Abstract
Drinking water contaminated with arsenic, a human carcinogen, is a worldwide health issue. An understanding of cellular signaling events in response to arsenic exposure and rational designing of strategies to reduce arsenic damages by modulating signaling events are important to fight against arsenic-induced diseases. Previously, we reported that activation of the Nrf2-mediated cellular defense pathway confers protection against toxic effects induced by sodium arsenite [As(III)] or monomethylarsonous acid [MMA(III)]. Paradoxically, arsenic has been reported to induce the Nrf2-dependent signaling pathway. Here, we report the unique mechanism of Nrf2 induction by arsenic. Similar to tert-butylhydroquinone (tBHQ) or sulforaphane (SF), arsenic induced the Nrf2-dependent response through enhancing Nrf2 protein levels by inhibiting Nrf2 ubiquitination and degradation. However, the detailed action of arsenic in Nrf2 induction is different from that of tBHQ or SF. Arsenic markedly enhanced the interaction between Keap1 and Cul3, subunits of the E3 ubiquitin ligase for Nrf2, which led to impaired dynamic assembly/disassembly of the E3 ubiquitin ligase and thus decreased its ligase activity. Furthermore, induction of Nrf2 by arsenic is independent of the previously identified C151 residue in Keap1 that is required for Nrf2 activation by tBHQ or SF. Distinct mechanisms of Nrf2 activation by seemingly harmful and beneficial reagents provide a molecular basis to design Nrf2-activating agents for therapeutic intervention.
Introduction
Human exposure to arsenic is primarily through drinking contaminated water (Tchounwou et al., 1999; Smith et al., 2000; Tchounwou et al., 2003). High doses of arsenic cause acute toxicity while chronic arsenic exposure results in a high incidence of tumors in the skin, lung, kidney, liver, and bladder in populations living in the geographic areas where arsenic concentration is high in drinking water. (Chen et al., 1992; Byrd et al., 1996; Tchounwou et al., 2003; Chen et al., 2004). Numerous studies have been performed in elucidating molecular events associated with arsenic-induced tumor formation or malignant transformation, both in animal and cell culture models. Results from these studies have revealed that arsenic induces global DNA hypomethylation and modulates gene expression profiles, which implicates a non-genotoxic mechanism of carcinogenesis and thus emphasizes the importance of investigating molecular signaling events elicited by arsenic exposure. Aberrant expressed genes can be classified into many categories such as stress response genes, hormone-related genes, cytokines, apoptotic genes, cell cycle regulatory genes, proteolytic genes, and proto-oncogenes (Kitchin, 2001; Yih et al., 2002; Zheng et al., 2003; Chen et al., 2004). In particular, genes important in controlling cell proliferation and transformation are aberrantly expressed during the arsenic-induce carcinogenesis (Shimizu et al., 1998; Chen et al., 2001; Hamadeh et al., 2002; Li et al., 2003; Liu et al., 2004; Benbrahim-Tallaa et al., 2005).
The transcription factor Nrf2 regulates an antioxidant response that defends cells from toxic and carcinogenic effects of environmental pollutants (Kobayashi and Yamamoto, 2006; Zhang, 2006; Kensler et al., 2007). Activity of Nrf2 is tightly regulated by Keap1 at multiple levels: (i) Keap1 is able to sense a disturbance in the cellular redox condition to modulate the Nrf2 signaling pathway accordingly. Several cysteine residues have been proposed to play a role in the sensing mechanism. We have demonstrated that mutation of C151 in Keap1 adequately blocks tBHQ or SF-induced activation of Nrf2 (Zhang and Hannink, 2003; Levonen et al., 2004; Wakabayashi et al., 2004). (ii) Functioning as an E3 ubiquitin ligase, Keap1 constantly targets Nrf2 for ubiquitination and degradation to maintain low constitutive levels of Nrf2 under basal conditions (Cullinan et al., 2004; Kobayashi et al., 2004; Zhang et al., 2004; Furukawa and Xiong, 2005). (iii) Upon induction, E3 ubiquitin ligase activity is inhibited, leading to decreased degradation of Nrf2 and enhanced nuclear translocation of Nrf2 (Zhang et al., 2004; Jain et al., 2005; Zhang, 2006). (iv) At the post induction stage, Keap1 facilitates Nrf2 nuclear export and its association with the cytoplasmic ubiquitination and degradation machinery to turn off the Nrf2 signal (Karapetian et al., 2005; Nguyen et al., 2005; Velichkova and Hasson, 2005; Sun et al., 2007). These multiple control mechanisms rendered by Keap1 ensure prompt removal of hazardous reactive oxygen species to maintain cellular redox homeostasis.
Ubiquitination of Nrf2 is regulated by the Keap1-Cul3 E3 ubiquitin ligase complex in which Keap1 functions as a substrate adaptor (Zhang, 2006). Thus, the activity of the Keap1-containing E3 ubiquitin ligase complex is critical in maintaining the protein level of Nrf2. The E3 ubiquitin ligase complex consists of Keap1, Cul3, Rbx1, and a ubiquitin-charged E2 (Figure 5 of the review article) (Zhang, 2006). The assembly and disassembly of the complex is a tightly regulated event that is controlled by many other protein or protein complexes. It is thought that the reason for the assembly/disassembly of the Cul-containing E3 ubiquitin complex is for many substrate adaptor proteins to share the common Cul-Rbx1 core complex. Proper assembly/disassembly of the Keap1-Cul3 E3 complex is important in controlling ubiquitination of Nrf2. Both increased and decreased association of Keap1 with Cul3, as demonstrated by modulating CAND1 levels with CAND1 si-RNA or CAND1 overexpression, impair the ubiquitination of Nrf2, leading to stabilization of Nrf2 (Lo and Hannink, 2006). In addition, Keap1 mutants that have increased association with Cul3 were shown to have a lower ability to target Nrf2 for ubiquitination (Zhang et al., 2004), further demonstrating the importance of having proper affinity of Keap1 for Cul3 to maintain dynamic assembly/disassembly of the E3 ligase complex.
Inorganic arsenic is the primary form of arsenic in drinking water. It is metabolized in the liver into organic metabolites including MMA(III) that is 20 times more potent than inorganic arsenic in eliciting toxic effects (Bredfeldt et al., 2006). Recently, we have reported that activation of the Nrf2 pathway confers protection against toxic effects induced by both sodium arsenite [As(III)] and monomethylarsonous acid [MMA(III)], demonstrating the feasibility of using Nrf2 activators for intervention of arsenic-induced damage in populations at high risk (Wang et al., 2007). Paradoxically, the Nrf2 pathway is also induced by toxic chemicals that evoke oxidative stress. Arsenic has been demonstrated to activate the Nrf2-dependent response in various cell types (Pi et al., 2003; He et al., 2006; Kimura et al., 2006; Massrieh et al., 2006). Interestingly, low dose arsenic exposure decreases the incidence of cancer in humans (Lamm et al., 2004; Snow et al., 2005). This low-dose protective mechanism of arsenic has been confirmed in experimental models (Pott et al., 1998; Romach et al., 2000; Bae et al., 2002; Mahata et al., 2004; Snow et al., 2005). We also observed an increased proliferation in response to low doses of arsenic exposure (Wang et al., 2007). It is possible that protection reported by exposure to low levels of arsenic is due to activation of the Nrf2-mediated defense response. However, this Nrf2-dependent protection may be overwhelmed at high levels of arsenic exposure, causing arsenic-induced damage. Currently, it is still unclear whether beneficial natural-occurring compounds and harmful oxidative stress activate this pathway in similar manners or through distinct mechanisms.
In the current work, the molecular mechanism of Nrf2 activation by arsenic was investigated. Our results clearly demonstrate that both As(III) and MMA(III) were able to activate Nrf2 by increasing association between Keap1 and Cul3, therefore disrupting the dynamic assembly/disassembly process of the Keap1-Cul3 E3 ubiquitin ligase complex. Reduced E3 ubiquitin ligase activity led to decreased degradation of Nrf2 and activation of the Nrf2 downstream effects. Furthermore, upregulation of Nrf2 by As(III) and MMA(III) was independent of the previously identified cysteine residue C151 in Keap1, which indicates a distinct mechanism by which As(III) and MMA(III) activate Nrf2 compared to other Nrf2 inducers, such as tBHQ and SF.
Materials and methods
Construction of recombinant DNA molecules
Expression plasmids for Keap1-WT, Keap1-C151S, CBD-tagged version of Keap1-WT and Keap1-C151S, HA-Nrf2, HA-Cul3, HA-Cul3-K712R, and Flag-CAND1 have been described (Zhang and Hannink, 2003; Zhang et al., 2004; Lo and Hannink, 2006). The ARE TATA-Inr luciferase reporter plasmid, named mGST-ARE-Luc, was constructed by insertion of a 41 bp ARE sequence from the promoter of the mouse GST-Ya gene into a cloning site of pGL4.22-luc2CP-Puro (Promega).
Chemicals, cell culture, transfection, and induction
Sodium arsenite, tBHQ, and SF were purchased from Sigma. MMA(III) (99% purity) was synthesized in the Synthetic Chemistry Core at the University of Arizona. All chemicals were dissolved in H2O. MDA-MB-231 cells were purchased from ATCC and maintained in Eagle’s minimal Essential medium (MEM) in the presence of 10% fetal bovine serum (FBS). UROtsa cells were generously provided by Drs. Mary Ann and Donald Sens (University of North Dakota). UROtsa cells were grown in DMEM medium enriched with 5% FBS according to the original condition described (Petzoldt et al., 1995). All mammalian cells were incubated at 37° C in a humidified incubator containing 5% CO2. Transfections were performed with Lipofectamine Plus (Invitrogen) according to the manufacturer’s instructions. DNA amounts in each transfection were kept constant by the addition of empty pcDNA3 plasmid.
Reporter gene assay
Plasmids for mGST-ARE-Luc, hRluc/TK (pGL4.74-hRluc-TK, Promega), Nrf2, and Keap1, were cotransfected into MDA-MB-231 cells followed by treatment with Nrf2 inducers for 16 h. Firefly and renilla luciferase activities were measured using the Promega dual-luciferase reporter gene assay system. Firefly luciferase activity was normalized to renilla luciferase activity. All luciferase reporter gene assays were performed three times and the statistical significance of the data was determined by the student t-test and labeled in the figure with asterisks.
Real time RT-PCR
The PCR condition and Taqman probes and primers for Nrf2, NQO1, HO-1, and GAPDH were reported previously (Wang et al., 2007). Briefly, Taqman probes were from the universal probe library (Roche): hNrf2 (#70), hNQO1 (#87), hHO-1 (#25), and hGAPDH (#25). Oligos used for primers were synthesized by IDT.
hNrf2: forward (acacggtccacagctcatc) and reverse (tgtcaatcaaatccatgtcctg);
hNQO1: forward (atgtatgacaaaggacccttcc) and reverse (tcccttgcagagagtacatgg);
hHO-1: forward (aactttcagaagggccaggt) and reverse (ctgggctctccttgttgc);
hGAPDH: forward (ctgacttcaacagcgacacc) and reverse (tgctgtagccaaattcgttgt).
The real-time PCR condition was as follows: one cycle of initial denaturation (95°C for 10 min), 40 cycles of amplification (95° C for 10 sec and 60° C for 20 sec), and a cooling program (50° C for 5 sec). Mean crossing point (Cp) values and standard deviations were determined. Crossing point values were normalized to the respective crossing point values of the hGAPDH reference gene. Data is presented as a fold change in gene expression in treated cells compared to nontreated cells. Triplicate samples were included and the statistical significance of the data was determined by the student t-test.
Antibodies, immunoprecipitation, and immunoblot analysis
Antibodies against Nrf2 (Santa Cruz), Keap1 (Santa Cruz), chitin binding domain (New England Biolabs), α-tubulin (Santa Cruz), and the Myc and HA epitopes (Covance) were purchased from commercial sources. For detection of protein expression in total cell lysates, cells were lysed in sample buffer (50 mM Tris-HCl [pH 6.8], 2% SDS, 10% Glycerol, 100 mM dithiothreitol (DTT), 0.1% bromophenol blue). For experiments with transient transfection, cells were harvested 48 h post-transfection. For immunoprecipitation assays, cells were lysed in RIPA buffer (10 mM sodium phosphate [pH 8.0], 150 mM NaCl, 1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS) containing 1 mM DTT, 1 mM phenylmethylsulfonyl fluoride (PMSF), and protease inhibitor cocktail (Sigma). Cell lysates were pre-cleared with protein A beads and incubated with 2 µg of affinity-purified antibodies for 2 h at 4° C, followed by incubation at 4° C with protein A-agarose beads for 2 h. After washing with RIPA buffer, immunoprecipitated complexes were eluted in sample buffer by boiling for 4 minutes, electrophoresed through SDS-polyacrylamide gels, and subjected to immunoblot analysis. Generally, 50 µg of protein was used for immunoblot analysis and 500 ug of protein was used for immunoprecipitation/immunoblot analysis. The Bradford protein assay (Bio-Rad) was used for measurement of protein concentration. For the measurement of the Nrf2 half-life, UROtsa cells were either left untreated or treated with As(III) or MMA(III) for 4 h. 50 µM cycloheximide was added to block protein synthesis. Total cell lysates were collected at different time points and subjected to immunoblot analysis. The relative intensity of bands was quantified by the ChemiDoc CRS gel documentation system (Bio-Rad) and quantified using Quantity One software.
Ubiquitination of Nrf2
To detect ubiquitinated Nrf2 in vivo, cells were transfected with expression vectors for HA-ubiquitin, Keap1, and Gal4-Neh2. The transfected cells were exposed to 10 µM MG132 (Sigma) for 4 h. Cells were lysed by boiling in a buffer containing 2% SDS, 150 mM NaCl, 10 mM Tris-HCl and 1 mM DTT. This rapid lysis procedure inactivated cellular ubiquitin hydrolases and therefore preserved ubiquitin-conjugates present in cells prior to lysis. Protein-protein interactions, including association of Nrf2 with Keap1, were also disrupted by this lysis procedure. For immunoprecipitation, these lysates were diluted five-fold in buffer lacking SDS and incubated with anti-Gal4 antibodies. Anti-Gal4 immunoprecipitates were analyzed by immunoblot with antibodies directed against the HA epitope.
Results
Both As(III) and MMA(III) enhance the transcriptional activity and the protein level of ectopically expressed Nrf2
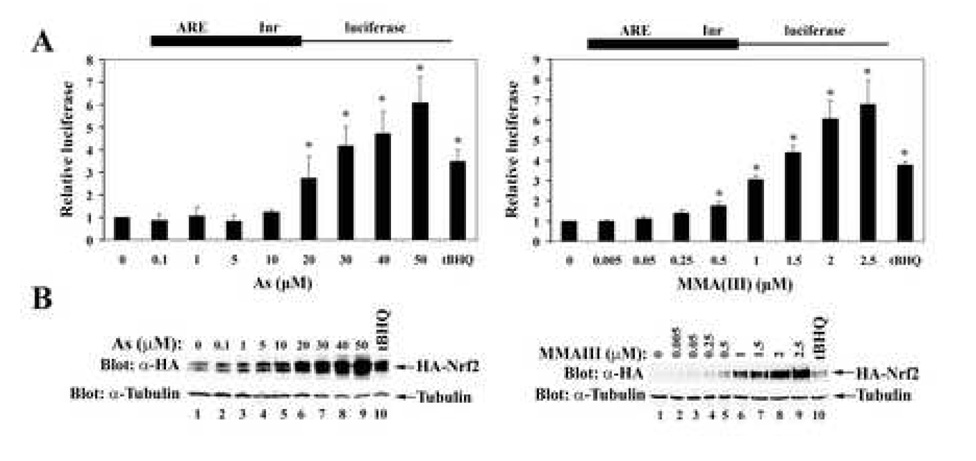
Upregulation of the Nrf2-mediated cellular antioxidant response by As(III) and MMA(III) was determined using an ARE-dependent firefly luciferase reporter gene assay in transient transfected MDA-MB-231 cells following As(III) or MMA(III) treatment. It has been demonstrated that Nrf2 is maximally induced by tBHQ or SF in MDA-MB-231 cells and thus this cell line was used for testing activation of Nrf2 by arsenic. As(III) induced the activity of Nrf2 in a concentration-dependent manner (Figure 1A, left panel) without obvious cell death. Induction was detected at a concentration as low as 20 µM and the fold of induction was significantly higher with 30–50 µM As(III) compared to that with 50 µM tBHQ. Likewise, MMA(III) activated the activity of Nrf2 in a concentration-dependent manner with approximately 10–20 times greater potency compared to As(III) (Figure 1A, right panel). Previous work has demonstrated that tBHQ and SF activate Nrf2 through stabilization of the Nrf2 protein. Therefore, aliquots of lysates from the luciferase reporter gene assay were subjected to immunoblot analysis with an HA antibody for detection of the steady-state levels of Nrf2. Both As(III) and MMA(III) enhanced the steady-state level of Nrf2 in a concentration-dependent manner (Figure 1B, both left and right panels). For immunoblot assays, endogenous α-tubulin was included in all experiments to show equal loading of proteins.
Figure 1.
Both As(III) and MMA(III) enhance the transcriptional activity and the protein level of ectopically expressed Nrf2. (A) The Nrf2-dependent transcriptional activity was determined in MDA-MB-231 cells cotransfected with plasmids containing an ARE-firefly luciferase, TK-renilla luciferase, Nrf2, and Keap1. The transfected cells were exposed to the indicated concentrations of either As(III) (left panel) or MMA(III) (right panel) for 16 h prior to analysis of firefly and renilla luciferase activities. 50 µM tBHQ was included as a positive control. The data represent the mean of three independent experiments with duplicate samples in each experiment. (B) A small aliquot of lysates was used for immunoblot analysis with anti-HA and anti-α-tubulin antibodies for detection of HA-Nrf2 and endogenous α-tubulin.
As(III) and MMA(III) activate the endogenous Nrf2-dependent antioxidant response
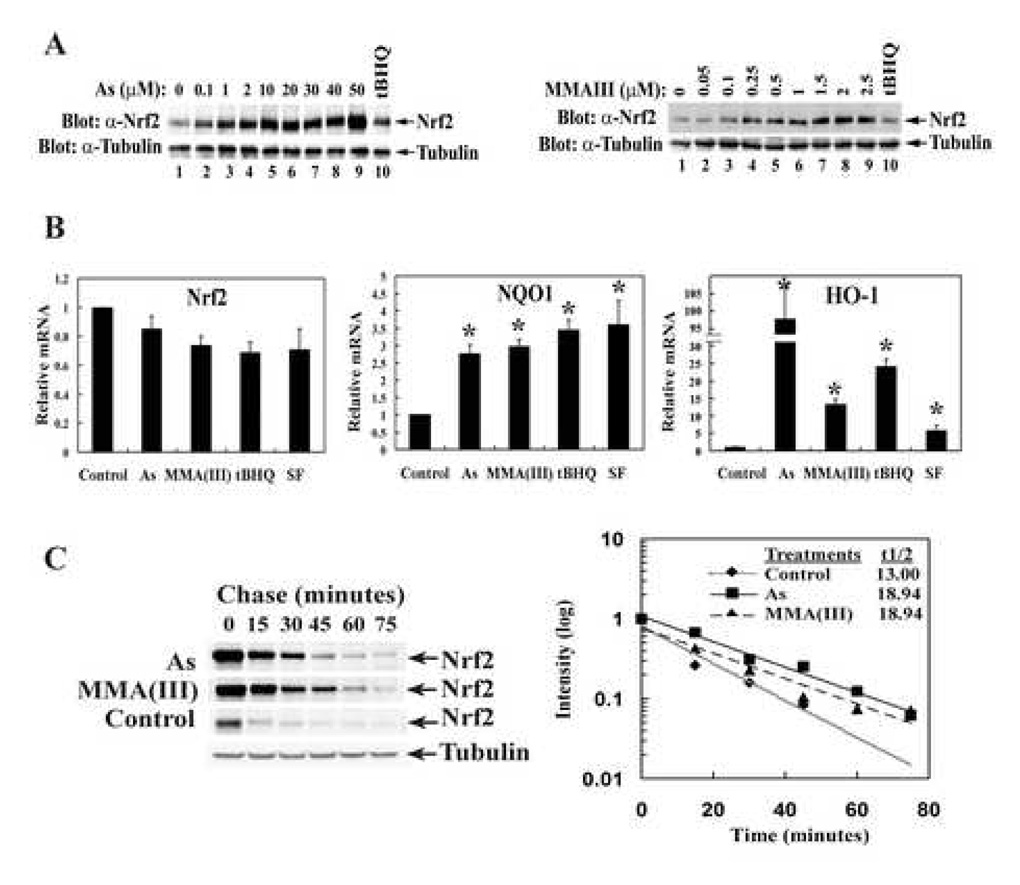
Next, the ability of As(III) and MMA(III) in activating the endogenous Nrf2 signaling pathway was tested in UROtsa cells, since the bladder is one of the target organs for arsenic. UROtsa cells were treated with As(III) or MMA(III), and the endogenous Nrf2 level, as well as mRNA expression of HO-1 and NQO1, two Nrf2 target genes, were determined by immunoblot and real-time RT-PCR analysis. Not surprisingly, levels of the endogenous Nrf2 protein in UROtsa cells were increased in a concentration-dependent manner by As(III) or MMA(III) treatments (Figure 2A, both left and right panels). To understand whether increased levels of the Nrf2 protein in response to As(III) or MMA(III) is a result of upregulation of Nrf2 transcription, real time RT-PCR was performed using mRNA extracted from UROtsa cells treated with H2O, As(III), MMA(III), tBHQ, or SF. Nrf2 mRNA levels remained constant (Figure 2B, Nrf2 panel), indicating that As(III) and MMA(III) may act in a similar way as tBHQ and SF to block degradation of the Nrf2 protein. Next, half-life of the endogenous Nrf2 protein under both untreated and treated conditions were measured by cycloheximide/immunoblot analysis. The half-life of Nrf2 under non-treated conditions was 9 min. However, following As(III) or MMA(III) treatment, the half-life of Nrf2 increased to 19 min (Figure 2C). The transcriptional activation of the Nrf2 downstream target genes were also measured by real-time RT-PCR. The mRNA levels of NQO1 and HO-1 were induced by all treatments and the induction of HO-1 by As(III) was more dramatic (Figure 2B, NQO1 and HO-1 panels).
Figure 2.
As(III) and MMA(III) activate the endogenous Nrf2-dependent antioxidant response. (A) UROtsa cells were treated with the indicated concentrations of As(III) (left panel) or MMA(III) (right panel) for 16 h. The endogenous Nrf2 and α-tubulin protein levels were detected by immunoblot analysis with anti-Nrf2 and anti-α-tubulin antibodies. (B) UROtsa cells were treated for 16 h with 10 µM As(III), 1 µM MMA(III), 50 µM tBHQ, or 7.5 µM SF. The mRNA levels of Nrf2, HO-1, and NQO1, were determined by real-time PCR. (C) UROtsa cells were either left untreated (control) or treated with 20 µM As(III) or 2 µM MMA(III) for 4 h. Cycloheximide (50 µM) was added and cell lysates were collected at the indicated time points and subjected to immunoblot analysis with anti-Nrf2 and anti-α-tubulin antibodies. The relative intensities of the Nrf2 bands were quantified by the ChemiDoc XRS gel documentation system from Bio-Rad and plotted on a semi-log scale. The half-life of Nrf2 in each group was indicated.
As(III) and MMA(III) have no effect on the Nrf2-Keap1 complex, but enhance the Cul3-Keap1 interaction
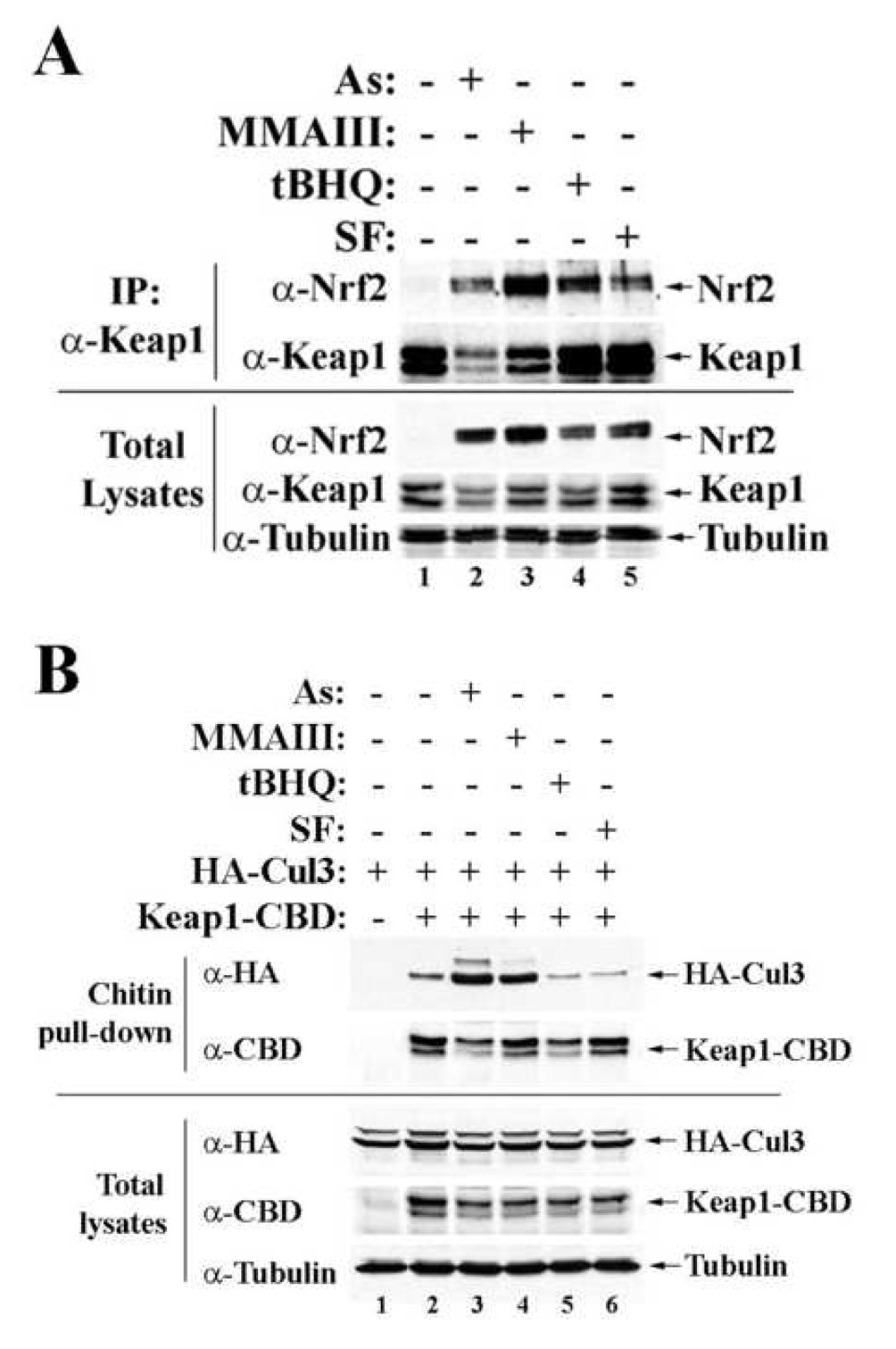
To further understand the action of As(III) and MMA(III) in Nrf2 stabilization, the interactions of Nrf2 with Keap1 and Keap1 with Cul3, were assessed by immunoprecipitation analysis. To assess the Nrf2-Keap1 interaction, the endogenous Nrf2 and Keap1 proteins were measured using lysates from MDA-MB-231 cells treated with the indicated chemicals. There were still significant amounts of Keap1-associated Nrf2 following treatment with As(III), MMA(III), tBHQ, or SF, indicating that none of the chemicals disrupt the Nrf2-Keap1 complex (Figure 3A, top two panels), consistent with our previous findings with tBHQ or SF (Zhang et al., 2004). As expected, the Nrf2 protein level was low in the untreated sample, whereas chemical treatment induced Nrf2 protein levels without significant change in Keap1 levels (Figure 3A, bottom three panels). Assembly/disassembly of Keap1 with Cul3 is important for the ligase function of the Keap1-Cul3 E3 complex and therefore ubiquitination of Nrf2. Next, the ability of these chemicals in modulating interaction of Keap1 with Cul3 was assessed in transfected cells under different treatment conditions. MDA-MB-231 cells were cotransfected with expression vectors for HA-Cul3 and Keap1-CBD. The cell lysates were subjected to a pulldown assay using chitin beads, followed by immunoblot analysis. Consistent with our previous finding, association of Cul3 with Keap1 was reduced slightly following tBHQ or SF treatment (Figure 3B, top α-HA panel, compare lane 2 with lanes 5 and 6). Surprisingly, there was an increase in the affinity of Keap1 for Cul3 in response to As(III) or MMA(III) treatment as more Cul3 proteins were copurified with Keap1 in samples treated with As(III) or MMA(III) (Figure 3B, top α-HA panel, compare lane 2 with lanes 3 and 4). Equal expression of HA-Cul3 or Keap1-CBD was observed when small aliquots of total lysates were analyzed with anti-HA, anti-CBD, and anti-α-tubulin antibodies (Figure 3B, bottom three panels). These results suggest that As(III) and MMA(III) inhibit the activity of the Keap1-Cul3 E3 ubiquitin ligase by interfering with the dynamic assembly/disassembly of Keap1 with the Cul3-containing core complex.
Figure 3.
As(III) and MMA(III) have no effect on the Nrf2-Keap1 complex, but enhance the Cul3-Keap1 interaction. (A) Interaction of endogenous Nrf2 and Keap1 was measured by immunoprecipitation assay in MDA-MB-231 cells treated with H2O, 20 µM As(III), 2 µM MMA(III), 100 µM tBHQ, or 15 µM SF for 4 h. The Keap1 immunoprecipitates were blotted with anti-Nrf2 and anti-Keap1 antibodies (top two panels). Small aliquots of total lysates were subjected to immunoblot analysis with anti-Nrf2, anti-Keap1, and anti-α-tubulin antibodies (Bottom three panels). (B) Keap1-CBD and HA-Cul3 cotransfected MDA-MB-231 cells were treated similarly with the indicated chemicals. The Keap1 complexes were purified using chitin beads and subjected to immunoblot analysis with anti-HA and anti-CBD antibodies for detection of Cul3 and Keap1 (top two panels). Small aliquots of total lysates were immunoblotted with anti-HA for detecting Cul3, anti-CDB for detecting Keap1, and anti-α-tubulin for loading control (bottom three panels).
As(III)- or MMA(III)-induced activation of Nrf2 is independent of C151 in Keap1
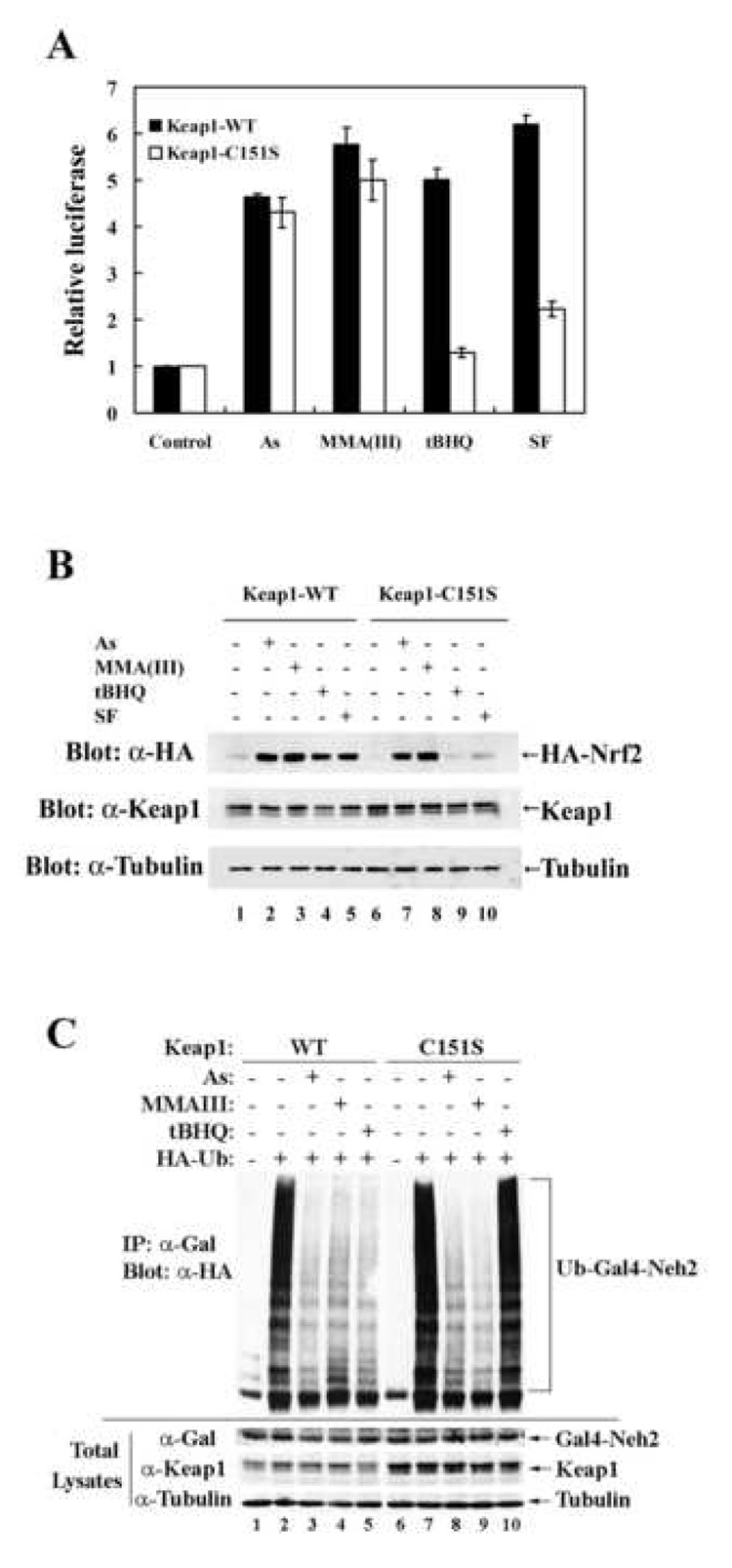
We have reported previously that the cysteine residue (C151) in Keap1 is required for activation of Nrf2 by tBHQ or SF (Zhang and Hannink, 2003). Thus, the necessity of this residue in Nrf2 activation by As(III) and MMA(III) was tested using the ARE-luciferase reporter gene assay system. In the presence of Keap1-WT, Nrf2 activity was substantially enhanced by As(III), MMA(III), tBHQ, and SF (Figure 4A, black bars). Consistent with our previous report, mutation of C151 in Keap1 blocked activation of Nrf2 by tBHQ or SF (Figure 4A, compare 1st white bar with last two white bars). Interestingly, this mutation had no effect on As(III)- or MMA(III)-induced activation of Nrf2 (Figure 4A, 2nd and 3rd white bars). Aliquots of cell lysates were subjected to immunoblot analysis with anti-HA and anti-Keap1 antibodies for detection of Nrf2 and Keap1. In Keap1-WT cotransfected cells, all treatments increased the steady-state levels of Nrf2 and the protein level of Nrf2 correlated well with the luciferase activity (Figure 4B, α-HA panel, lanes 1–5). As expected, induction of the Nrf2 protein by tBHQ or SF was blocked in Keap1-C151S cotransfected cells while this mutation had no effect on Nrf2 induction by As(III) or MMA(III) (Figure 4B, α-HA panel, compare lanes 4 and 5 with lanes 9 and 10; lanes 2 and 3 with lanes 7 and 8).
Figure 4.
As(III)- and MMA(III)-induced activation of Nrf2 is independent of C151 in Keap1. (A) The ARE luciferase reporter gene assay was performed in the same way as described in Figure 1A, except that Keap1-C151S, rather than Keap1-WT, was cotransfected in half of the samples. The transfected cells were treated for 16 h with 10 µM As(III), 1 µM MMA(III), 50 µM tBHQ, or 7.5 µM SF prior to the measurement of firefly and renilla luciferase activities. (B) The same lysates from A were used for immunoblot analysis with anti-HA, anti-Keap1, and anti-α-tubulin antibodies. (C) In vivo ubiquitination assay was performed in MDA-MB-231 cells cotransfected with expression vectors for HA-ubiquitin and Gal4-Neh2, and an expression vector for either Keap1-WT or Keap1-C151S. The transfected cells were treated for 4 h with H2O, 20 µM As(III), 2 µM MMA(III), or 100 µM tBHQ, along with 10 µM MG132. The cells were lysed and proteins were denatured by heating. The Gal4-Neh2 proteins were immunoprecipitated with anti-Nrf2 and immunoblotted with anti-HA for detection of ubiquitin-conjugated Gal4-Neh2 proteins (top panel). Small aliquots of samples were subjected to immunoblot analysis with anti-Gal, anti-Keap1, and anti-α-tubulin antibodies (bottom three panels).
As demonstrated previously, tBHQ and SF stabilize Nrf2 through inhibition of the Keap1-mediated ubiquitination of Nrf2. To further understand activation of Nrf2 by As(III) and MMA(III), in vivo ubiquitination of Nrf2 was carried out in cells cotransfected with expression vectors for Gal4-Neh2 fusion protein, HA-ubiquitin, and either Keap1-WT or Keap1-C151S. Since Neh2 contains seven lysine residues that have been identified as the ubiquitin-conjugating sites of Nrf2, ubiquitination of the Gal4-Neh2 fusion protein was used as an index of Nrf2 ubiquitination (Zhang et al., 2004). In the presence of Keap1-WT, ubiquitination of Gal4-Neh2 was completely blocked by all the treatments (Figure 4C, top panel, compare lane 2 with lanes 3–5). Amazingly, As(III) and MMA(III) were still able to block ubiquitination of Nrf2 while tBHQ lost its ability to inhibit ubiquitination of Gal4-Neh2 when Keap1-C151S was cotransfected (Figure 4C, top panel, lanes 7–10). Equal expression levels of Gal4-Neh2 and Keap1 were shown in the bottom three panels of Figure 4C. Collectively, these data consistently demonstrate that activation of Nrf2, by both As(III) and MMA(III), is independent of C151 in Keap1 and implicate that structurally diverse Nrf2 inducers activate Nrf2 through distinct mechanisms.
Discussion
Both As(III) and MMA(III) strongly induced the Nrf2-mediated antioxidant response in the human bladder urothelium, which is consistent with the reported activation of Nrf2 by As(III) in other cell types (Pi et al., 2003; He et al., 2006; Kimura et al., 2006; Massrieh et al., 2006). Activation of the Nrf2 pathway by arsenic is primary through prolonged half life of Nrf2, resulting in increased steady-state levels of Nrf2. Further investigation of the detailed actions of As(III) and MMA(III) in upregulating the Nrf2 pathway revealed a distinct mechanism of Nrf2 activation by As(III) and MMA(III). As illustrated, As(III) and MMA(III) had no effect on the interaction of Nrf2 with Keap1, which supports our previous conclusion that tBHQ or SF does not dissociate the Keap1-Nrf2 complex (Zhang et al., 2004). Surprisingly, both As(III) and MMA(III) inhibited the Keap1-dependant E3 ubiquitin ligase by augmenting the association of Keap1 with Cul3, which is in sharp contrast with the enhanced dissociation of Keap1 with Cul3 in response to tBHQ and SF. Keap1, functioning as a substrate adaptor for Nrf2, is constantly undergoing assembly/disassembly with the Cul3-Rbx1 core enzyme (Lo and Hannink, 2006). Under the redox balanced conditions, Keap1 brings Nrf2 into the Cul3-Rbx1 E3 ubiquitin ligase complex for ubiquitination and degradation to maintain low constitutive levels of Nrf2 (Cullinan et al., 2004; Kobayashi et al., 2004; Zhang et al., 2004; Furukawa and Xiong, 2005). The cycle of association/dissociation of Keap1 with Cul3 is very important in controlling the levels of Nrf2 and has to be tightly regulated. Any disturbance in the interaction between Keap1 and Cul3 results in compromised assembly of the active E3 ubiquitin ligase, and therefore activation of the Nrf2 pathway. We have shown that mutations in the BTB domain of Keap1 (125A3 or 162A3) increased its affinity for Cul3 and decreased its ability in targeting Nrf2 for ubiquitination and degradation (Zhang et al., 2004). This scenario resembles the currently observed As(III) or MMA(III)-treated condition in which the dynamic cycles of assembly/disassembly are disrupted by the enhanced affinity of Keap1 for Cul3. The stronger association of Keap1 with Cul3 prevents Keap1 from bringing in another Nrf2 molecule into the complex for degradation. In addition, increased association of Keap1 with the core enzyme results in Keap1 selfubiquitination (Zhang et al., 2004). Together, these findings reveal a model by which both increased and decreased affinity of a substrate adaptor for the Cul-containing core complex reduce the activity of Cul-containing E3 ubiquitin ligases. How arsenic enhances the binding of Keap1 to the Cul3-Rbx1 core complex requires further investigation.
In this report, we provide strong evidence that As(III) and MMA(III) are able to activate the Nrf2-mediated antioxidant response in human bladder urothelial cells, indicating that activation of Nrf2 by arsenic is not limited to a particular cell type. More significantly, we provide evidence that As(III) and MMA(III) activate the Nrf2 pathway through a novel mechanism, as demonstrated by the striking finding that induction of Nrf2 by As(III) and MMA(III) is independent of C151 in Keap1 while this cysteine residue is required for activation of Nrf2 by tBHQ and SF. It is likely that tBHQ and SF may directly act on the thiol group of Keap1 by a C151-dependent mechanism, as many electrophilic agents have been demonstrated to directly alkylate C151 in Keap1 in vitro (Eggler et al., 2007; Luo et al., 2007). In contrast, As(III) and MMA(III) may not interact with cysteine residues in Keap1 directly, rather, they affect the cycles of assembly/disassembly of the Keap1-Cul3 E3 ubiquitin ligase complex by enhancing the interaction between Keap1 and Cul3. Currently, the reason for different activating mechanisms by diverse chemicals is not clear. It is possible that structurally diverse chemicals activate different subsets of Nrf2 downstream target genes to elicit specific effects according to the chemical nature of Nrf2 inducers. In conclusion, our results clearly demonstrate the existence of distinct mechanisms in Nrf2 activation by structurally diverse Nrf2-activators. Distinct mechanisms of Nrf2 activation by seemingly harmful and beneficial reagents provide a molecular basis to design Nrf2-activating agents for therapeutic intervention.
Acknowledgments
This study was supported by the NIH grants ES015010-01 and American Cancer Society RSG-07-154-01-CNE.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bae DS, Hanneman WH, Yang RS, Campain JA. Characterization of gene expression changes associated with MNNG, arsenic, or metal mixture treatment in human keratinocytes: application of cDNA microarray technology. Environ Health Perspect. 2002;110 Suppl 6:931–941. doi: 10.1289/ehp.02110s6931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benbrahim-Tallaa L, Waterland RA, Styblo M, Achanzar WE, Webber MM, Waalkes MP. Molecular events associated with arsenic-induced malignant transformation of human prostatic epithelial cells: aberrant genomic DNA methylation and K-ras oncogene activation. Toxicol Appl Pharmacol. 2005;206:288–298. doi: 10.1016/j.taap.2004.11.017. [DOI] [PubMed] [Google Scholar]
- Bredfeldt TG, Jagadish B, Eblin KE, Mash EA, Gandolfi AJ. Monomethylarsonous acid induces transformation of human bladder cells. Toxicol Appl Pharmacol. 2006;216:69–79. doi: 10.1016/j.taap.2006.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrd DM, Roegner ML, Griffiths JC, Lamm SH, Grumski KS, Wilson R, Lai S. Carcinogenic risks of inorganic arsenic in perspective. Int Arch Occup Environ Health. 1996;68:484–494. doi: 10.1007/BF00377874. [DOI] [PubMed] [Google Scholar]
- Chen CJ, Chen CW, Wu MM, Kuo TL. Cancer potential in liver, lung, bladder and kidney due to ingested inorganic arsenic in drinking water. Br J Cancer. 1992;66:888–892. doi: 10.1038/bjc.1992.380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Li S, Liu J, Diwan BA, Barrett JC, Waalkes MP. Chronic inorganic arsenic exposure induces hepatic global and individual gene hypomethylation: implications for arsenic hepatocarcinogenesis. Carcinogenesis. 2004;25:1779–1786. doi: 10.1093/carcin/bgh161. [DOI] [PubMed] [Google Scholar]
- Chen H, Liu J, Zhao CQ, Diwan BA, Merrick BA, Waalkes MP. Association of c-myc overexpression and hyperproliferation with arsenite-induced malignant transformation. Toxicol Appl Pharmacol. 2001;175:260–268. doi: 10.1006/taap.2001.9253. [DOI] [PubMed] [Google Scholar]
- Cullinan SB, Gordan JD, Jin J, Harper JW, Diehl JA. The Keap1-BTB protein is an adaptor that bridges Nrf2 to a Cul3-based E3 ligase: oxidative stress sensing by a Cul3-Keap1 ligase. Mol Cell Biol. 2004;24:8477–8486. doi: 10.1128/MCB.24.19.8477-8486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggler AL, Luo Y, van Breemen RB, Mesecar AD. Identification of the Highly Reactive Cysteine 151 in the Chemopreventive Agent-Sensor Keap1 Protein is Method-Dependent. Chem Res Toxicol. 2007 doi: 10.1021/tx700217c. [DOI] [PubMed] [Google Scholar]
- Furukawa M, Xiong Y. BTB protein Keap1 targets antioxidant transcription factor Nrf2 for ubiquitination by the Cullin 3-Roc1 ligase. Mol Cell Biol. 2005;25:162–171. doi: 10.1128/MCB.25.1.162-171.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamadeh HK, Trouba KJ, Amin RP, Afshari CA, Germolec D. Coordination of altered DNA repair and damage pathways in arsenite-exposed keratinocytes. Toxicol Sci. 2002;69:306–316. doi: 10.1093/toxsci/69.2.306. [DOI] [PubMed] [Google Scholar]
- He X, Chen MG, Lin GX, Ma Q. Arsenic induces NAD(P)H-quinone oxidoreductase I by disrupting the Nrf2 × Keap1 × Cul3 complex and recruiting Nrf2 × Maf to the antioxidant response element enhancer. J Biol Chem. 2006;281:23620–23631. doi: 10.1074/jbc.M604120200. [DOI] [PubMed] [Google Scholar]
- Jain AK, Bloom DA, Jaiswal AK. Nuclear import and export signals in control of Nrf2. J Biol Chem. 2005;280:29158–29168. doi: 10.1074/jbc.M502083200. [DOI] [PubMed] [Google Scholar]
- Karapetian RN, Evstafieva AG, Abaeva IS, Chichkova NV, Filonov GS, Rubtsov YP, Sukhacheva EA, Melnikov SV, Schneider U, Wanker EE, Vartapetian AB. Nuclear oncoprotein prothymosin alpha is a partner of Keap1: implications for expression of oxidative stress-protecting genes. Mol Cell Biol. 2005;25:1089–1099. doi: 10.1128/MCB.25.3.1089-1099.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kensler TW, Wakabayashi N, Biswal S. Cell survival responses to environmental stresses via the Keap1-Nrf2-ARE pathway. Annu Rev Pharmacol Toxicol. 2007;47:89–116. doi: 10.1146/annurev.pharmtox.46.120604.141046. [DOI] [PubMed] [Google Scholar]
- Kimura A, Ishida Y, Hayashi T, Wada T, Yokoyama H, Sugaya T, Mukaida N, Kondo T. Interferon-gamma plays protective roles in sodium arsenite-induced renal injury by up-regulating intrarenal multidrug resistance-associated protein 1 expression. Am J Pathol. 2006;169:1118–1128. doi: 10.2353/ajpath.2006.060024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitchin KT. Recent advances in arsenic carcinogenesis: modes of action, animal model systems, and methylated arsenic metabolites. Toxicol Appl Pharmacol. 2001;172:249–261. doi: 10.1006/taap.2001.9157. [DOI] [PubMed] [Google Scholar]
- Kobayashi A, Kang MI, Okawa H, Ohtsuji M, Zenke Y, Chiba T, Igarashi K, Yamamoto M. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol Cell Biol. 2004;24:7130–7139. doi: 10.1128/MCB.24.16.7130-7139.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi M, Yamamoto M. Nrf2-Keap1 regulation of cellular defense mechanisms against electrophiles and reactive oxygen species. Adv Enzyme Regul. 2006;46:113–140. doi: 10.1016/j.advenzreg.2006.01.007. [DOI] [PubMed] [Google Scholar]
- Lamm SH, Engel A, Kruse MB, Feinleib M, Byrd DM, Lai S, Wilson R. Arsenic in drinking water and bladder cancer mortality in the United States: an analysis based on 133 U.S. counties and 30 years of observation. J Occup Environ Med. 2004;46:298–306. doi: 10.1097/01.jom.0000116801.67556.8f. [DOI] [PubMed] [Google Scholar]
- Levonen AL, Landar A, Ramachandran A, Ceaser EK, Dickinson DA, Zanoni G, Morrow JD, Darley-Usmar VM. Cellular mechanisms of redox cell signalling: role of cysteine modification in controlling antioxidant defences in response to electrophilic lipid oxidation products. Biochem J. 2004;378:373–382. doi: 10.1042/BJ20031049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Gorospe M, Barnes J, Liu Y. Tumor promoter arsenite stimulates histone H3 phosphoacetylation of proto-oncogenes c-fos and c-jun chromatin in human diploid fibroblasts. J Biol Chem. 2003;278:13183–13191. doi: 10.1074/jbc.M300269200. [DOI] [PubMed] [Google Scholar]
- Liu J, Xie Y, Ward JM, Diwan BA, Waalkes MP. Toxicogenomic analysis of aberrant gene expression in liver tumors and nontumorous livers of adult mice exposed in utero to inorganic arsenic. Toxicol Sci. 2004;77:249–257. doi: 10.1093/toxsci/kfh055. [DOI] [PubMed] [Google Scholar]
- Lo SC, Hannink M. CAND1-mediated substrate adaptor recycling is required for efficient repression of Nrf2 by Keap1. Mol Cell Biol. 2006;26:1235–1244. doi: 10.1128/MCB.26.4.1235-1244.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y, Eggler AL, Liu D, Liu G, Mesecar AD, van Breemen RB. Sites of alkylation of human keap1 by natural chemoprevention agents. J Am Soc Mass Spectrom. 2007;18:2226–2232. doi: 10.1016/j.jasms.2007.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahata J, Ghosh P, Sarkar JN, Ray K, Natarajan AT, Giri AK. Effect of sodium arsenite on peripheral lymphocytes in vitro: individual susceptibility among a population exposed to arsenic through the drinking water. Mutagenesis. 2004;19:223–229. doi: 10.1093/mutage/geh022. [DOI] [PubMed] [Google Scholar]
- Massrieh W, Derjuga A, Blank V. Induction of endogenous Nrf2/small maf heterodimers by arsenic-mediated stress in placental choriocarcinoma cells. Antioxid Redox Signal. 2006;8:53–59. doi: 10.1089/ars.2006.8.53. [DOI] [PubMed] [Google Scholar]
- Nguyen T, Sherratt PJ, Nioi P, Yang CS, Pickett CB. Nrf2 controls constitutive and inducible expression of ARE-driven genes through a dynamic pathway involving nucleocytoplasmic shuttling by Keap1. J Biol Chem. 2005;280:32485–32492. doi: 10.1074/jbc.M503074200. [DOI] [PubMed] [Google Scholar]
- Petzoldt JL, Leigh IM, Duffy PG, Sexton C, Masters JR. Immortalisation of human urothelial cells. Urol Res. 1995;23:377–380. doi: 10.1007/BF00698738. [DOI] [PubMed] [Google Scholar]
- Pi J, Qu W, Reece JM, Kumagai Y, Waalkes MP. Transcription factor Nrf2 activation by inorganic arsenic in cultured keratinocytes: involvement of hydrogen peroxide. Exp Cell Res. 2003;290:234–245. doi: 10.1016/s0014-4827(03)00341-0. [DOI] [PubMed] [Google Scholar]
- Pott WA, Benjamin SA, Yang RS. Antagonistic interactions of an arsenic-containing mixture in a multiple organ carcinogenicity bioassay. Cancer Lett. 1998;133:185–190. doi: 10.1016/s0304-3835(98)00229-8. [DOI] [PubMed] [Google Scholar]
- Romach EH, Zhao CQ, Del Razo LM, Cebrian ME, Waalkes MP. Studies on the mechanisms of arsenic-induced self tolerance developed in liver epithelial cells through continuous low-level arsenite exposure. Toxicol Sci. 2000;54:500–508. doi: 10.1093/toxsci/54.2.500. [DOI] [PubMed] [Google Scholar]
- Shimizu M, Hochadel JF, Fulmer BA, Waalkes MP. Effect of glutathione depletion and metallothionein gene expression on arsenic-induced cytotoxicity and c-myc expression in vitro. Toxicol Sci. 1998;45:204–211. doi: 10.1006/toxs.1998.2539. [DOI] [PubMed] [Google Scholar]
- Smith AH, Lingas EO, Rahman M. Contamination of drinking-water by arsenic in Bangladesh: a public health emergency. Bull World Health Organ. 2000;78:1093–1103. [PMC free article] [PubMed] [Google Scholar]
- Snow ET, Sykora P, Durham TR, Klein CB. Arsenic, mode of action at biologically plausible low doses: What are the implications for low dose cancer risk? Toxicol Appl Pharmacol. 2005;207:557–564. doi: 10.1016/j.taap.2005.01.048. [DOI] [PubMed] [Google Scholar]
- Sun Z, Zhang S, Chan JY, Zhang DD. Keap1 controls postinduction repression of the Nrf2-mediated antioxidant response by escorting nuclear export of Nrf2. Mol Cell Biol. 2007;27:6334–6349. doi: 10.1128/MCB.00630-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchounwou PB, Patlolla AK, Centeno JA. Carcinogenic and systemic health effects associated with arsenic exposure--a critical review. Toxicol Pathol. 2003;31:575–588. doi: 10.1080/01926230390242007. [DOI] [PubMed] [Google Scholar]
- Tchounwou PB, Wilson B, Ishaque A. Important considerations in the development of public health advisories for arsenic and arsenic-containing compounds in drinking water. Rev Environ Health. 1999;14:211–229. doi: 10.1515/reveh.1999.14.4.211. [DOI] [PubMed] [Google Scholar]
- Velichkova M, Hasson T. Keap1 regulates the oxidation-sensitive shuttling of Nrf2 into and out of the nucleus via a Crm1-dependent nuclear export mechanism. Mol Cell Biol. 2005;25:4501–4513. doi: 10.1128/MCB.25.11.4501-4513.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakabayashi N, Dinkova-Kostova AT, Holtzclaw WD, Kang MI, Kobayashi A, Yamamoto M, Kensler TW, Talalay P. Protection against electrophile and oxidant stress by induction of the phase 2 response: fate of cysteines of the Keap1 sensor modified by inducers. Proc Natl Acad Sci U S A. 2004;101:2040–2045. doi: 10.1073/pnas.0307301101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XJ, Sun Z, Chen W, Eblin KE, Gandolfi JA, Zhang DD. Nrf2 protects human bladder urothelial cells from arsenite and monomethylarsonous acid toxicity. Toxicol Appl Pharmacol. 2007;225:206–213. doi: 10.1016/j.taap.2007.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yih LH, Peck K, Lee TC. Changes in gene expression profiles of human fibroblasts in response to sodium arsenite treatment. Carcinogenesis. 2002;23:867–876. doi: 10.1093/carcin/23.5.867. [DOI] [PubMed] [Google Scholar]
- Zhang DD. Mechanistic studies of the Nrf2-Keap1 signaling pathway. Drug Metab Rev. 2006;38:769–789. doi: 10.1080/03602530600971974. [DOI] [PubMed] [Google Scholar]
- Zhang DD, Hannink M. Distinct cysteine residues in Keap1 are required for Keap1-dependent ubiquitination of Nrf2 and for stabilization of Nrf2 by chemopreventive agents and oxidative stress. Mol Cell Biol. 2003;23:8137–8151. doi: 10.1128/MCB.23.22.8137-8151.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang DD, Lo SC, Cross JV, Templeton DJ, Hannink M. Keap1 is a redox-regulated substrate adaptor protein for a Cul3-dependent ubiquitin ligase complex. Mol Cell Biol. 2004;24:10941–10953. doi: 10.1128/MCB.24.24.10941-10953.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng XH, Watts GS, Vaught S, Gandolfi AJ. Low-level arsenite induced gene expression in HEK293 cells. Toxicology. 2003;187:39–48. doi: 10.1016/s0300-483x(03)00025-8. [DOI] [PubMed] [Google Scholar]