of aberrant STAT3 in immuno-responsiveness in bone marrow progenitor cells [18], and in the study that showed that the phosphopeptide-mediated inhibition of STAT3 function led to an increased sensitivity of polyamine-depleted cells to apoptosis [19].
The exploration of peptidomimetic and phosphotyrosine mimic approaches in an effort to ameliorate some of the problems with peptides and extend the development efforts led to the identification of ISS 610, a peptidomimetic analog of the tripeptide, PY*L [20], which was derived from the original hexapeptide, PY*LKTK. Structurally similar to PY*L, ISS 610 exhibited properties that mirrored the inhibitory mode and the effect of the lead phosphopeptide in disrupting STAT3 DNA-binding activity [20]. In vitro IC50 improved from 235 μM (for PY*LKTK) to 42 μM for the inhibition of STAT3 DNA-binding activity. Additionally, there was significant increase in selectivity, with the in vitro IC50 for STAT1 as 310 μM and for STAT5 as 285 μM [20]. Notwithstanding, its intracellular activity remained at 1 mm, in part due to the poor membrane permeability imposed by the presence of the pTyr moiety. The observed intracellular effects included the blockade of constitutive STAT3 activity and function, and the selective growth inhibition and induction of apoptosis of v-Src-transformed mouse fibroblasts and the human breast carcinoma cells that harbor persistent STAT3 activity [20]. Although ISS 610 is significantly smaller in molecular size compared to the PY*LKTK-mts, per its tripeptide parent and its lack of the mts appendage, its main drawback is the poor membrane permeability.
More recent studies extending the peptidomimetic approach employed computational modeling to probe the binding of ISS 610 to the STAT3 SH2 domain. This led to a generation of novel oxazole-based small molecules as STAT3 inhibitors, which have increased membrane permeability and a reduced peptidic character [12]. One of these oxazoles, S3I-M2001, blocked STAT3 DNA binding in vitro with an activity similar to that of ISS 610 (IC50 of 79 μM) by disrupting STAT3:STAT3 dimerization [12]. However, there was no gain in selectivity, which was a twofold preference for STAT3 over STAT1 (IC50 of 159 μM). While the cell-free activity of S3I-M2001 represents no gain from that of ISS 610, the improvement with S3I-M2001 is seen in its intracellular activity. It induced significant inhibitory effects on the STAT3 phosphorylation and activation, nuclear accumulation, and transcriptional activity in the NIH3T3/v-Src fibroblasts and human breast carcinoma cells at 50 – 100 μM concentrations. This level of activity represents a nearly 10-fold gain over that of ISS 610. In the same concentration range, S3I-M2001 showed minimal effect on JAK and Src phosphorylation in NIH3T3/v-Src fibroblasts and human breast carcinoma cells. Biological effects of S31-M2001 on cancer cells were observed at 100 μM, including the inhibition of cell growth, survival and metastasis in vitro of the transformed mouse fibroblasts (NIH3T3/v-Src) and the human breast and pancreatic carcinoma models that harbor aberrant STAT3. Furthermore, in vivo evaluation of S3I-M2001 showed a significant induction of the regression of human breast carcinoma (MDA-MB-231) xenografts at 5 – 20 mg/kg. The encouraging initial observations with the S3I-M2001 raise the possibility that a novel small-molecule STAT3 inhibitor can be derived based on targeting the STAT3 SH2 domain that shows appreciable antitumor effects in whole animals. However, more remains to be done with the oxazole class of compounds to derive second and perhaps third generations of analogs of suitable characteristics to facilitate clinical development.
The phosphopeptide approach to inhibit aberrant STAT3 activity was further pursued in a separate study that identified the STAT3-inhbitory peptide sequence, Y*LPQTV and several modified versions, including hydrocinnamoyl-Tyr(PO3H2)-Leu- cis-3,4-methanoPro-Gln-NHBn [21,22]. Studies indicate these peptides have good activities. Compared with the preceding agents, these other compounds inhibited STAT3 DNA-binding activity in vitro at the sub-micromolar concentrations (IC50 values of 0.15 – 0.29 μM) by a similar mechanism of disrupting STAT3 dimerization. While the in vitro STAT3-inhibitory activities are encouraging, the main limitations to in vivo usage are the peptidic nature, and the presence of the pTyr and its potential impact on membrane permeability. It will be of interest to determine the intracellular activity, biological effects on malignant cells, and antitumor effects in vivo, and to define the mechanisms of action for these peptides. However, the physicochemical nature of these molecules presents some challenges to in vivo testing.
Short peptides, called peptide aptamers, from a randomized peptide library [23] have been identified and shown to specifically interact with the STAT3 dimerization domain containing the critical pTyr residue. The interaction leads to the inhibition of STAT3 DNA binding and transcriptional activity [24]. There are perceived differences between the modes of action of the peptide aptamers and those of the preceding peptide inhibitors of STAT3, although both strategies disrupt STAT3 dimerization and block DNA-binding activity. Based on the mode of inhibition, peptide aptamers appear to function in a manner similar to the STAT3 SH2 domain, although they are different in sequences from the SH2 domain [23,24]. This is because the purported peptide aptamer-interacting region of the STAT3 protein is made up of the nearly 100 amino acid residues flanking the Tyr705STAT3. These data suggest that the peptide aptamers might block the binding of the STAT3 SH2 domain to the pTyr705 sequence, thereby blocking dimerization. Thus, while the other STAT3 dimerization disruption peptides interact with the SH2 domain by design, the STAT3-inhibitory peptide aptamers are mimics of the SH2 domain that interact with the SH2 domain-binding site around the pTyr705STAT3. In terms of the biological and in vivo effects of the peptide aptamer molecules that interact with the STAT3 dimerization domain, these are yet to be studied or reported.
In summary, some progress has been made with the peptidic approaches to anti-STAT3 drug discovery. The main limitations of the peptide and peptidomimetic inhibitory approaches remain the low cell permeability, stability, and the consequential low biological activities. The molecules presented above will need to undergo redesign to derive nonpeptidic small-molecule analogs of improved physicochemical properties, which will have high affinity for STAT3, show high selectivity and specificity of effects, and possess increased membrane permeability, as well as good intracellular stability to facilitate preclinical investigation and clinical development.
2.1.1.2 Nonpeptidic chemical inhibitors targeting the STAT3 SH2 domain
There has been recent increase in the efforts to identify nonpeptide, novel small-molecule STAT3 inhibitors. Nonpeptidic small-molecules are cell-permeable and represent a rather more attractive approach to inhibit aberrant STAT3 activity in cancer cells. The use of a structure-based computational modeling of the STAT3 SH2 domain bound with the phosphopeptide in the virtual database screening in two independent studies identified two chemical probes as inhibitors of STAT3 [11,25]. The small-molecule, STA-21 selectively inhibited STAT3 DNA-binding activity in vitro at 20 μM by disrupting STAT3:STAT3 dimerization, and at 20 – 30 μM suppressed STAT3-mediated gene transcription and induced cell growth inhibition and apoptosis through the caspase pathway of human breast carcinoma and rhabdomyosarcoma model cells harboring constitutively active STAT3 [25,26]. More recently, derivatives of STA-21, identified as compounds 1 and 2 [27], have been reported to exhibit a similar STAT3-inhibitory activity profile. These molecules were also effective against the proliferation of prostate carcinoma cell lines.
Another small-molecule STAT3 inhibitor more recently identified is S3I-201 (a.k.a. NSC 74859) [11]. This agent selectively abrogated STAT3 DNA-binding activity in vitro with an IC50 of 86 μM by blocking the formation of STAT3:STAT3 dimers, and was more than 3-fold selective for STAT3 over STAT1. Intracellular activity was observed at 100 μM and included the inhibition of STAT3-dependent gene transcription, and the blockade of the proliferation and survival of human breast carcinoma cells that harbor constitutive STAT3 activity. On the mechanism underlying the biological effects, S3I-201 repressed the expression in human breast tumor cells, and the viral Src-transformed mouse fibroblasts of Bcl-xL, survivin, and cyclin D1, which are some of the known STAT3-regulated genes. Moreover, S3I-201 showed a remarkable antitumor effect in MDA-MB-231 human breast tumor xenografts, inducing tumor regression at 5 mg/kg in nude mice models. These reported studies [11,25-27] highlight a few of the most promising STAT3-inhibitory chemical entities that exhibit a good antitumor efficacy in xenografts. For STA-21 and its derivatives, and S3I-201, the medium to high micromolar activities necessitate medicinal chemistry work to derive analogs of enhanced activities, selectivity, and specificity of effects, thereby facilitating their preclinical and eventually clinical development.
Another nonpeptidic small-molecule inhibitor of STAT3 was discovered by random screening of a collection of 17,298 compounds. Stattic (STAT three-inhibitory compound) [28] inhibited the binding of the phosphopeptide, GY*LPQTV-NH2 to the STAT3 SH2 domain in vitro with an IC50 of 5.1 μM. Moreover, Stattic selectively blocked the IL-6-induced STAT3 activation, nuclear accumulation and DNA-binding activity in human HepG2 hepatoma cells at 20 μM, inhibited the persistent phosphorylation of STAT3 (Tyr705) in the human breast cancer lines, MDA-MB-435 and MDA-MB-231, and induced the apoptosis of same cells. Key issues remain to be addressed, including the molecular mechanisms underlying the inhibition of STAT3 and the consequent induction of biological effects. Also, it is unknown what the in vivo activity and effects of the Stattic compound will be in human tumor xenografts. Such an in vivo study remains to be performed and reported and will provide the initial assessment of the potential therapeutic utility of the Stattic molecule. More recently, catechol (1,2-dihydroxybenzene)-containing compounds [29] were identified through computational modeling and virtual screening as potential STAT3 inhibitors that target the SH2 domain. Experimentally, these agents inhibited the STAT3 DNA-binding activity in vitro, with compound 8 having an IC50 of 106 μM [29]. No additional evaluations of effects of catechol compounds have been reported.
Overall, the nonpeptide small molecules represent some of the most encouraging agents as potent STAT3 inhibitors with a good potential for development into novel anti-STAT3 therapeutics. In contrast to the peptide or peptidomimetic-derived molecules, a better part of these compounds are their physicochemical properties and their activity levels. However, selectivity and specificity of effects are key issues that remain to be clearly defined. Also, though promising, these nonpeptide small-molecules may not be close to clinical development, given the moderate to high micromolar activities. These molecules may yet require medicinal chemistry work to further optimize their structures for improved activity, selectivity and in vivo efficacy, and other pharmacological properties. One of the key issues probably influencing the success of the afore-mentioned inhibitory modalities relates to the fact that the targeting site is a protein – protein interaction. However, with each new report of a STAT3 inhibitor targeting the dimerization event comes an increasing optimism that specific modalities may be realized that could provide a viable anti-STAT3 candidate agent to be added to the armamentarium of anticancer weapons.
2.1.2 Strategy 2: inhibitors targeting the DNA-binding domain of STAT3
Given that the transcriptional activity of STAT3 requires the physical interaction of its DNA-binding domain with the consensus DNA-binding sequence in the promoter regions of responsive genes (Figure 1, site C), the disruption of the protein:DNA interactions has the potential to inhibit STAT3-dependent gene transcription and thus block its tumor-promoting functions. Nonetheless, there has been far less emphasis on targeting the STAT3:DNA-binding interface for drug design. A few reports have surfaced, but no concerted, strategic plans directed at this site have been reported in the literature.
A class of platinum (IV) compounds has been reported that potentially block the DNA-binding activity of STAT3. The agent, IS3 295, blocked STAT3 DNA-binding activity with an IC50 of 1.4 μM [30]. The exact mode of inhibition of STAT3 is not clearly understood, although the initial evidence suggests a potential interaction with the STAT3 DNA-binding domain [30], in turn disrupting the binding to the STAT3-responsive DNA sequences. Inhibition of intra-cellular STAT3 activation was found to involve the reduction of pTyr705STAT3 level. Moreover, IS3 295 inhibited STAT3-dependent gene transcription, including the induction of cyclin D1 and Bcl-xL, blocked cell proliferation and malignant transformation, and induced apoptosis of NIH3T3/v-Src mouse fibroblasts and human breast carcinoma cells harboring constitutive STAT3 activity at 10 μM [30].
Additional platinum (IV) compounds, CPA-1, CPA-7, and platinum (IV) tetrachloride [31], were separately identified that inhibited STAT3 DNA-binding with similar activity, diminished the pTyr705STAT3 level and STAT3-mediated gene regulation, and induced cell growth inhibition and apoptosis of human breast, lung and prostate cancer cells harboring constitutively active STAT3 at low micromolar levels. The in vitro levels of activity against STAT3 activation were significantly better for the platinum (IV) complexes compared with the peptide-based or the nonpeptide small molecules discussed in the preceding sections. However, selectivity for STAT3 over STAT1 was modestly 2.5- to 4-fold and is similar.
Of these platinum (IV) complexes, only CPA-7 has been evaluated for efficacy in vivo. The initial in vivo study showed CPA-7 to induce the regression of CT26 mouse colon cancer model at 5 mg/kg, an efficacy that should be sufficiently good to justify the evaluation of the remaining platinum (IV) complexes. Given that the CPA-7 and its class of agents share structural resemblance to the clinical-approved drug, Cisplatin, there is justification based on the good in vitro activities and the in vivo effects to pursue further investigation to determine their clinical potential and for deriving suitable therapeutic doses. For the platinum (IV) complexes, efforts need to be directed at establishing the pharmacological and toxicity profiles.
It is important to note that the platinum (IV) complexes exhibit a different activity profile with respect to STAT3 from that of cisplatin, a platinum (II) complex with no measurable inhibitory effect against STAT3 activity [30,31]. In summary, the overall in vitro and in vivo activities of the CPA-7 and the platinum (IV) complexes warrant their investigation in line with pursuing their preclinical investigation and clinical development.
A tetrahydro-isobenzofuranone derivative isolated from ascomycete was reported to block STAT3 DNA-binding activity. Galiellalactone selectively inhibited IL-6-induced STAT3 DNA-binding and transcriptional activity without affecting STAT3 phosphorylation in HepG2 hepatoma cells [32]. Though galiellalactone is presumed to exert its effect by interacting with the STAT3 DNA-binding domain, it is not entirely clear how this agent inhibits the binding of the STAT3 dimers to the DNA consensus sequence, an important question that needs addressing. On effects, the inhibition of STAT3 activity by 25 μM galiellalactone led to the downregulation of STAT3-dependent gene expression and the induction of apoptosis of the DU145 and PC-3 human prostate cancer cells harboring persistently active STAT3. Moreover, this agent induced the growth inhibition of human prostate (DU145) tumor xenografts delivered at 3 mg/kg per day intraperitoneally [33], a good in vivo activity and effect. More studies are needed to assess further the therapeutic suitability of this agent and establish a pharmacologic and toxicity profile.
Recently, 20-mer peptide aptamers have also been identified to specifically interact with the DNA-binding domain of STAT3 and in turn inhibit STAT3 DNA-binding and transcriptional activities [24]. Random peptide aptamer sequence was constructed as a fusion protein for expression and for functional analysis. In reported studies, purified recombinant peptide aptamer, tagged with a protein transduction motif of nine arginines and fused with thioredoxin as a scaffold protein [23,24], was cell-permeable and selectively induced cell growth inhibition and apoptosis, with a 0.27 μM concentration inducing a 50% growth inhibition of human U266 multiple myeloma and glioblastoma cells, and mouse B16 melanoma cells harboring constitutively active STAT3 [24,34].
With the published level of activity, peptide aptamers as inhibitors of STAT3 represent one of the effective approaches to disrupt STAT3 functions in vitro compared with others already discussed. One of the main issues is the physical characteristics of the modality and how adaptable the approach is for widespread application. While the studies support the conceptual basis for the use of recombinant peptide aptamers as inhibitors of STAT3, limitations for in vivo usage include the physicochemical properties, stability and the permeability of the fusion protein. Further evaluations on permeability and in vivo bioavailability of the recombinant peptide aptamers will be needed.
2.1.3 Strategy 3: oligonucleotide approaches to inhibit STAT3 signaling
Approaches developed to inhibit target gene expression based on oligonucleotide technology include antisense RNA, small interfering RNA (siRNA), and decoy oligodeoxynucleotide (ODN). The knockdown of the STAT3 protein by antisense RNA or siRNA approaches has been demonstrated in studies, which showed the induction of tumor cell apoptosis and tumor regression [35-37]. Those studies provide the conceptual framework to derive siRNA-based agents targeting STAT3 as therapeutic modalities.
Decoy ODN strategy has been used not only as a powerful research tool in gene regulation in vitro and in vivo, but also as a novel strategy in gene therapy. Decoy ODN has been demonstrated to inhibit the activity of several target transcription factors, including nuclear factor- κB [38,39], AP-1 [40], and E2F [41]. Synthetic double-stranded ODN mimics the consensus sequence of the cis-element, which has highly specific affinity to the target transcription factor. After introduction into cells, exogenous decoy ODN would compete for the binding of the targeted transcription factor with the endogenous cis-elements and block the authentic cis – trans interaction, resulting in the downregulation of the transcription factor-mediated gene transcription (Figure 1, site C) [41]. In vitro and in vivo investigations have shown that the application of STAT3 decoy ODN of the sequence of 5′-CATTTCCCGTAAATC-3′ blocked STAT3-dependent transcription of such genes as cyclin D1, c-Myc, Bcl-xL, and survivin, leading to reduced proliferation and induction of apoptosis of a variety of human tumor cell lines, including lung, liver, prostate, head and neck carcinoma cells, as well as the regression in xenografts of the human head and neck carcinoma cells and A549 non-small cell lung cancer cells at a dose of 25 μg/day [42-47]. This approach holds promise in terms of deriving novel therapeutic strategies against the many human cancers harboring aberrant STAT3. Though the clinical trials of the decoy ODN against E2F was approved by the US FDA in 2001, the long-term clinical outcome has yet to be established [48].
Recently, a new class of ODN inhibitors against STAT3 was developed, designated as G-quartet ODN [49]. These are G-rich ODN with the ability to form inter- and intra-molecular four-stranded G-quartet structures. While the exact mode of inhibition of STAT3 is somehow obscure, computational modeling studies indicate that the G-quartet ODN can specifically insert between the SH2 domains of STAT3 dimers via interactions with the amino acids residues 638 – 652 of STAT3 (Figure 1, site B), thereby disrupting STAT3:STAT3 dimers, inhibiting the binding to DNA, and hence selectively blocking STAT3 activity, without affecting the other members of the STAT family [50]. The G-quartet ODN, T40214, selectively abrogated IL-6-induced STAT3 phosphorylation and DNA-binding activity in vitro with an IC50 of 5 μM [51]. The application of T40214 in vivo at 5 – 10 mg/kg every 2 days in tumor xenografts of human breast, lung, prostate, head and neck cancer cell lines markedly inhibited STAT3-mediated gene transcription and induced tumor cell apoptosis and tumor regression [13,14,51]. The G-quartet approach therefore has the potential to generate a novel candidate anticancer therapeutic modality that targets aberrant STAT3 function insofar as molecular details of the mechanism of action is understood and the selectivity for STAT3 over other proteins and specificity of effects are fulfilled. The physicochemical properties of the G-quartet may also need to be optimized to facilitate preclinical investigation and clinical development.
Altogether, the ODN approach represents a good conceptual basis for deriving novel anti-STAT therapeutic agents, and the substantive in vitro and in vivo data provide support for serious consideration of this strategy. Selectivity and specificity of effects are some of the advantages of this approach over other modalities. While the proof of concept is supported by the existing studies, the remaining important issues relate to deriving anti-STAT3 oligonucleotide agents of optimum physicochemical properties that will have a suitable bioavailability profile, a low toxicity, and good pharmacological properties.
2.1.4 Strategy 4: inhibitors targeting STAT3 N-terminal domain
The N-terminal domain of STAT3 protein comprises nearly 130 amino acids and contains eight helices [52,53]. The N-terminal domain mediates the protein – protein interactions in the binding of STAT3 dimers to DNA sites and in the assembly of the transcriptional machinery, including the interactions between two STAT3 dimers in the tetramer formation and with other transcriptional factors and regulators [2,52,53].
Recently, a category of short peptides, which are derived from the helices of the N-terminal domain of the STAT3 protein, especially helix-2, specifically recognized and bound to STAT3 but not other STAT members, leading to the inhibition of STAT3 transcriptional activity without affecting the phosphorylation status [53]. The cell-permeable form of these peptides, generated by the fusion with Penetratin (a protein transduction motif of the sequence of RQIKIWFPNRR-Nle-KWKK-NH2), selectively induced cell growth inhibition and apoptosis of human MDA-MB-231, MDA-MB-435 and MCF-7 breast cancer cells, with a 50% growth inhibition at low microM level (< 10 μM). However, details of the mechanisms of action of the peptides, their interaction with the N-terminal domain of STAT3, and how this affects STAT3 functions, remain to be investigated. This report [53] represents the first to describe such an approach, identifying one additional strategy in the discovery of potential anti-STAT3 candidate agents as novel anticancer therapeutic modality. The challenge of effectively targeting protein – protein interactions for drug discovery and development could similarly impact upon this approach.
2.2 Strategies that indirectly inhibit the upstream components of the STAT3 activation
The ligand – receptor interaction at the extracellular surface links the receptor to the STAT3 pathway via the activities of Tyr kinases. Inhibitors of the upstream intracellular Tyr kinases provide indirect, albeit effective, mechanisms to attenuate the constitutive activation of STAT3 signaling in malignant cells. Besides the existing Tyr kinases inhibitors, such as the EGFR and Src inhibitors that block aberrant STAT3 signaling, a variety of other small molecules have been reported that indirectly block STAT3 signaling via their effects on the upstream Tyr kinases (Figure 1, site A). For example, JSI-124 is a cucurbitacin that indirectly inhibits the activation of STAT3 in vitro at 10 μM [54]. Indirectly, this molecule markedly suppressed STAT3 phosphorylation in the v-Src-transformed mouse fibroblasts, human A549 lung carcinoma cells, and MDA-MB-468 breast carcinoma cells that harbor constitutively-active STAT3 by inhibiting an upstream kinase. The exact identify of the Tyr kinase, however remains unknown. The application of JSI-124 at 10 μM induced a loss of viability and growth inhibition, and apoptosis of v-Src-transformed mouse fibroblasts, human breast, lung, lymphoma, glioma, and colon carcinoma cells [54-57]. At a dose of 1 mg/kg/day, JSI-124 inhibited growth of tumor models of A549 human non-small cell lung cancer, v-Src-transformed mouse fibroblasts and human breast cancer tumors in nude mice [54]. Moreover, JSI-124 promoted the antitumor immune response of dendritic cells [58].
Other cucurbitacin analogs have been discovered, as well as other agents that reportedly induce antitumor activity by a similar mechanism [59-61]. Of these, withacnistin, a natural product and the single major component of the agent available in the NCI DTP Repository, NSC135075, which was initially identified erroneously as cucurbitacin Q, selectively inhibited STAT3 phosphorylation and activation, resulting in the induction of tumor cell apoptosis and growth inhibition of human lung and breast tumor xenografts [59]. Literature reports suggest that cucurbitacin and analogs may provide effective mechanisms to disrupt the aberrant activation of the STAT3 signaling pathway. Although a general assessment of the cucurbitacin and withacnistin molecules suggests that they target the JAK/STAT pathway, it remains unknown how these agents inhibit STAT3 activation. For example, withacnistin had no measurable effect on the kinases, JAK2, Src, Akt, Erk, and JNK activities. Nonetheless, their relatively useful activity makes them worth pursuing further.
Other modalities that have been reported to indirectly modulate STAT3 signaling include the receptor Tyr kinase inhibitors, tyrphostins [62], the JAK kinase inhibitors, such as AG490, WP1066 [63,64] and TG101209 [65], and resveratrol [66] and indirubin [67,68], which potentially target the Src kinase, as well as curcumin [69,70], which reportedly inhibits the JAK – STAT pathway. Mechanisms of action in relation to STAT3 inhibition for some of these agents remain to be clearly defined. Also, the possible general effects on many signal transduction pathways, which have been corroborated for these agents and suggest a less specific mechanism of action [71-73], highlight the limitations of their use as modalities that specifically inhibit STAT3 signaling.
3. Expert opinion
It has been 13 years since the original discovery of the association of constitutive STAT3 activation with malignant transformation [74]. Since then, a large number of studies have been undertaken for the validation of STAT3 as a cancer drug target [2,9,75,76], and substantial efforts have poured into the discovery of novel STAT3 inhibitors [e.g., [77]]. There is a large number of STAT3 inhibitors reported to date and noted herein, which is a reflection of the enhanced activity in this arena in recent years. Of these inhibitors, a few show good activity in terms of the inhibition of STAT3 biological functions and the associated antitumor cell effects, as well as the inhibition of tumor growth in the mouse models of human tumors. These inhibitors are mostly at the experimental stage and not in the form considered to be suitable for clinical utility.
There is no evidence in the published literature of a suitable STAT3 inhibitor that is near clinical development. Since much of the efforts at STAT3 drug discovery is directed at disrupting the STAT3:STAT3 dimerization, the slow progress of obtaining suitable STAT3 inhibitors for preclinical investigation and for clinical development could be attributed to the challenge of targeting protein – protein interactions, given the large surface area of the target, and hence the difficulty of identifying a small molecule that sufficiently disrupts the interaction. Nonetheless, recent successes relating to the disruption of the Bcl-xL and Bax interaction [78-80] offer some ray of hope that STAT3 inhibitors that target the dimerization event could be developed sooner rather than later for clinical application. On the other hand, not much progress has been made with regard to modalities that target other aspects of STAT3 activity and function, such as the DNA-binding interface or the oligonucleotide approach. Part of the reason for the lack of great progress in these other areas is the fact that far less attention has been given to these areas compared with the focus on dimerization disruption. Thus, more efforts may be needed on the targeting of other sites in the STAT3 activation process in order to achieve rapid success.
The ultimate and obvious question is what the overall response will be to STAT3 inhibitors as novel anticancer agents, and how well patients will respond. The outcome in patients cannot be addressed until a suitable anti-STAT3 agent becomes available in the clinic. Thus the need remains high for suitable and effective STAT3 inhibitors for clinical application in cancer patients, as well as for a clinical validation of the STAT3 protein in patients whose tumors harbor aberrant STAT3. Clinically appropriate STAT3 inhibitors will also provide the tools to interrogate tumor tissues with the goal of deriving clinically useful biomarkers with a sensitivity profile that correlates with the blockade of aberrant STAT3 in human tumors. Such biomarkers could serve a useful platform for identifying patients who might benefit from an anti-STAT3 therapy.
Auspiciously, diverse conceptual bases via several of the leading compounds have been proposed. The expectation is that these leading agents will facilitate the design of novel small-molecule inhibitors of STAT3 suitable for a potential clinical application. Structural analytical tools, such as NMR analysis coupled with computational modeling, should facilitate the critical evaluation of the protein, such as of the DNA-binding interface and the SH2 domain bound to small molecules, in order to derive information on key structural determinants that can provide the basis for establishing new paradigms in the design of novel small molecules that effectively and selectively disrupt STAT3 DNA-binding activity and dimerization.
On a final note, now might be the time for a critical re-evaluation of our overall approaches to targeting STAT3 and for developing new models for disrupting the protein in order to accomplish the goal of delivering clinically useful direct STAT3 inhibitors as novel anticancer agents in a timely manner.
Acknowledgments
We thank all colleagues and members of our laboratory for stimulating discussions. This work was supported by the National Cancer Institute Grant CA106439 (JT).
Footnotes
Declaration of interest The authors state no conflict of interest and have received no payment in preparation of this manuscript.
Bibliography
Papers of special note have been highlighted as either of interest (•) or of considerable interest (••) to readers.
- 1.Darnell JE., Jr STATs and gene regulation. Science. 1997;277(5332):1630–5. doi: 10.1126/science.277.5332.1630. [DOI] [PubMed] [Google Scholar]
- 2.Turkson J. STAT proteins as novel targets for cancer drug discovery. Expert Opin Ther Targets. 2004;8(5):409–22. doi: 10.1517/14728222.8.5.409. [DOI] [PubMed] [Google Scholar]
- 3.Schroder M, Kroeger KM, Volk HD, et al. Preassociation of nonactivated STAT3 molecules demonstrated in living cells using bioluminescence resonance energy transfer: a new model of STAT activation? J Leukoc Biol. 2004;75(5):792–7. doi: 10.1189/jlb.1003496. [DOI] [PubMed] [Google Scholar]
- 4.Sehgal PB. Paradigm shifts in the cell biology of STAT signaling. Semin Cell Dev Biol. 2008;19:329–40. doi: 10.1016/j.semcdb.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu L, McBride KM, Reich NC. STAT3 nuclear import is independent of tyrosine phosphorylation and mediated by importin-alpha3. Proc Natl Acad Sci USA. 2005;102(23):8150–5. doi: 10.1073/pnas.0501643102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haura EB, Turkson J, Jove R. Mechanisms of disease: Insights into the emerging role of signal transducers and activators of transcription in cancer. Nat Clin Pract Oncol. 2005;2(6):315–24. doi: 10.1038/ncponc0195. [DOI] [PubMed] [Google Scholar]
- 7.Kortylewski M, Kujawski M, Wang T, et al. Inhibiting Stat3 signaling in the hematopoietic system elicits multicomponent antitumor immunity. Nat Med. 2005;11(12):1314–21. doi: 10.1038/nm1325. [DOI] [PubMed] [Google Scholar]
- 8.Kortylewski M, Yu H. Stat3 as a potential target for cancer immunotherapy. J Immunother. 2007;30(2):131–9. doi: 10.1097/01.cji.0000211327.76266.65. [DOI] [PubMed] [Google Scholar]
- 9.Yu H, Jove R. The STATs of cancer: new molecular targets come of age. Nat Rev Cancer. 2004;4(2):97–105. doi: 10.1038/nrc1275. [DOI] [PubMed] [Google Scholar]; •• A critical review of the molecular mechanisms of Stat3-mediated tumorigenesis.
- 10.Darnell JE. Validating Stat3 in cancer therapy. Nat Med. 2005;11(6):595–6. doi: 10.1038/nm0605-595. [DOI] [PubMed] [Google Scholar]
- 11.Siddiquee K, Zhang S, Guida WC, et al. Selective chemical probe inhibitor of Stat3, identified through structure-based virtual screening, induces antitumor activity. Proc Natl Acad Sci USA. 2007;104(18):7391–6. doi: 10.1073/pnas.0609757104. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• One of the examples of nonpeptidic inhibitors of Stat3 by disruption of Stat3 dimerization.
- 12.Siddiquee KA, Gunning PT, Glenn M, et al. An oxazole-based small-molecule Stat3 inhibitor modulates Stat3 stability and processing and induces antitumor cell effects. ACS Chem Biol. 2007;2(12):787–98. doi: 10.1021/cb7001973. [DOI] [PubMed] [Google Scholar]
- 13.Jing N, Zhu Q, Yuan P, et al. Targeting signal transducer and activator of transcription 3 with G-quartet oligonucleotides: a potential novel therapy for head and neck cancer. Mol Cancer Ther. 2006;5(2):279–86. doi: 10.1158/1535-7163.MCT-05-0302. [DOI] [PubMed] [Google Scholar]
- 14.Weerasinghe P, Garcia GE, Zhu Q, et al. Inhibition of Stat3 activation and tumor growth suppression of non-small cell lung cancer by G-quartet oligonucleotides. Int J Oncol. 2007;31(1):129–36. [PubMed] [Google Scholar]
- 15.Turkson J, Ryan D, Kim JS, et al. Phosphotyrosyl peptides block Stat3-mediated DNA binding activity, gene regulation, and cell transformation. J Biol Chem. 2001;276(48):45443–55. doi: 10.1074/jbc.M107527200. [DOI] [PubMed] [Google Scholar]; •• First proof-of-concept for inhibition of Stat3 by peptide derivatives.
- 16.McMurray JS. A new small-molecule Stat3 inhibitor. Chem Biol. 2006;13(11):1123–4. doi: 10.1016/j.chembiol.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 17.Vultur A, Cao J, Arulanandam R, et al. Cell-to-cell adhesion modulates Stat3 activity in normal and breast carcinoma cells. Oncogene. 2004;2315:2600–16. doi: 10.1038/sj.onc.1207378. [DOI] [PubMed] [Google Scholar]
- 18.Wang T, Niu G, Kortylewski M, et al. Regulation of the innate and adaptive immune responses by Stat-3 signaling in tumor cells. Nat Med. 2004;10(1):48–54. doi: 10.1038/nm976. [DOI] [PubMed] [Google Scholar]; • First evidence of a role of Stat3 in regulation of immune responses in tumor cells.
- 19.Bhattacharya S, Ray RM, Johnson LR. STAT3-mediated transcription of Bcl-2, Mcl-1 and c-IAP2 prevents apoptosis in polyamine-depleted cells. Biochem J. 2005;392(Pt 2):335–44. doi: 10.1042/BJ20050465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Turkson J, Kim JS, Zhang S, et al. Novel peptidomimetic inhibitors of signal transducer and activator of transcription 3 dimerization and biological activity. Mol Cancer Ther. 2004;3(3):261–9. [PubMed] [Google Scholar]; •• First evidence for peptidomimetics as Stat3 inhibitors by disruption of Stat3 dimerization.
- 21.Ren Z, Cabell LA, Schaefer TS, McMurray JS. Identification of a high-affinity phosphopeptide inhibitor of Stat3. Bioorg Med Chem Lett. 2003;13(4):633–6. doi: 10.1016/s0960-894x(02)01050-8. [DOI] [PubMed] [Google Scholar]
- 22.Coleman DRT, Ren Z, Mandal PK, et al. Investigation of the binding determinants of phosphopeptides targeted to the SRC homology 2 domain of the signal transducer and activator of transcription 3. Development of a high-affinity peptide inhibitor. J Med Chem. 2005;48(21):6661–70. doi: 10.1021/jm050513m. [DOI] [PubMed] [Google Scholar]
- 23.Buerger C, Nagel-Wolfrum K, Kunz C, et al. Sequence-specific peptide aptamers, interacting with the intracellular domain of the epidermal growth factor receptor, interfere with Stat3 activation and inhibit the growth of tumor cells. J Biol Chem. 2003;278(39):37610–21. doi: 10.1074/jbc.M301629200. [DOI] [PubMed] [Google Scholar]
- 24.Nagel-Wolfrum K, Buerger C, Wittig I, et al. The interaction of specific peptide aptamers with the DNA binding domain and the dimerization domain of the transcription factor Stat3 inhibits transactivation and induces apoptosis in tumor cells. Mol Cancer Res. 2004;2(3):170–82. [PubMed] [Google Scholar]
- 25.Song H, Wang R, Wang S, Lin J. A low-molecular-weight compound discovered through virtual database screening inhibits Stat3 function in breast cancer cells. Proc Natl Acad Sci USA. 2005;102(13):4700–5. doi: 10.1073/pnas.0409894102. [DOI] [PMC free article] [PubMed] [Google Scholar]; • First example of the nonpeptidic chemical inhibitors of Stat3 via targeting its SH2 domain.
- 26.Chen CL, Loy A, Cen L, et al. Signal transducer and activator of transcription 3 is involved in cell growth and survival of human rhabdomyosarcoma and osteosarcoma cells. BMC Cancer. 2007;7:111. doi: 10.1186/1471-2407-7-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bhasin D, Cisek K, Pandharkar T, et al. Design, synthesis, and studies of small molecule STAT3 inhibitors. Bioorg Med Chem Lett. 2008;18(1):391–5. doi: 10.1016/j.bmcl.2007.10.031. [DOI] [PubMed] [Google Scholar]
- 28.Schust J, Sperl B, Hollis A, et al. Stattic: a small-molecule inhibitor of STAT3 activation and dimerization. Chem Biol. 2006;13(11):1235–42. doi: 10.1016/j.chembiol.2006.09.018. [DOI] [PubMed] [Google Scholar]
- 29.Hao W, Hu Y, Niu C, et al. Discovery of the catechol structural moiety as a Stat3 SH2 domain inhibitor by virtual screening. Bioorg Med Chem Lett. 2008;18(18):4988–92. doi: 10.1016/j.bmcl.2008.08.032. [DOI] [PubMed] [Google Scholar]
- 30.Turkson J, Zhang S, Mora LB, et al. A novel platinum compound inhibits constitutive Stat3 signaling and induces cell cycle arrest and apoptosis of malignant cells. J Biol Chem. 2005;280(38):32979–88. doi: 10.1074/jbc.M502694200. [DOI] [PubMed] [Google Scholar]
- 31.Turkson J, Zhang S, Palmer J, et al. Inhibition of constitutive signal transducer and activator of transcription 3 activation by novel platinum complexes with potent antitumor activity. Mol Cancer Ther. 2004;3(12):1533–42. [PubMed] [Google Scholar]
- 32.Weidler M, Rether J, Anke T, Erkel G. Inhibition of interleukin-6 signaling by galiellalactone. FEBS Lett. 2000;484(1):1–6. doi: 10.1016/s0014-5793(00)02115-3. [DOI] [PubMed] [Google Scholar]
- 33.Hellsten R, Johansson M, Dahlman A, et al. Galiellalactone is a novel therapeutic candidate against hormone-refractory prostate cancer expressing activated Stat3. Prostate. 2008;68(3):269–80. doi: 10.1002/pros.20699. [DOI] [PubMed] [Google Scholar]
- 34.Borghouts C, Kunz C, Delis N, Groner B. Monomeric recombinant peptide aptamers are required for efficient intracellular uptake and target inhibition. Mol Cancer Res. 2008;6(2):267–81. doi: 10.1158/1541-7786.MCR-07-0245. [DOI] [PubMed] [Google Scholar]
- 35.Gao L, Zhang L, Hu J, et al. Down-regulation of signal transducer and activator of transcription 3 expression using vector-based small interfering RNAs suppresses growth of human prostate tumor in vivo. Clin Cancer Res. 2005;11(17):6333–41. doi: 10.1158/1078-0432.CCR-05-0148. [DOI] [PubMed] [Google Scholar]
- 36.Konnikova L, Kotecki M, Kruger MM, Cochran BH. Knockdown of STAT3 expression by RNAi induces apoptosis in astrocytoma cells. BMC Cancer. 2003;3:23. doi: 10.1186/1471-2407-3-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ling X, Arlinghaus RB. Knockdown of STAT3 expression by RNA interference inhibits the induction of breast tumors in immunocompetent mice. Cancer Res. 2005;65(7):2532–6. doi: 10.1158/0008-5472.CAN-04-2425. [DOI] [PubMed] [Google Scholar]
- 38.Son G, Iimuro Y, Seki E, et al. Selective inactivation of NF-kappaB in the liver using NF-kappaB decoy suppresses CCl4-induced liver injury and fibrosis. Am J Physiol Gastrointest Liver Physiol. 2007;293(3):G631–9. doi: 10.1152/ajpgi.00185.2007. [DOI] [PubMed] [Google Scholar]
- 39.Bezzerri V, Borgatti M, Nicolis E, et al. Transcription factor oligodeoxynucleotides to NF-kappaB inhibit transcription of IL-8 in bronchial cells. Am J Respir Cell Mol Biol. 2008;39(1):86–96. doi: 10.1165/rcmb.2007-0176OC. [DOI] [PubMed] [Google Scholar]
- 40.Moriyama I, Ishihara S, Rumi MA, et al. Decoy oligodeoxynucleotide targeting activator protein-1 (AP-1) attenuates intestinal inflammation in murine experimental colitis. Lab Invest. 2008;88(6):652–63. doi: 10.1038/labinvest.2008.38. [DOI] [PubMed] [Google Scholar]
- 41.Tomita N, Ogihara T, Morishita R. Transcription factors as molecular targets: molecular mechanisms of decoy ODN and their design. Curr Drug Targets. 2003;4(8):603–8. doi: 10.2174/1389450033490803. [DOI] [PubMed] [Google Scholar]
- 42.Leong PL, Andrews GA, Johnson DE, et al. Targeted inhibition of Stat3 with a decoy oligonucleotide abrogates head and neck cancer cell growth. Proc Natl Acad Sci USA. 2003;100(7):4138–43. doi: 10.1073/pnas.0534764100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barton BE, Murphy TF, Shu P, et al. Novel single-stranded oligonucleotides that inhibit signal transducer and activator of transcription 3 induce apoptosis in vitro and in vivo in prostate cancer cell lines. Mol Cancer Ther. 2004;3(10):1183–91. [PubMed] [Google Scholar]
- 44.Aggarwal BB, Sethi G, Ahn KS, et al. Targeting signal-transducer-and-activator-of-transcription-3 for prevention and therapy of cancer: modern target but ancient solution. Ann NY Acad Sci. 2006;1091:151–69. doi: 10.1196/annals.1378.063. [DOI] [PubMed] [Google Scholar]
- 45.Sun X, Zhang J, Wang L, Tian Z. Growth inhibition of human hepatocellular carcinoma cells by blocking STAT3 activation with decoy-ODN. Cancer Lett. 2008;262(2):201–13. doi: 10.1016/j.canlet.2007.12.009. [DOI] [PubMed] [Google Scholar]
- 46.Zhang X, Zhang J, Wang L, et al. Therapeutic effects of STAT3 decoy oligodeoxynucleotide on human lung cancer in xenograft mice. BMC Cancer. 2007;7:149. doi: 10.1186/1471-2407-7-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xi S, Gooding WE, Grandis JR. In vivo antitumor efficacy of STAT3 blockade using a transcription factor decoy approach: implications for cancer therapy. Oncogene. 2005;24(6):970–9. doi: 10.1038/sj.onc.1208316. [DOI] [PubMed] [Google Scholar]
- 48.Morishita R, Aoki M, Ogihara T. Does gene therapy become pharmacotherapy? Exp Physiol. 2005;90(3):307–13. doi: 10.1113/expphysiol.2005.030403. [DOI] [PubMed] [Google Scholar]
- 49.Jing N, Li Y, Xu X, et al. Targeting Stat3 with G-quartet oligodeoxynucleotides in human cancer cells. DNA Cell Biol. 2003;22(11):685–96. doi: 10.1089/104454903770946665. [DOI] [PubMed] [Google Scholar]; • First evidence of G-rich oligonucleotides as Stat3 inhibitors.
- 50.Zhu Q, Jing N. Computational study on mechanism of G-quartet oligonucleotide T40214 selectively targeting Stat3. J Comput Aided Mol Des. 2007;21(1011):641–8. doi: 10.1007/s10822-007-9147-6. [DOI] [PubMed] [Google Scholar]; • First proof-of-principle for G-rich oligonucleotides as Stat3 inhibitors by interfering Stat3 dimerization.
- 51.Jing N, Li Y, Xiong W, et al. G-quartet oligonucleotides: a new class of signal transducer and activator of transcription 3 inhibitors that suppresses growth of prostate and breast tumors through induction of apoptosis. Cancer Res. 2004;64(18):6603–9. doi: 10.1158/0008-5472.CAN-03-4041. [DOI] [PubMed] [Google Scholar]
- 52.Becker S, Groner B, Muller CW. Three-dimensional structure of the Stat3beta homodimer bound to DNA. Nature. 1998;394(6689):145–51. doi: 10.1038/28101. [DOI] [PubMed] [Google Scholar]
- 53.Timofeeva OA, Gaponenko V, Lockett SJ, et al. Rationally designed inhibitors identify STAT3 N-domain as a promising anticancer drug target. ACS Chem Biol. 2007;2(12):799–809. doi: 10.1021/cb700186x. [DOI] [PubMed] [Google Scholar]; •• First proof-of-concept for inhibition of Stat3 function via targeting its N-terminal domain.
- 54.Blaskovich MA, Sun J, Cantor A, et al. Discovery of JSI-124 (cucurbitacin I), a selective Janus kinase/signal transducer and activator of transcription 3 signaling pathway inhibitor with potent antitumor activity against human and murine cancer cells in mice. Cancer Res. 2003;63(6):1270–9. [PubMed] [Google Scholar]
- 55.van Kester MS, Out-Luiting JJ, von dem Borne PA, et al. Cucurbitacin I inhibits Stat3 and induces apoptosis in Sezary cells. J Invest Dermatol. 2008;128(7):1691–5. doi: 10.1038/sj.jid.5701246. [DOI] [PubMed] [Google Scholar]
- 56.Shi X, Franko B, Frantz C, et al. JSI-124 (cucurbitacin I) inhibits Janus kinase-3/signal transducer and activator of transcription-3 signalling, downregulates nucleophosmin-anaplastic lymphoma kinase (ALK), and induces apoptosis in ALK-positive anaplastic large cell lymphoma cells. Br J Haematol. 2006;135(1):26–32. doi: 10.1111/j.1365-2141.2006.06259.x. [DOI] [PubMed] [Google Scholar]
- 57.Su Y, Li G, Zhang X, et al. JSI-124 Inhibits Glioblastoma Multiforme Cell Proliferation through G2/M Cell Cycle Arrest and Apoptosis Augment. Cancer Biol Ther. 2008;7(8):1243–9. doi: 10.4161/cbt.7.8.6263. [DOI] [PubMed] [Google Scholar]
- 58.Nefedova Y, Nagaraj S, Rosenbauer A, et al. Regulation of dendritic cell differentiation and antitumor immune response in cancer by pharmacologic-selective inhibition of the janus-activated kinase 2/signal transducers and activators of transcription 3 pathway. Cancer Res. 2005;65(20):9525–35. doi: 10.1158/0008-5472.CAN-05-0529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sun J, Blaskovich MA, Jove R, et al. a selective STAT3 activation inhibitor with potent antitumor activity. Oncogene. 2005;24(20):3236–45. doi: 10.1038/sj.onc.1208470. [DOI] [PubMed] [Google Scholar]
- 60.Liu T, Zhang M, Zhang H, et al. Combined antitumor activity of cucurbitacin B and docetaxel in laryngeal cancer. Eur J Pharmacol. 2008;587(13):78–84. doi: 10.1016/j.ejphar.2008.03.032. [DOI] [PubMed] [Google Scholar]
- 61.Tannin-Spitz T, Grossman S, Dovrat S, et al. Growth inhibitory activity of cucurbitacin glucosides isolated from Citrullus colocynthis on human breast cancer cells. Biochem Pharmacol. 2007;73(1):56–67. doi: 10.1016/j.bcp.2006.09.012. [DOI] [PubMed] [Google Scholar]
- 62.Levitzki A. Tyrphostins: tyrosine kinase blockers as novel antiproliferative agents and dissectors of signal transduction. FASEB J. 1992;6(14):3275–82. doi: 10.1096/fasebj.6.14.1426765. [DOI] [PubMed] [Google Scholar]
- 63.Ferrajoli A, Faderl S, Van Q, et al. WP1066 disrupts Janus kinase-2 and induces caspase-dependent apoptosis in acute myelogenous leukemia cells. Cancer Res. 2007;67(23):11291–9. doi: 10.1158/0008-5472.CAN-07-0593. [DOI] [PubMed] [Google Scholar]
- 64.Iwamaru A, Szymanski S, Iwado E, et al. A novel inhibitor of the STAT3 pathway induces apoptosis in malignant glioma cells both in vitro and in vivo. Oncogene. 2007;26(17):2435–44. doi: 10.1038/sj.onc.1210031. [DOI] [PubMed] [Google Scholar]
- 65.Pardanani A, Hood J, Lasho T, et al. TG101209, a small molecule JAK2-selective kinase inhibitor potently inhibits myeloproliferative disorder-associated JAK2V617F and MPLW515L/K mutations. Leukemia. 2007;21(8):1658–68. doi: 10.1038/sj.leu.2404750. [DOI] [PubMed] [Google Scholar]
- 66.Kotha A, Sekharam M, Cilenti L, et al. Resveratrol inhibits Src and Stat3 signaling and induces the apoptosis of malignant cells containing activated Stat3 protein. Mol Cancer Ther. 2006;5(3):621–9. doi: 10.1158/1535-7163.MCT-05-0268. [DOI] [PubMed] [Google Scholar]
- 67.Nam S, Buettner R, Turkson J, et al. Indirubin derivatives inhibit Stat3 signaling and induce apoptosis in human cancer cells. Proc Natl Acad Sci USA. 2005;102(17):5998–6003. doi: 10.1073/pnas.0409467102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lee MJ, Kim MY, Mo JS, et al. Indirubin-3′ -monoxime, a derivative of a Chinese anti-leukemia medicine, inhibits Notch1 signaling. Cancer Lett. 2008;265(2):215–25. doi: 10.1016/j.canlet.2008.02.013. [DOI] [PubMed] [Google Scholar]
- 69.Natarajan C, Bright JJ. Curcumin inhibits experimental allergic encephalomyelitis by blocking IL-12 signaling through Janus kinase-STAT pathway in T lymphocytes. J Immunol. 2002;168(12):6506–13. doi: 10.4049/jimmunol.168.12.6506. [DOI] [PubMed] [Google Scholar]
- 70.Bharti AC, Donato N, Aggarwal BB. Curcumin (diferuloylmethane) inhibits constitutive and IL-6-inducible STAT3 phosphorylation in human multiple myeloma cells. J Immunol. 2003;171(7):3863–71. doi: 10.4049/jimmunol.171.7.3863. [DOI] [PubMed] [Google Scholar]
- 71.Kagialis-Girard S, Mialou V, Chebel A, et al. Inhibition of normal lymphocyte proliferation by Indirubin-3′ -monoxime: a multifactorial process. Leuk Lymphoma. 2007;48(3):605–15. doi: 10.1080/10428190601059696. [DOI] [PubMed] [Google Scholar]
- 72.Gonzales AM, Orlando RA. Curcumin and resveratrol inhibit nuclear factor-kappaB-mediated cytokine expression in adipocytes. Nutr Metab (Lond) 2008;5:17. doi: 10.1186/1743-7075-5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mackenzie GG, Queisser N, Wolfson ML, et al. Curcumin induces cell-arrest and apoptosis in association with the inhibition of constitutively active NF-kappaB and STAT3 pathways in Hodgkin's lymphoma cells. Int J Cancer. 2008;123(1):56–65. doi: 10.1002/ijc.23477. [DOI] [PubMed] [Google Scholar]
- 74.Yu CL, Meyer DJ, Campbell GS, et al. Enhanced DNA-binding activity of a Stat3-related protein in cells transformed by the Src oncoprotein. Science. 1995;269(5220):81–3. doi: 10.1126/science.7541555. [DOI] [PubMed] [Google Scholar]
- 75.Bowman T, Garcia R, Turkson J, Jove R. STATs in oncogenesis. Oncogene. 2000;19(21):2474–88. doi: 10.1038/sj.onc.1203527. [DOI] [PubMed] [Google Scholar]
- 76.Al Zaid Siddiquee K, Turkson J. STAT3 as a target for inducing apoptosis in solid and hematological tumors. Cell Res. 2008;18(2):254–67. doi: 10.1038/cr.2008.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fletcher S, Turkson J, Gunning PT. Molecular approaches towards the inhibition of the signal transducer and activator of transcription 3 (Stat3) protein. Chem Med Chem. 2008;3(8):1159–68. doi: 10.1002/cmdc.200800123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang S, Yang D, Lippman ME. Targeting Bcl-2 and Bcl-XL with nonpeptidic small-molecule antagonists. Semin Oncol. 2003;30(5 Suppl 16):133–42. doi: 10.1053/j.seminoncol.2003.08.015. [DOI] [PubMed] [Google Scholar]
- 79.Mohammad RM, Goustin AS, Aboukameel A, et al. Preclinical studies of TW-37, a new nonpeptidic small-molecule inhibitor of Bcl-2, in diffuse large cell lymphoma xenograft model reveal drug action on both Bcl-2 and Mcl-1. Clin Cancer Res. 2007;13(7):2226–35. doi: 10.1158/1078-0432.CCR-06-1574. [DOI] [PubMed] [Google Scholar]
- 80.Mohammad RM, Wang S, Aboukameel A, et al. Preclinical studies of a nonpeptidic small-molecule inhibitor of Bcl-2 and Bcl-X(L) [(-)-gossypol] against diffuse large cell lymphoma. Mol Cancer Ther. 2005;4(1):13–21. [PubMed] [Google Scholar]