Abstract
Motivation: A challenging problem after a genome-wide association study (GWAS) is to balance the statistical evidence of genotype–phenotype correlation with a priori evidence of biological relevance.
Results: We introduce a method for systematically prioritizing single nucleotide polymorphisms (SNPs) for further study after a GWAS. The method combines evidence across multiple domains including statistical evidence of genotype–phenotype correlation, known pathways in the pathologic development of disease, SNP/gene functional properties, comparative genomics, prior evidence of genetic linkage, and linkage disequilibrium. We apply this method to a GWAS of nicotine dependence, and use simulated data to test it on several commercial SNP microarrays.
Availability: A comprehensive database of biological prioritization scores for all known SNPs is available at http://zork.wustl.edu/gin. This can be used to prioritize nicotine dependence association studies through a straightforward mathematical formula—no special software is necessary.
Contact: ssaccone@wustl.edu
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
Genome-wide association studies (GWAS) are hypothesis-free and are aimed at the discovery of novel variants that influence human disease. However, these often ignore the wealth of biological information available, such as disease-specific biochemical pathways, known functional properties of single nucleotide polymorphisms (SNPs), comparative genomics, prior evidence of genetic linkage, and linkage disequilibrium (LD). We introduce a systematic method for combining information across multiple domains when selecting SNPs for further study after a GWAS. We then implement this method using a combined GWAS (Bierut et al., 2007) and candidate gene (Saccone et al., 2007) study of nicotine dependence, and use simulations to test the method on several commercial microarrays.
A challenging problem after a GWAS is determining how to pursue the many identified and potentially real SNP associations. If there are insufficient resources to test all potential associations, such as all SNPs with P ≤ 0.05, then the SNPs must be prioritized. Even if there are unlimited genotyping resources, or a second full GWAS of all SNPs is possible in the replication sample, the final result will be a number of replications, which must then be prioritized for costly functional studies. This problem is compounded by the fact that each replicated SNP may be in LD with many other potential causal variants. Because these LD proxies will have similar association results, the prioritization scheme cannot rely on statistical evidence of association alone.
Studies will often use biological data to guide the prioritization process. For example, genes in biochemical pathways related to the disease can be given greater weight. Failure to do this systematically, however, can have adverse effects. The biological importance of a gene may be artificially inflated if post hoc rationalization is used instead of establishing a priori biological relevance (Chanock et al., 2007).
We introduce a method of selecting SNPs for further study after a GWAS that gives greater weight to biologically relevant SNPs via an a priori, systematically defined algorithm. We combine the statistical evidence of association with measures of biological relevance to rank SNPs for further study. We do this in such a way that is it clear how each component influences the prioritization process. Our method is designed for maximum interpretability in order to viably incorporate a broad array of genomic annotation and biological information.
2 METHODS
We are interested in the following question: given a SNP, a phenotype, and a biological database, what can we say about the connection between the SNP and the phenotype? We introduce a novel method of assigning each SNP a numeric prioritization score representing the extent of biological relevance.
2.1 Prioritizing SNPs using genomic information networks
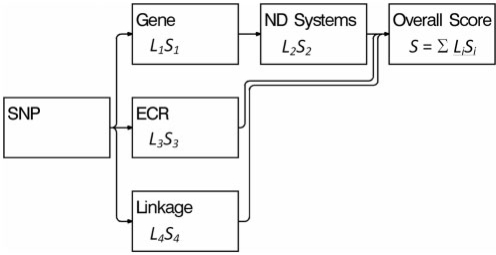
A genomic information network (GIN) is a directed graph, where the nodes correspond to features from a biological database (Fig. 1). The GIN represents a process: it begins with a SNP and ends in the terminal node with the determination of its overall prioritization score S. The overall score is a cumulative measure of biological relevance obtained by combining information across multiple domains. For example, if a SNP is in a gene then it will link to the gene node, which will increase the overall score. If that gene is part of a biological system related to the disease, this will further increase the score. The GIN in Figure 1 has a node labeled ND Systems, which represents gene systems related to nicotine dependence.
Fig. 1.
The model for a genomic information network. The symbols beneath the names of the nodes represent the prioritization score Si and the link index Li.
With the overall score S determined, we rank SNPs from a GWAS for further study by P/10S where S is the overall score and P is the associated P-value. The P-value should strictly be a measure of genotype–phenotype correlation, and should not incorporate biology. We call the term 10S the weight, and P/10S the weighted P-value(Roeder et al., 2006). The scores represent orders of magnitude; a change of 1 in the score corresponds to an order of magnitude change in the weighted P-value. The weighted P-values are not an assessment of genotype-phenotype correlation; their only purpose is to rank SNPs for further study.
Equivalently, if we let m be the mean weight among the SNPs being prioritized, and define the normalized weight to be w=10S/m, then using the normalized weights instead of 10S does not change the rankings. Normalized weights are more interpretable because they give a sense of scale relative to a particular SNP set.
The initial node in the GIN corresponds to a SNP, and connects to the primary nodes. To minimize redundancy, we do not permit links between the primary nodes. For example, the gene node is not allowed to link to the linkage region node. This prevents linkage evidence from being counted twice when a gene resides in a linkage region.
We assign each node that is strictly between the initial and terminal nodes a score Si representing a priori biological relevance. The evidence may depend on the phenotype, such as a known biochemical pathway, or be independent of the phenotype, such as known functional properties of the SNP. Table 1 shows a summary of the scores used for our nicotine dependence GIN. We considered genic SNPs to be an order of magnitude more relevant than non-genic SNPs, and therefore assigned the gene node a score of 1. In general, we use the dbSNP criteria for a SNP to be in a gene, which means within 2 Kb of the 5′ end or 500 bp of the 3′ end of the transcript.
Table 1.
A summary of the scores used to assess the biological relevance of various objects in the genomic information network
| Node | Score |
|---|---|
| Gene | 1.0 |
| ND Systemsa | |
| Tier 1 Genes | 1.5 |
| Tier 2 Genes | 1.0 |
| Tier 3 Genes | 0.5 |
| ECRb | P2/2 |
| Linkagec | 0.5 |
aGene systems related to nicotine dependence.
bEvolutionary conserved region, P=human/mouse conservation percentage.
cGenomic regions with prior evidence of genetic linkage.
If the gene is known to be relevant to nicotine dependence, the gene node links to the ND Systems node. We defined biologically relevant gene systems and categories for nicotine dependence through an expert committee within the NIDA Genetics Consortium (http://zork.wustl.edu/nida). These categories were divided into three tiers (Table S1), with Tier one genes receiving the highest priority for further study. We considered Tier 2 genes, the basic neurotransmitter systems, to be an order of magnitude more relevant than arbitrary genes. Tiers 1 and 3 were then scored a half order higher and lower, respectively, than Tier 2. The primary sources for gene data were KEGG GENES and KEGG BRITE (Kanehisa et al., 2004).
In the node labeled ECR, we prioritized human/mouse standard evolutionary conserved regions (ECRs) from ECRbase (Loots and Ovcharenko, 2007) with a conservative scoring of P2/2, so that the maximum score for the conservation node is a half order of magnitude and drops off rapidly as P decreases. In addition to a priori biological evidence, we also included prior evidence of genetic linkage. In the node labeled Linkage, we prioritized SNPs in a previously identified linkage region for heavy smoking (Saccone et al., 2007) using a conservative half order of magnitude. Future iterations of the method may use a variable prioritization as a function of the LOD score, but this will require a more precise adjustment for multiple testing and corrections for errors between the genetic and physical maps.
We assign each node a link index Li—a non-negative number indicating the strength of the link to the node. The default value of the link index is 1. For example, in the gene node, if the link is weak, such as a SNP being in LD with a gene rather than actually in the gene, then the link index is less than 1. If the link is strong, such as a SNP being a non-synonymous coding change in a gene, then the link index is greater than 1. The link indices scale the scores of the corresponding nodes, so that overall prioritization score has the formula S=∑LiSi.
Table 2 shows a summary of the link indices. For example, we considered a non-synonymous SNP to be twice as relevant as an intronic SNP. Therefore, when a non-synonymous SNP links to a gene, the link index is 2. Other coding variants have a link index of 1.5. Since we did not have detailed information on 3′/5′ promoter regions, we used a conservative link index of 1.25. The locus designation is used by dbSNP to mean that the SNP is within 2.5 Kb of the 5′ or 500 bp of 3′ end of the transcript, but is not actually in the transcript. All SNP annotation was derived from dbSNP build 126 (http://www.ncbi.nlm.nih.gov/projects/SNP).
Table 2.
A summary of the link indices used in the genomic information network. A link index will scale the score of a node depending on the strength of the link to that node. For example, a SNP linking to a gene through LD rather than actually being in the gene is considered a weaker connection, and the score is therefore scaled down proportionally to r2
| Node | Nature of Link | Link Index |
|---|---|---|
| Gene | Coding – nonsynonymous | 2.00 |
| Gene | Coding – synonymous | 1.50 |
| Gene | 3′/5′ promoter | 1.25 |
| Gene | Intron | 1.00 |
| Gene | Locus | 0.75 |
| All | LD | S → S…r2/2 |
2.2 Incorporating LD proxies
A SNP will link to the gene node if it is in the gene, or is in LD with another SNP in the gene. In the latter case we call the second SNP an LD proxy. An LD proxy need not be in the original set of genotyped SNPs. This feature is useful, because if a low-scoring genotyped SNP is in strong LD with a high-scoring non-genotyped SNP, the genotyped SNP will receive a higher score. This prevents us from missing an association signal that is potentially due to a highly biologically relevant but non-genotyped SNP.
We measured LD by estimating r2 in the European American sample from public release 21 of the HapMap Project (http://www.hapmap.org). This was done using the program HaploView (Barrett et al., 2005) for all markers with a minor allele frequency (MAF) greater than or equal to 10% which were within 500 Kb. We considered LD to be a valid link to a node if r2 ≥ 0.5. If the link from a SNP to a node was through an LD proxy, then we scored the node based on the properties of the proxy. However, we reduced the link index by a factor of r2/2 (bottom row of Table 2), because we prefer to have SNPs in a gene rather than in LD with a gene.
We used the following algorithm to determine whether to use a proxy, and to select a particular proxy when there is more than one. We required that if an LD proxy is used, the same proxy SNP must be used for each node. Because the goal is to identify the most biologically relevant representative, we selected from among the original SNP and all potential proxies, the SNP with the maximum overall score. Table S2 shows the details of this process for rs16969968 (see Figure S1 for a graphical view), which was recently reported to be associated with nicotine dependence (Saccone et al., 2007). While many other LD proxies were considered for this SNP, rs16969968 itself was the most biologically relevant.
2.3 Availability
To make our method available for other association studies of nicotine dependence, we computed prioritization scores for all SNPs in dbSNP build 126. The resulting set of ∼11.4 million SNPs is available at http://zork.wustl.edu/gin. Other studies can utilize these data by ranking SNPs by P/10S, where P is the P-value from their study and S is the score from the database.
3 APPLICATION TO GWAS DATA
3.1 Nicotine dependence
We implemented our prioritization method using data from the NicSNP nicotine dependence study. This study used both GWAS (Bierut et al., 2007) and candidate gene (Saccone et al., 2007) strategies with a sample of 1050 nicotine dependent cases and 879 non-dependent smokers, all of European descent. In the GWAS, 2.4 million SNPs were tested for association using pooled genotyping in roughly half of the sample, and the top 40 000 signals were then individually genotyped in the full sample. In the candidate gene study, approximately 4000 SNPs covering 348 genes were individually genotyped.
We computed prioritization scores for the 33 918 SNPs that passed quality control measures from the combined GWAS and candidate gene studies. Table 3 shows the top 10 SNPs ranked by the weighted P-value P/w. No LD proxies were needed for the top 10 signals; each was found to be the most biologically relevant among all potential proxies. Also, no SNPs in the top 10 were in the linkage region used for the GIN.
Table 3.
The top 10 prioritized SNPs from the NicSNP GWAS and candidate gene study ranked by weighted P-value (P/w)
| SNP | Chr | Pos (bp) | Gene | Tier | Function | Mouse Conserva (%) | P-value (P) | N. Weightb (w) | W. P-valuec (P/w) | Rank by P | Rank by P/w |
|---|---|---|---|---|---|---|---|---|---|---|---|
| rs16969968 | 15 | 76 669,980 | CHRNA5 | 1 | Nonsynonymous | 86 | 6.4E−04 | 2.8E+02 | 2.3E−06 | 199 | 1 |
| rs1051730 | 15 | 76 681 394 | CHRNA3 | 1 | Synonymous | 91 | 9.9E−04 | 9.9E+01 | 1.0E−05 | 267 | 2 |
| rs6474413 | 8 | 42 670 221 | CHRNB3 | 1 | Locus | – | 9.4E−05 | 6.8E+00 | 1.4E−05 | 33 | 3 |
| rs578776 | 15 | 76 675 455 | CHRNA3 | 1 | 3′/5′ promoter | – | 3.1E−04 | 2.1E+01 | 1.4E−05 | 123 | 4 |
| rs4142041 | 10 | 68 310 957 | CTNNA3 | – | Intron | – | 5.6E−06 | 3.8E−01 | 1.5E−05 | 2 | 5 |
| rs999 | 6 | 32 261 864 | PBX2 | – | 3′/5′ promoter | – | 1.4E−05 | 6.8E−01 | 2.1E−05 | 3 | 6 |
| rs12623467 | 2 | 51 078 593 | NRXN1 | – | Intron | – | 1.5E−05 | 3.8E−01 | 3.9E−05 | 4 | 7 |
| rs2836823 | 21 | 39 286 525 | – | – | – | – | 1.5E−06 | 3.8E−02 | 4.0E−05 | 1 | 8 |
| rs12380218 | 9 | 79 125 480 | VPS13A | – | Intron | – | 2.1E−05 | 3.8E−01 | 5.5E−05 | 6 | 9 |
| rs2673931 | 5 | 135 717 335 | TRPC7 | – | Intron | 72 | 3.9E−05 | 6.9E−01 | 5.6E−05 | 8 | 10 |
aHuman/Mouse evolutionary conserved region – the percentage of conservation.
bNormalized Weight.
cWeighted P-value.
The top ranked SNP from our prioritization methods was the non-synonymous coding SNP rs16969968 in the nicotinic receptor gene CHRNA5. This was the fifth strongest signal reported in the NicSNP candidate gene study (Saccone et al., 2007), where it was highlighted as the most promising SNP for further study. By original P-value, it ranked 199 out of all 33 918 SNPs from the combined GWAS and candidate gene studies. Table S3 shows the details of how the rankings were determined for the top 10 prioritized SNPs.
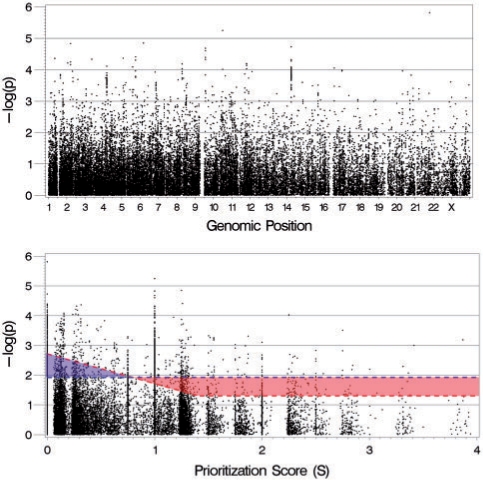
To obtain a broader view of our prioritization scheme and its application to the NicSNP study, we plot the original (non-weighted) P-values against genomic position (Fig. 2, top), and against the overall prioritization score S (Fig. 2, bottom). The latter plot, which we call the P*S plot, introduces a useful way to visualize the GWAS results together with the SNP prioritization scores. The biologically relevant signals are in the upper right-hand region of the P*S plot. The most extreme member of this region is rs16969968, our most highly prioritized SNP. At the other end of the plot, the cluster over S=0 corresponds to SNPs which are not in or near genes, nor in linkage or conserved regions, and are not in LD with r2≥ 0.5 for any such region within 500 Kb.
Fig. 2.
The results of the combined NicSNP GWAS and candidate gene studies. (Top) The original P-values plotted against genomic position. (Bottom) The original P-values plotted against the prioritization score. Signals above the dashed red line correspond to the top 1536 SNPs by weighted P-value with (uncorrected) P ≤ 0.05 (the GIN prioritization method). Signals above the horizontal blue dashed correspond to the top 1536 SNPs by non-weighted P-value (the straight P-value method). The red-shaded knife-shaped and blue-shaded triangular regions show the difference between these two methods.
To select SNPs for replication, we chose the top 1536 (4.5%) SNPs by weighted P-value where P ≤ 0.05 (the number 1536 is used for technical reasons because this reflects the optimal number of SNPs on an Illumina chip in order to reduce genotyping costs). Here, P is the original P-value, and is not corrected for multiple testing. We imposed the condition P ≤ 0.05 to prevent SNPs with little evidence of association from being selected for replication.
The top 1536 SNPs by weighted P-value where (uncorrected) P ≤ 0.05 correspond to signals above the red dashed line in the bottom graph of Figure 2. This dashed line is divided into sloped and horizontal segments intersecting at S=1.4. Because the horizontal segment corresponds to P=0.05, and extends from S=1.4, it follows all SNPs in gene systems relevant to nicotine dependence with P ≤ 0.05 would be selected for further study; the GIN assigns an overall score of at least 1.5 to these SNPs. In the sloped segment, a gradient threshold is used for lower priority genomic regions; the P-value threshold for selection becomes smaller, and therefore stricter, with decreasing biological relevance.
If the top 1536 SNPs were selected using an uncorrected, non-weighted P-value ranking, the condition would be P ≤ 0.012; these SNPs lie above the blue dashed line (Fig. 2, bottom). We refer to this strategy as the ‘straight P-value’ method. If we use our prioritization method over the straight P-value method, the 527 SNPs in the blue shaded triangular region in Figure 2 are traded for the 527 SNPs in red shaded knife-shaped region. The SNPs in the triangular region have a mean P-value of 0.007, which ranges from 0.002 to 0.012, and the maximum prioritization score is 0.79. Therefore none of the SNPs in the triangular region are in genes, because genic SNPs have a score of at least 1. In fact, more than half of them (266/527) have a score of 0, and therefore are not even in LD with r2 ≥ 0.5 for any gene, conserved region or linkage region within 500 Kb.
The red shaded knife-shaped region contains the 527 SNPs we gain using the prioritization method over the straight P-value method. They include the coding SNP rs4953 (P=0.016) in the nicotinic receptor gene CHRNB3, and the non-synonymous SNP rs1805065 (P=0.038) in the neurotransmitter transporter gene SLC6A2. The mean P-value in the knife-shaped region is 0.02, and ranges from 0.012 to 0.05.
The condition P ≤ 0.05 was used in the prioritization process to prevent SNPs with little evidence of association from being selected for replication. In general, we could consider the condition P ≤ T for other thresholds T. Note that the number of SNPs being selected for replication is chosen independently of T. The condition P ≤ T is a filter that is applied to the top SNPs ranked by weighted P-value. In the NicSNP study we selected the top 1536 SNPs ranked by weighted P-value and filtered by the condition P ≤ 0.05.
The choice of threshold T is an important step in the prioritization process. In the bottom of Figure 2, the threshold T corresponds to the lower horizontal boundary of the knife-shaped region. As T approaches the straight P-value threshold of 0.012, the triangular and knife-shaped regions collapse to the straight line P=0.012. In Table S4 we show the effect of changing the threshold T in the NicSNP study. The results are not very sensitive to changes in T, except when T approaches 0.012. As T approaches 0.012, however, the number of SNPs in the triangular and knife-shaped regions becomes small, and therefore has a reduced impact on the set of replication SNPs overall.
The goal of our prioritization method is to ensure that potentially true associations in biologically relevant regions are selected for replication. The use of the threshold T =0.05 is consistent with that goal, as this is a traditional threshold for significance prior to correcting for multiple testing. However, smaller (or larger) thresholds could be used so that a larger (or smaller, respectively) proportion of SNPs with lower prioritization scores are selected (see the fourth column of Table S4), and the exact choice of T depends on the particular goals of a study.
In summary, the prioritization method preferentially selects SNPs with increased biological relevance, where in this case there is on average about a half order of magnitude difference in P-values for these 527 SNPs compared to the straight P-value strategy. In general, the difference between these two strategies will depend on the number of SNPs being selected for follow up, and the distribution of scores among the SNPs being prioritized.
3.2 A sensitivity analysis
To determine the sensitivity of the prioritization results to the scores assigned to the nodes, we scaled the scores of each node by a factor F which varied from 0 to 5 (Fig. S2). Given that the mean prioritization score applied to the NicSNP study was 0.7 ± 0.6 and ranged from 0 to 3.9, this is a substantial range of factors. We limited these tests to the 4528 SNPs where P ≤ 0.05 to be consistent with our SNP selection strategy. We replaced the scores Si of each node with FSi and recomputed the rankings by weighted P-value (this is equivalent to fixing the score Si and scaling the link index Li). We then measured the Pearson correlation coefficient between the rankings before and after scaling. The correlation coefficient is then plotted against the scaling factor F in Figure S2.
In one case we explored the effect when all the nodes are scaled uniformly (the ‘All Nodes’ plot in Fig. S2), so that the scaling is the same for all SNPs. However, we also explored the case where a different scaling factor F was used for different SNPs. The other four plots in Figure S2 correspond to the case where one node at a time is scaled. For example, the curve labeled ‘Linkage Node’ represents the case where only the scores of the Linkage Node are scaled. It is important to test this because not all diseases have been studied with family data, and therefore linkage results may not be available.
When F =1 the rankings are unchanged and the correlation is 1. When testing an individual node, setting F=0 corresponds to dropping that node from the GIN. In the ‘All Nodes’ test, setting F=0 corresponds to the original non-weighted P-values.
We found that the results were not very sensitive to the scores. The rankings were more sensitive when the scores were scaled down, although the correlation did not drop below 0.8 until F was below 0.2. Scaling up had much less of an effect. Even when we scaled up to a very large factor of 5, the correlation stayed above 0.87. The results were similar when, instead of correlation, we looked specifically at how many SNPs remained in the top 1536 weighted rankings after scaling. For example, 76% of these would still have been selected even when F was set to 5 in and we scaled all the nodes uniformly. The conclusion is that moderate adjustments to the scores will not result in substantial changes in the rankings. This is relevant because the scoring system depends on the particular preferences of a study.
3.3 Simulated data on commercial SNP microarrays
To compare the biological prioritization and straight P-value strategies on larger SNP sets, we used 1000 simulations of uniformly distributed P-values on five commercial SNP microarrays: the Affymetrix Genome-Wide Human SNP Array 5.0 and 6.0 (http://www.affymetrix.com), and the Illumina HumanHap 300-Duo, 610-Quad and Human1M (http://www.illumina.com). Using our nicotine dependence GIN, we simulated studies that follow up on 0.01%, 0.1% and 1% of the original number of SNPs, for further study (Table S5). The method of selecting SNPs is the same as that for the NicSNP data, where the top SNPs ranked by the weighted P-values with P ≤ 0.05 are selected.
The performance of our prioritization method is better than the straight P-value approach when the disease variant shows evidence of biological relevance. For example, SNPs in Tier 1 genes for nicotine dependence will be selected at P ≤ 0.05 when following up on at least 0.1% of the SNPs on any of these microarrays (see Table S5). The difference between the two strategies is more pronounced when following up on fewer SNPs. The mean P-values for SNPs selected by biological prioritization over the straight P-value method differed by roughly a half order of magnitude when following up on 1% of the SNPs, and roughly 1.5 orders of magnitude for a follow up of 0.01%.
The results are significantly influenced by the distribution of scores on an array; we report this for the Affymetrix 6.0 and Illumina 1M arrays in Figure S3, which shows the increased genic coverage by Illumina. Also, in Table S6, we show the effect of varying the condition P ≤ T when following up on 0.1% of the SNPs for the Affymetrix and Illumina 1M arrays. Similar to the NicSNP case, the results do not appear to be very sensitive to changes in T, except when T approaches the straight P-value threshold of 0.001, where the GIN prioritization and straight P-value methods rapidly converge. With the condition P ≤ 0.05, the difference in mean P-value between the two methods is about a one order of magnitude, but drops to a half order of magnitude when the condition P ≤ 0.005 is used. Hence, the threshold T can be varied in order to adjust the weight given to biologically relevant SNPs, thereby accommodating the particular goals of a study.
3.4 The most biologically relevant known common SNPs for nicotine dependence
In order to gauge biological relevance in a more global context, we scored all 2.5 million common (MAF ≥ 10%) SNPs relative to the HapMap European American sample. Table 4 shows the top 10 most biologically relevant SNPs ranked by their prioritization scores for nicotine dependence. There are all non-synonymous in Tier 1 genes. Six of these 10 SNPs were genotyped in the NicSNP study. Of these six SNPs, rs16969968 had a P-value of 6.4E—04 and was our top SNP for follow up. The SNP rs2266780 in the nicotine metabolizing gene FMO3 with a P-value of 0.053 may have increased interest because of the biological prioritization.
Table 4.
The top ten nicotine dependence prioritization scores out of all known common SNPs
| SNP | Gene | Mouse Conserv.a (%) | MAF (%) | Overall Score | NicSNP P-value |
|---|---|---|---|---|---|
| rs2266782 | FMO3 | 90 | 38 | 3.90 | 5.1E−01 |
| rs6265 | BDNF | 89 | 18 | 3.90 | 8.6E−01 |
| rs2020862 | FMO2 | 89 | 24 | 3.90 | – |
| rs2307492 | FMO2 | 89 | 15 | 3.90 | 4.2E−01 |
| rs2266780 | FMO3 | 87 | 17 | 3.88 | 5.3E−02 |
| rs3756669 | UGT3A1 | 87 | 10 | 3.88 | – |
| rs16969968 | CHRNA5 | 86 | 42 | 3.87 | 6.4E−04 |
| rs676823 | GPR109A | 82 | 21 | 3.84 | – |
| rs1798192 | GPR109B | 82 | 38 | 3.84 | – |
| rs2454727 | GPR109B | 82 | 34 | 3.84 | – |
aHuman/Mouse evolutionary conserved region – the percentage of conservation
3.5 The impact on findings with unknown biology
An important question is whether known associations in the literature would have been missed using our method because they lack biological relevance. One example is the confirmed type 2 diabetes association of the SNP rs10811661 (Saxena et al., 2007). This SNP is 125 Kb from the nearest gene on chromosome 9p, and is not in LD with any genic SNP within 500 Kb. The prioritization score for rs10811661 is 0.11; it is not 0 because it is in LD with a human/mouse conserved region. The P-value for rs10811661 from the initial GWAS by Saxena and colleagues was 3.6E—05. After initial genotyping on the Affymetrix 500K microarray, this SNP was among the top 107 SNPs (0.02%) ranked by straight P-value; these 107 SNPs were then genotyped in a replication study. To determine if this SNP would have been selected by our prioritization method, still configured for nicotine dependence, we performed 1000 simulations on the comparable Affymetrix 5.0 array.
We found that on average the smallest P-value that would not have been selected by our prioritization method when following up on 0.02% of the SNPs would be 3.2E—05. While this is inconclusive because of the different phenotypes and the fact that the numbers are close, it is possible that rs10811661 would not have been selected as part of the 107 SNPs. However, 0.02% is a relatively small follow-up, and simulations show that increasing this to 0.1% would guarantee that SNPs with P≤ 1.2E—04 (−log(P)=3.9, see the second row of Table S5) would be selected by our method.
4 DISCUSSION
In order to highlight biologically relevant associations from a GWAS, which may otherwise be obscured by the large number of tests, we have developed a method for incorporating a broad array of genomic data into the prioritization of SNPs for further study. In this instance we targeted nicotine dependence, but applications to other phenotypes are straightforward: one need only specify a different set of biologically relevant genes. Because investigators are typically drawn to signals in biologically relevant genomic regions, our method of establishing a priori hypotheses will protect against post hoc reasoning and legitimize the prioritization process (Chanock et al., 2007).
The SNP rs16969968 ranked number one in our prioritization of the NicSNP results (Table 3) and was the seventh most biologically relevant out of all common HapMap SNPs (Table 4). After initially being reported by the NicSNP study (Saccone et al., 2007), there is now extensive evidence from independent datasets that this SNP is associated with nicotine dependence and closely related smoking phenotypes (Berrettini et al., 2008; Bierut et al., 2008; Thorgeirsson et al., 2008) and lung cancer (Amos et al., 2008; Hung et al., 2008). With the exception of Hung and colleagues, who genotyped and reported an association at rs16969968, this SNP was not genotyped in these studies, but the reported associations were with LD proxies for rs16969968. It is very interesting to see convincing, replicated evidence of association for a SNP with such strong biological relevance. However, it is still not clear to what extent known biology will predict variants that influence disease. Our prioritization method is not designed to act as a predictor, but to preferentially select biologically relevant signals when resources are limited, either for genotyping or for functional studies in the laboratory.
In Table 3, six of the top 10 SNPs, as ranked by the prioritization method, are also among the top 10 SNPs as ranked by the original non-weighted P-value. Furthermore, these 10 SNPs would have been selected by the straight P-value method as long as the top 300 SNPs ranked by straight P-value were followed up. Therefore, an important question is how many of these 10 SNPs are true associations. At the time of writing, there are multiple published replications for the NicSNP result for rs16969968 in independent samples, either by directly genotyping this SNP itself, or through strong LD proxies (Berrettini et al., 2008; Bierut et al., 2008; Thorgeirsson et al., 2008). The distinct NicSNP result at rs578776 also shows evidence of published replication through an LD proxy (Berrettini et al., 2008; Thorgeirsson et al., 2008). Also at the time of writing, there have been two other nicotine dependence or smoking quantity GWAS other than NicSNP (Berrettini et al., 2008; Thorgeirsson et al., 2008), but the complete results (P-values) for these studies are not available at this time. However, an important future task will be to determine how true associations fare in the GIN prioritization selection method compared with the straight P-value method. For example, in Figure 2, do the true associations tend to occur in (or above) the knife-shaped region, or in (or above) the triangular region? More generally, it will be interesting to study the distribution of prioritization scores among all known true associations for any disease. This will require the configuration of new GINs for other diseases for which a GWAS has been conducted.
Our GIN prioritization method offers one particular strategy for prioritizing various forms of a priori evidence. Different studies will have different preferences for incorporating this kind of data. Our scoring system is flexible, and can be configured to accommodate a variety of objectives.
Other methods have been proposed for prioritizing SNPs once various parameters, analogous to our prioritization scores, have been established (Chen et al., 2007, Curtis et al., 2007; Lewinger et al., 2007). However, it is unclear how to go from one parameter system to the other, and therefore difficult to compare methodologies. This will be studied in future iterations of the method.
There are many other forms of genomic annotation and biological data that could be incorporated into a GIN. For example, the change in amino acid for the top ranked SNP rs16969968, which changes residue 398 of the protein, from aspartic acid (encoded by the G allele) to asparagine (encoded by A, the risk allele), results in a change in the charge of the amino acid of the α5 subunit (Cserzo et al., 1997). It would be straightforward to adjust the link index of the gene node in order to incorporate this additional data into the GIN. There are many other publicly available resources on SNP functional properties (Jegga et al., 2007; Jiang et al., 2007; Lee and Shatkay, 2007; Wang et al., 2006; Yuan et al., 2006), and tools for nominating and prioritizing genes biologically relevant to a disease (Adie et al., 2006; Gaulton et al., 2007; Masotti et al., 2007).
No matter how many databases we incorporate, our method will always be limited to using known biology. There may be unknown biological mechanisms driving these associations, and these may fail to be discovered if too much emphasis is placed on current biological knowledge. For example, while the gene node in the GIN prioritizes SNPs using the dbSNP criteria of being within 2 Kb of the 5′ end and 500 bp of the 3′ end of a gene, it has been demonstrated that some genes have regulatory regions as far as 8 kb upstream (Blackwood and Kadonaga, 1998). However, the basic premise of this method is to lead with the strongest biological information available while allowing the more significant signals to be included for further study, even if they reside in regions of apparently low biological relevance. We believe this is a practical procedure for resource-limited situations.
Our method does not incorporate information regarding the number of potential associations detected in or near a gene. For example, it is known that even for the single-gene disorder of cystic fibrosis, there are over 500 different mutant alleles (Zielenski and Tsui, 1995). It would be useful to integrate an additional mechanism into the prioritization process that somehow gives additional weight to genes with multiple SNP associations (the number of associations would have to be corrected for LD). This will be studied in future iterations of the GIN prioritization method.
A GWAS cannot viably detect complex interactions between genes due to low statistical power after adjustment for a staggering number of tests. There are now many public databases that provide data on biochemical pathways and metabolic networks (Altman, 2007; Arakawa et al., 2005; Harris et al., 2004; Karp et al., 2005; Mi et al., 2007; Vastrik et al., 2007; von Mering et al., 2007). In future iterations of the method the GIN model will be generalized to prioritize tests of gene–gene interaction, and will incorporate these databases to elucidate the intricate genetic structure of complex disease (Thomas, 2005, 2006a, 2006b).
5 CONCLUSION
We developed a novel method for incorporating a broad array of biological data across multiple domains into the prioritization of SNPs after a GWAS. In this instance we targeted nicotine dependence, but applications to other diseases are straightforward—one need only specify a different, appropriate set of biologically relevant genes. Because investigators are typically drawn to signals in biologically relevant genomic regions, our method of establishing a priori hypotheses will protect against post hoc reasoning. This method will continue to evolve with the growth and expansion of biological databases.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Anne M. Bowcock and Gary D. Stormo of the Department of Genetics at Washington University School of Medicine for their helpful contributions during the preparation of this manuscript. We are also grateful to Weimin Duan for his assistance in managing our local genomic annotation databases. Finally, we thank the anonymous reviewers for their helpful contributions to the manuscript. In memory of Theodore Reich, founding Principal Investigator of COGEND, we are indebted to his leadership in the establishment and nurturing of COGEND and acknowledge with great admiration his seminal scientific contributions to the field.
Funding: This work was supported by American Cancer Society (IRG5801050), the National Cancer Institute (P01CA89392), the National Institute on Drug Abuse (R01DA019963, R56DA12854, K02DA021237, K01DA015129 and N01DA07079), the National Human Genome Research Institute (U01HG004422), the National Institute of Alcohol Abuse and Alcoholism (U10AA008401), and the National Institute of Mental Health (U24MH068457). The NicSNP project is a collaborative research group and part of the NIDA Genetics Consortium, and NicSNP genotyping was performed by Perlegen Sciences under NIDA Contract HHSN271200477471C. Phenotypic and genotypic data for the NicSNP project are stored in the NIDA Center for Genetic Studies (NCGS) at http://zork.wustl.edu/.
Conflicts of Interest: Drs SF Saccone, LJ Bierut, AM Goate and JP Rice are listed as inventors on a patent (US 20070258898) held by Perlegen Sciences, Inc., covering the use of certain SNPs in determining the diagnosis, prognosis and treatment of addiction. Dr NL Saccone is the spouse of Dr SF Saccone, who is listed as an inventor on the aforementioned patent. Dr Bierut has served as a consultant to Pfizer.
REFERENCES
- Adie EA, et al. SUSPECTS: enabling fast and effective prioritization of positional candidates. Bioinformatics. 2006;22:773–774. doi: 10.1093/bioinformatics/btk031. [DOI] [PubMed] [Google Scholar]
- Altman RB. PharmGKB: a logical home for knowledge relating genotype to drug response phenotype. Nat. Genet. 2007;39:426. doi: 10.1038/ng0407-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amos CI, et al. Genome-wide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25.1. Nat. Genet. 2008;40:616–622. doi: 10.1038/ng.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arakawa K, et al. KEGG-based pathway visualization tool for complex omics data. In Silico Biol. 2005;5:419–423. [PubMed] [Google Scholar]
- Barrett JC, et al. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Berrettini W, et al. alpha-5/alpha-3 nicotinic receptor subunit alleles increase risk for heavy smoking. Mol. Psychiatry. 2008;13:368–373. doi: 10.1038/sj.mp.4002154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierut LJ, et al. Novel genes identified in a high-density genome wide association study for nicotine dependence. Hum. Mol. Genet. 2007;16:24–35. doi: 10.1093/hmg/ddl441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierut LJ, et al. Am. J. Psychiatry. 2008. Variants in nicotinic receptors and risk for nicotine dependence. (published online Dec 1, 2007; doi:2010.1176/appi.ajp.2007.01234567) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwood EM, et al. Going the distance: a current view of enhancer action. Science. 1998;281:60–63. doi: 10.1126/science.281.5373.60. [DOI] [PubMed] [Google Scholar]
- Chanock SJ, et al. Replicating genotype–phenotype associations. Nature. 2007;447:655–660. doi: 10.1038/447655a. [DOI] [PubMed] [Google Scholar]
- Chen GK, et al. Enriching the analysis of genomewide association studies with hierarchical modeling. Am. J. Hum. Genet. 2007;81 doi: 10.1086/519794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cserzo M, et al. Prediction of transmembrane alpha-helices in prokaryotic membrane proteins: the dense alignment surface method. Protein Eng. 1997;10:673–676. doi: 10.1093/protein/10.6.673. [DOI] [PubMed] [Google Scholar]
- Curtis D, et al. A pragmatic suggestion for dealing with results for candidate genes obtained from genome wide association studies. BMC Genet. 2007;8:20. doi: 10.1186/1471-2156-8-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaulton KJ, et al. Acomputational system to select candidate genes for complex human traits. Bioinformatics. 2007;23:1132–1140. doi: 10.1093/bioinformatics/btm001. [DOI] [PubMed] [Google Scholar]
- Harris MA, et al. The gene ontology (GO) database and informatics resource. Nucleic Acids Res. 2004;32:D258–D261. doi: 10.1093/nar/gkh036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung RJ, et al. A susceptibility locus for lung cancer maps to nicotinic acetylcholine receptor subunit genes on 15q25. Nature. 2008;452:633–637. doi: 10.1038/nature06885. [DOI] [PubMed] [Google Scholar]
- Jegga AG, et al. PolyDoms: a whole genome database for the identification of non-synonymous coding SNPs with the potential to impact disease. Nucleic Acids Res. 2007;35:D700–D706. doi: 10.1093/nar/gkl826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang R, et al. Sequence-based prioritization of nonsynonymous singlenucleotide polymorphisms for the study of disease mutations. Am. J. Hum. Genet. 2007;81:346–360. doi: 10.1086/519747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, et al. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004;32:D277–D280. doi: 10.1093/nar/gkh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karp PD, et al. Expansion of the BioCyc collection of pathway/genome databases to 160 genomes. Nucleic Acids Res. 2005;33:6083–6089. doi: 10.1093/nar/gki892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee PH, et al. F-SNP: computationally predicted functional SNPs for disease association studies. Nucleic Acids Res. 2007;36:D820–D824. doi: 10.1093/nar/gkm904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewinger JP, et al. Hierarchical Bayes prioritization of marker associations from a genome-wide association scan for further investigation. Genet. Epidemiol. 2007;31:871–882. doi: 10.1002/gepi.20248. [DOI] [PubMed] [Google Scholar]
- Loots G, Orcharenko I. ECRbase: database of evolutionary conserved regions, promoters, and transcription factor binding sites in vertebrate genomes. Bioinformatics. 2007;23:122–124. doi: 10.1093/bioinformatics/btl546. [DOI] [PubMed] [Google Scholar]
- Masotti D, et al. TOM: enhancement and extension of a tool suite for in silico approaches to multigenic complex disorders. Bioinformatics. 2007;24:428–429. doi: 10.1093/bioinformatics/btm588. [DOI] [PubMed] [Google Scholar]
- Mi H, et al. PANTHER version 6: protein sequence and function evolution data with expanded representation of biological pathways. Nucleic Acids Res. 2007;35:D247–D252. doi: 10.1093/nar/gkl869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeder K, et al. Using linkage genome scans to improve power of association in genome scans. Am. J. Hum. Genet. 2006;78:243–252. doi: 10.1086/500026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccone SF, et al. Cholinergic nicotinic receptor genes implicated in a nicotine dependence association study targeting 348 candidate genes with 3713 SNPs. Hum. Mol. Genet. 2007a;16:36–49. doi: 10.1093/hmg/ddl438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccone SF, et al. Genetic linkage to chromosome 22q12 for a heavy-smoking quantitative trait in two independent samples. Am. J. Hum. Genet. 2007b;80:856–866. doi: 10.1086/513703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena R, et al. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316:1331–1336. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- Thomas DC. The need for a systematic approach to complex pathways in molecular epidemiology. Cancer Epidemiol. Biomarkers Prev. 2005;14:557–559. doi: 10.1158/1055-9965.EPI-14-3-EDB. [DOI] [PubMed] [Google Scholar]
- Thomas DC. Are we ready for genome-wide association studies? Cancer Epidemiol. Biomarkers Prev. 2006a;15:595–598. doi: 10.1158/1055-9965.EPI-06-0146. [DOI] [PubMed] [Google Scholar]
- Thomas DC. High-volume “-omics” technologies and the future of molecular epidemiology. Epidemiology. 2006b;17:490–491. doi: 10.1097/01.ede.0000229950.29674.68. [DOI] [PubMed] [Google Scholar]
- Thorgeirsson TE, et al. A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature. 2008;452:638–642. doi: 10.1038/nature06846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vastrik I, et al. Reactome: a knowledge base of biologic pathways and processes. Genome Biol. 2007;8:R39. doi: 10.1186/gb-2007-8-3-r39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Mering C, et al. STRING 7–recent developments in the integration and prediction of protein interactions. Nucleic Acids Res. 2007;35:D358–D362. doi: 10.1093/nar/gkl825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ET, et al. Global landscape of recent inferred Darwinian selection for Homo sapiens. Proc. Natl Acad. Sci. USA. 2006;103:135–140. doi: 10.1073/pnas.0509691102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan HY, et al. FASTSNP: an always up-to-date and extendable service for SNP function analysis and prioritization. Nucleic Acids Res. 2006;34:W635–W641. doi: 10.1093/nar/gkl236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielenski J, et al. Cystic fibrosis: genotypic and phenotypic variations. Annu. Rev. Genet. 1995;29:777–807. doi: 10.1146/annurev.ge.29.120195.004021. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.