Abstract
Translesion DNA synthesis (TLS) of damaged DNA templates is catalyzed by specialized DNA polymerases. To probe the cellular TLS mechanism, a host-vector system consisting of mouse fibroblasts and a replicating plasmid bearing a single DNA adduct was developed. This system was used to explore the TLS mechanism of a heptanone-etheno-dC (H-εdC) adduct, an endogenous lesion produced by lipid peroxidation. In wild-type cells, H-εdC almost exclusively directed incorporation of dT and dA. Whereas knockout of the Y family TLS polymerase genes, Polh, Polk, or Poli, did not qualitatively affect these TLS events, inactivation of the Rev3 gene coding for a subunit of polymerase ζ or of the Rev1 gene abolished TLS associated with dA, but not dT, insertion. The analysis of results of the cellular studies and in vitro TLS studies using purified polymerases has revealed that the insertion of dA and dT was catalyzed by different polymerases in cells. While insertion of dT can be catalyzed by polymerase η, κ, and ι, insertion of dA is catalyzed by an unidentified polymerase that cannot catalyze extension from the resulting dA terminus. Therefore, the extension from this terminus requires the activity of polymerase ζ-REV1. These results provide new insight into how cells use different TLS pathways to overcome a synthesis block.
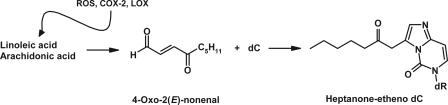
Endogenous reactive chemicals such as reactive oxygen species and certain lipid peroxidation products are thought to contribute significantly to aging, age-related degenerative diseases, and cancer (1–4). Cellular DNA is one of their targets, and replication of un-repaired DNA damage introduces mutations into the genome, thereby contributing to the aforementioned biological effects. Heptanone-etheno-dC (H-εdC)2 (Fig. 1) is one of the substituted etheno-base DNA adducts generated by 4-oxo-2(E)-nonenal, a bifunctional electrophilic lipid peroxidation product derived from both arachidonic acid and linoleic acid (5–8). This adduct, as well as H-εdG, serves as a biomarker of lipid peroxidation-mediated DNA damage (5). H-εdC and H-εdG were both found in the DNA of polyps from a cyclooxygenase-2 up-regulated Min mouse, a colorectal cancer mouse model (9). Our previous study has shown that H-εdC blocks DNA synthesis and highly miscodes in human cells (10), raising the possibility that cyclooxygenase-2-mediated lipid peroxidation contributes to colorectal carcinogenesis in Min mice through the formation of DNA adducts. Due to its strong genotoxicity and physiological significance, H-εdC is an attractive DNA lesion for the mechanistic study of mammalian translesion DNA synthesis (TLS).
FIGURE 1.
Formation and structure of heptanone-etheno-dC (H-εdC). ROS, reactive oxygen species; COX-2, cyclooxygenase-2; and LOX, lipoxygenase.
Recently, it was revealed that various specialized DNA polymerases are actively engaged in DNA synthesis across a DNA lesion (11, 12). The catalytic sites of these polymerases are much more spacious than those of replicative polymerases so that they can accommodate a modified template base and an incoming nucleotide (13–15). Accordingly, their fidelity of DNA synthesis is compromised on undamaged template DNA (16–19). These specialized polymerases overcome the blocking effect of a DNA lesion by sacrificing the fidelity of DNA synthesis. They are widely distributed in various living organisms (20), and hence TLS is considered a general response to un-repaired DNA damage. In mammalian cells, polymerase η, polymerase κ, polymerase ι, and REV1 are found. They are structurally related and form a new family of DNA polymerases named the Y family (20). Polymerase η is known to be important for preventing sunlight-induced skin cancer (21), but the physiological roles of polymerase κ and polymerase ι have not yet been clarified. Among the four Y-family polymerases, REV1 is unique in its ability to act as a 2′-deoxycytidyl transferase (22). Although this transferase activity is suspected to play a role in TLS of abasic residues during somatic hypermutation of immunoglobulin genes (23, 24), it does not appear to be important for TLS of other types of DNA damage. This is because (i) the insertion of dC rarely occurs opposite UV-induced lesions, although REV1 is vital in UV mutagenesis, (ii) a catalytically inactive mutant can still support TLS (25), and (iii) a catalytically active mutant cannot always support UV mutagenesis (26). Although the function of REV1 in TLS has not yet been revealed clearly, it is known to cooperate with polymerase ζ (27). Polymerase ζ (28) is another translesion-specialized polymerase, which belongs to the B family (27), and consists of two subunits of REV3 (catalytic subunit) and REV7 (accessory subunit). This polymerase is thought to play a role in extending a primer opposite a lesion rather than in inserting a nucleotide opposite a lesion (29, 30).
A large number of in vitro TLS studies on various DNA lesions have reported the abilities of specialized polymerases to catalyze TLS with varying efficiencies and fidelities. However, it is not clear whether these activities have any significance to TLS across a given lesion in cellular DNA. One of a few examples known in mammalian cells is the critical role of polymerase η to conduct accurate TLS across UV-induced cis-syn cyclobutane pyrimidine dimers (21). Polymerase κ appears to play a critical role in TLS across benzo[a]pyrene DNA adducts (31, 32).
Here, we report the development of a host-vector system that can be used to study cellular roles of specialized polymerases in TLS across a defined DNA adduct. This system was applied to a mechanistic analysis of TLS across an H-εdC adduct. This study has revealed that two distinct erroneous TLS pathways operate on this adduct in cells: induction of H-εdC → dT transitions is dependent on polymerase ζ and REV1, whereas that of H-εdC → dA transversions is not. Possible mechanisms for these two TLS events are proposed based on the results of the cellular and in vitro TLS experiments.
EXPERIMENTAL PROCEDURES
Construction of a Vector
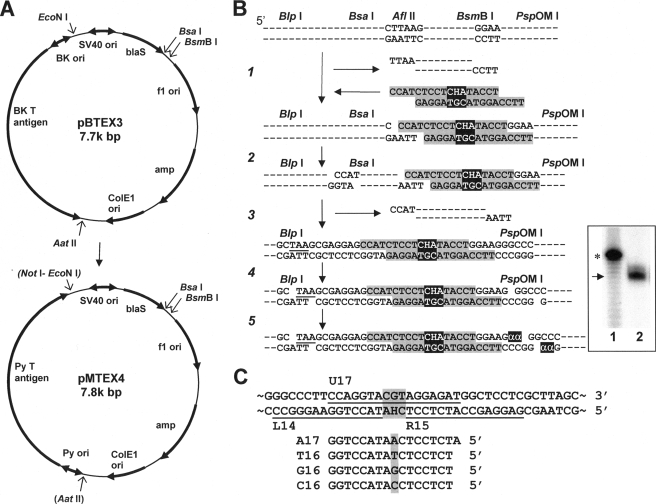
The replicating vector used for experiments conducted in the mouse embryonic fibroblast (MEF) lines was constructed by replacing the replication cassette consisting of the replication origin and the T antigen gene of BK virus in pBTEX3 (33) with a replication cassette from the mouse polyoma (Py) virus (Fig. 2A). To accomplish this replacement, the AatII-NotI fragment (3.3k bp) of pAmp-Py-lac (34) containing the replication cassette was ligated to the AatII-EcoNI fragment (4.5k bp) of pBTEX3. The NotI and EcoNI ends were made into blunt ends using a filling-in reaction catalyzed by the Klenow enzyme, which were then ligated. A BsmBI site overlapping the unique AatII site was destroyed by digesting with AatII followed by trimming of 3′ overhangs by T4 DNA polymerase and subsequent ligation. The new plasmid, pMTEX4, contains blasticidin S- and ampicillin-resistant genes, Py T antigen gene, and four origins (f1, ColE1, SV40, and Py) of replication.
FIGURE 2.
Experimental procedures. A, conversion of pBTEX3 to pMTEX4. amp, ampicillin resistance gene; BK, BK virus; blaS, blasticidin S resistance gene; ori, replication origin; Py, mouse polyoma virus; SV40, simian virus 40; (AatII) and (NotI-EcoNI), these sites were destroyed. B, construction and characterization of modified DNA containing a site-specific H-εdC. “H” represents H-εdC. Three base mismatches (CHA/TGC) are highlighted. “α”is[α-32P]dGMP. Inset: lane 1,40 mer (*) with a ladder of failure oligomers; lane 2, sample. An arrow indicates 35-mer. C, oligonucleotide probes used for a progeny analysis. L14 and R15 probes confirm the presence of oligonucleotide inserts. U17 probe detects progeny derived from an unmodified strand. A17, T16, G16, and C16 probes identify targeted TLS events.
Construction of Site-specifically Modified Plasmid
The synthesis, purification, and characterization of oligonucleotides containing H-εdC has been described previously (10). A modified 17-mer, 5′-CCATCTCCTCHATACCT, where H represents H-εdC, and its complementary 17-mer, 5′-TTCCAGGTACGTAGGAG, were annealed to form 4-nucleotide overhangs on both ends and mismatches on both sides of and opposite the adduct. The strategy for incorporating this duplex oligonucleotide is shown in Fig. 2B. In step 1, pMTEX4 was digested with AflII and BsmBI, and a large fragment was purified using a QIAquick PCR purification kit (Qiagen). The 5′-phosphorylated duplex oligonucleotide was ligated to the digested vector at the BsmBI-cleaved site at 4 °C overnight (step 1) followed by digestion with BsaI (step 2). Following purification of a large fragment, DNA was subjected to self-ligation to form closed circular DNA (step 3). The ligation mixture was re-digested with AflII to remove any residual parental plasmid. Closed circular constructs containing a site-specific, single H-εdC residue were purified by ultracentrifugation in a CsCl-ethidium bromide solution. The amount of modified construct was quantified by a UV spectrophotometer. This resulted in modified DNA, in which the H-εdC adduct was located 19 nucleotides downstream of a stop codon (TAA in Fig. 2B) of the blasticidin S-resistant gene.
To confirm that ligation of oligonucleotides had been accomplished successfully, the purified construct was digested with BlpI and PspOMI (step 4) and then labeled with [α-32P]dGTP and Klenow enzyme (step 5). Labeled fragments were separated in a denaturing 20% polyacrylamide gel and detected by using a Storm PhosphorImager (Amersham Biosciences). This procedure provided a labeled 35-mer (Fig. 2B, inset).
Translesion Experiments in Gene Knockout MEFs
MEFs—The immortalized MEFs used in this study were kind gifts from F. Hanaoka (Osaka University, Polh–/–) (35), H. Ohmori (Kyoto University, Polk–/–) (31), R. Woodgate (National Institutes of Health, Poli–/–) (36), and R. Wood (University of Pittsburg, Rev3–/–) (37). The development of Rev1–/– mice has been described previously (23). Two independent immortalized Rev1–/– MEF lines (SCDE2 and SCDE4) were established from embryos of 14.5 gestation day by spontaneous immortalization.
Introduction of Modified Plasmid into MEFs and Recovery of Progeny Plasmid—Cells were cultured in Dulbecco's modified Eagle's medium supplemented with fetal bovine serum (10%), penicillin (100 units/ml), and streptomycin (100 μg/ml) under 5% CO2 at 37 °C. Cells (1 × 106) were plated in a 25-cm2 flask and cultured overnight, after which they were transfected overnight with 500 ng of a modified construct by the FuGENE6 (Roche Applied Science) method according to the manufacturer's instruction. The next day, cells were detached by treating with trypsin-EDTA, seeded in a 150-cm2 flask, and cultured for 4 days. Progeny plasmids were recovered by the method of Hirt (38) and analyzed for translesional events as described below.
Analysis of Progeny Plasmids for Translesion Events—To recovered plasmids, 5 ng of pVgRXR (Invitrogen), which coded for Zeocin resistance, was added. This plasmid served as an internal control for DpnI digestion. The mixture was treated with DpnI (1 unit) for 1 h to remove nonreplicated input DNA and then used to transform Escherichia coli DH10BMax electrocompetent cells [F–, mcrA, Δ(mrr-hsdRMS-mcrBC), φ80lacZΔM15, ΔlacX74, deoR, recA1, endA1, araΔ139, Δ(ara, leu)7697, galU, galK, λ–, rpsL, nupG, tonA)] (Invitrogen) by an E. coli Pulser (Bio-Rad). Varying portions of a transformation mixture were plated on YT (1×) agar plates with ampicillin (100 μg/ml medium) and blasticidin S (50 μg/ml medium) or with Zeocin (25 μg/ml medium). Because the adduct was located close to the blasticidin S-resistance gene, transformants containing progeny plasmid with large deletions around the adduct site should not grow on a blasticidin-containing plate and were excluded from analysis. A marked reduction in the number of colonies on a Zeocin-containing plate assured efficient digestion of nonreplicated plasmid by DpnI. E. coli transformants were picked individually and subjected to oligonucleotide hybridization as described in detail previously (39). Oligonucleotide probes shown in Fig. 2C were used to determine the DNA sequence at the adduct site. L14 and R15 probes were used to confirm the presence of the oligonucleotide insert and to detect untargeted mutations and small deletions around the adduct site. Plasmids that did not hybridize to both L14 and R15 probes were omitted from a further analysis. U17 detected progeny derived from the unmodified complementary strand. A17, T16, G16, and C16 detected H-εdC → dA, H-εdC → dT, H-εdC → dG, and H-εdC → dC base substitutions, respectively. When plasmids did not hybridize to any of these four probes, DNA sequencing was conducted. More than 30 plasmids among those derived from TLS events were randomly selected for DNA sequencing to confirm the results of hybridization analyses. Thus, this strategy detected all types of events, including base substitutions, frameshifts, deletions, and insertions without any bias.
Complementation of REV1 Deficiency by the Expression of Human REV1 cDNA
A cDNA of the human REV1 (cREV1) gene, obtained from Origene, was cloned into pIRESneo2 (Clontech) using standard molecular biology techniques, including restriction enzyme digestion, PCR, and ligation, which created pIRESneo2(cREV1). The DNA sequence of a region amplified by PCR was verified by sequencing. SCDE4 cells were transfected with this plasmid by the FuGene6 method overnight and then seeded onto a 150-cm2 dish at varying percentages of transfected cells. The antibiotic G418 was added to dishes at 500 μg/ml. When G418-resistant colonies were formed, they were individually recovered by treating with trypsin-EDTA in a cylinder cup. Clonal cells were grown in the presence of G418 and screened for a transcript of the full length of cREV1. One of the clones [Rev1(SCDE4)+cREV1] was used in a complementation assay.
In Vitro TLS Studies Using Purified Polymerases
The 28-mer templates used were 5′-CTGCTCCTC(H/T)ATACCTACACGCTAGAAC, in which H represents H-εdC and T served as a control. The sequence shown in boldface was the same as that used in cellular experiments. The 18-mer primer, 5′-GTTCTAGCGTGTAGGTAT, was used in a nucleotide incorporation assay and a full TLS (incorporation and extension) assay; two 19-mer primers, 5′-GTTCTAGCGTGTAGGTAT(A/T), were used in assays for extension from dA and dT termini, respectively, which were located across from H-εdC. Saccharomyces cerevisiae polymerase ζ, REV1, and proliferating cell nuclear antigen were obtained from Enzymax (Lexington, KY). Sources of the other DNA polymerases have been described previously (40).
In experiments with polymerase η or polymerase κ, a reaction mixture (10 μl) contained 40 mm Tris-HCl (pH 8.0), 30 mm KCl, 5 mm MgCl2, 10 mm dithiothreitol, 250 μg/ml bovine serum albumin, and 40 nm 5′-32P-labeled primer annealed to a template. In experiments with polymerase ι, a reaction mixture (10 μl) contained 40 mm Tris-HCl (pH 8.0), 5 mm MgCl2, 10 mm β-mercaptoethanol, 250 μg/ml bovine serum albumin, 2.5% glycerol, and 40 nm of 5′-32P-labeled primer/template. Experiments with polymerase ζ (10 μl) contained 25 mm potassium phosphate (pH 7.0), 5 mm dithiothreitol, 100 μg/ml bovine serum albumin, 10% glycerol, and 40 nm of 5′-32P-labeled primer/template. The amount of each polymerase and the concentrations of dNTPs are indicated in the figure legends. Reactions were performed at 37 °C for 30 or 60 min and terminated by adding 10 μl of formamide dye mix (90% formamide, 0.01% xylene cyanol, 0.01% bromphenol blue, and 20 mm EDTA). Aliquots (5 μl) were subjected to electrophoresis in a denaturing 20% polyacrylamide gel at 2000 V for 2 h. Gel images were captured by a Storm scanner and analyzed by the ImageQuaNT software package (Amersham Biosciences).
RESULTS
To study the mechanism of mammalian TLS across an endogenous H-εdC adduct, a single H-εdC was site-specifically inserted into a plasmid vector. The modified vector was allowed to replicate in various MEF lines, in which one of the genes for TLS-specialized polymerases had been inactivated by gene targeting. This approach made it possible to evaluate the significance of each specialized polymerase in the TLS events observed in wild-type MEFs. Furthermore, in vitro TLS experiments using purified polymerases were conducted to complement cellular experiments.
DNA Synthesis Block by H-εdC—We placed the lesion in the middle of three consecutive base mismatches. This made it possible to determine the number of progeny plasmid derived from modified and unmodified strands; the ratio of progeny reflects the degree of DNA synthesis blocking. When there is no blocking, the ratio should be 50:50 as revealed with a control construct that had three base mismatches without a lesion (41). DNA repair (removal of a DNA lesion and the two flanking mismatches followed by a gap-filling synthesis) would convert the three nucleotide sequence of the modified strand to the sequence complementary to the unmodified strand, thus losing the strand tag. Because a DNA repair mechanism active on H-εdC is not known, the ratio obtained here in MEFs may not reflect a real blocking effect. However, the determined values are still useful in comparing the relative TLS efficiency among various MEFs. In fact, the detection of progeny derived from the H-εdC-containing strand at a substantial ratio implies that the single H-εdC lesion was not efficiently removed from our construct before TLS took place. The fraction (%) of progeny derived from the modified strand did not markedly decrease in any of gene knockout MEFs when compared with the wild-type MEFs (Table 1). The results indicate that none of the specialized polymerases dominates the entire TLS events.
TABLE 1.
Translesional events in various translesion-specialized DNA polymerase-deficient MEFs
|
Knockout gene
|
No. of progeny derived from
|
Nucleotide inserted opposite H-εdC
|
Other events
|
||||
|---|---|---|---|---|---|---|---|
| UMSa | MSa | T | A | C | G | ||
| None (wild type) | 208 (78)b | 59 (22) | 42 (71) | 13 (22) | 0 | 4 (7) | 0 |
| Polh (Expt 1) | 134 (75) | 44 (25) | 31 (70) | 11 (25) | 1 (2) | 1 (2) | 0 |
| Polh (Expt 2) | 187 (71) | 78 (29) | 47 (60) | 20 (26) | 2 (2) | 9 (12) | 0 |
| Polk | 244 (85) | 43 (15) | 26 (60) | 12 (28) | 3 (7) | 2 (5) | 0 |
| Poli | 140 (75) | 47 (25) | 21 (46) | 20 (43) | 0 | 5 (11) | 1c |
| Rev3 (Pol ζ) | 158 (84) | 29 (16) | 25 (86) | 0 | 0 | 4 (14) | 2d |
| Rev1 (SCDE4) | 191 (84) | 36 (16) | 34 (94) | 0 | 0 | 2 (6) | 0 |
| Rev1 (SCDE2) | 74 (78) | 21 (22) | 20 (95) | 1 (5) | 0 | 0 | 0 |
| Rev1 (SCDE4) + cREV1 | 224 (84) | 44 (16) | 29 (66) | 14 (32) | 0 | 0 | 1e |
UMS, unmodified strand; MS, modified strand.
The numbers in parentheses represent percentages.
TCHATA → TCATTA (H represents H-εdC and an underline indicates a sequence change).
TCHATA → TCATTA and TACTA.
TCHATA → TACTA.
Coding Specificity of H-εdC in MEFs—When L14 and R15 probes were used to confirm the presence of the oligonucleotide inserts, the number of E. coli transformants that did not give positive hybridization signals was small (<4%). These hybridization-negative plasmids most likely contained small deletions or untargeted point mutations in the vicinity of the adduct site. These plasmids were removed from further analysis. All plasmids included in Table 1 were positive in hybridization to both L14 and R15 probes. In wild-type MEFs, the insertion of dT opposite H-εdC was dominant (71%) followed by the insertion of dA (22%) and dG (7%). Thus, the overall miscoding frequency was 93% (Table 1). Similar results were obtained with polymerase η- and polymerase κ-deficient MEFs. In polymerase ι-deficient MEFs, the frequency (43%) of dA insertion was somewhat higher than in wild-type, polymerase η-, and polymerase κ-deficient MEFs, reaching close to the level (46%) of dT insertion. This suggests that polymerase ι plays a more important role than do polymerase η and polymerase κ in dT insertion. However, these results, together with the effects on TLS efficiencies (previous section), suggest that polymerase ι, polymerase η, and polymerase κ individually are not essential for TLS across the H-εdC lesion. In contrast, the coding events in MEFs deficient in REV1 or REV3 (polymerase ζ) were very different from those in the other MEFs. In these two MEF lines, TLS events accompanied by the insertion of dA opposite H-εdC disappeared, and most translesion events were associated with dT insertion (Table 1). The same phenomena were observed in two independent REV1-deficient cell lines, SCDE2 and SCDE4. TLS accompanied by dA insertion was restored by expressing hREV1 in SCDE4 (Table 1). These results indicate that REV1 and polymerase ζ play a critical role in the TLS associated with dA, but not dT, insertion, pointing out that these two TLS events are conducted by different mechanisms.
TLS Studies with Purified Polymerases—The above cellular studies have revealed a vital role of REV1 and polymerase ζ (REV3) in TLS associated with the insertion of dA opposite H-εdC. This result clearly indicates that the 2′-deoxycytidyl transferase activity of REV1 does not play any role and that REV1 exerts an unknown second function during this TLS event. Regarding the role of polymerase ζ, two possibilities are envisioned. First, polymerase ζ might catalyze the insertion of dA opposite H-εdC, and second, the polymerase might be essential to the extension from a dA terminus that is generated by another polymerase. Since mammalian polymerase ζ has not yet been purified, yeast polymerase ζ was employed to address these possibilities.
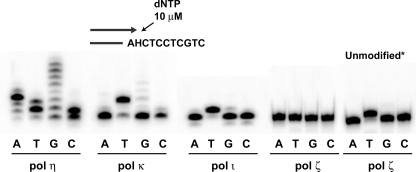
We examined in vitro the ability of polymerase ζ, as well as human polymerase η, polymerase κ, and polymerase ι, to insert a nucleotide opposite H-εdC. H-εdC was inserted into the same sequence context as that used in the cellular study. As shown in Fig. 3, polymerase ζ did not incorporate any of four nucleotides opposite H-εdC when 1 μl (71 ng, 352 fmol) of the original enzyme solution, as well as diluted solutions (data not shown), was used. The addition of yeast proliferating cell nuclear antigen (214 nm) to a reaction mixture had no effect on the nucleotide incorporation (data not shown). No incorporation of a nucleotide by polymerase ζ opposite this adduct was observed at dNTP concentrations of 10, 50, 100, 500, or 1000μm (data not shown). These results suggest that polymerase ζ cannot start DNA synthesis across from H-εdC. This is consistent with the generally accepted concept that polymerase ζ is poor at inserting a nucleotide opposite a lesion in DNA (29, 30). On the other hand, polymerase κ and polymerase ι inserted mostly dT, which was the major coding event in MEFs. Remarkably, polymerase η inserted any of four nucleotides and extended this insertion product with multiple dA or dG homopolymeric stretches.
FIGURE 3.
Nucleotide insertion opposite H-εdC by human Y-family polymerases and yeast polymerase ζ. Polymerase η (18.2 nm), polymerase κ (27.9 nm), polymerase ι (2.4 nm), and polymerase ζ (35.2 nm) were used in a 10-μl reaction mixture (refer to text). Reaction was conducted at 37 °C for 30 min. A half of reaction mixture was run in a denaturing 20% polyacrylamide gel. Unmodified*, represents a template with dA at the position of H-εdC.
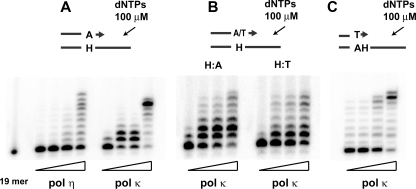
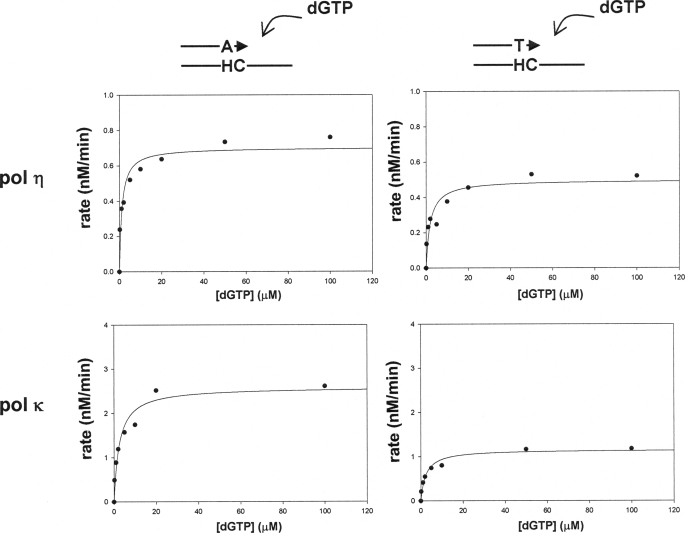
Because polymerase ζ cannot insert any nucleotide opposite H-εdC, this polymerase may specifically be involved in catalyzing the extension from a dA terminus generated by another polymerase. A current model (30) predicts that polymerase ζ-REV1 is recruited to a stalled site to take over a role in extension when an inserter polymerase cannot perform extension following nucleotide insertion. Given that the insertion of dA and dT opposite H-εdC is catalyzed by one polymerase, it is reasonable to assume that the dA terminus is more resistant to extension than is the dT terminus; consequently, the extension from the dA terminus requires polymerase ζ-REV1. We addressed this question by comparing the extension from the dT and dA termini, using pol η and pol κ, because they could catalyze extension from these termini in vitro (Fig. 4, A and B). A qualitative gel analysis with polymerase κ unexpectedly showed that extension from the dA terminus was more efficient than that from the dT terminus (Fig. 4B). To confirm this result, kinetic parameters of the extension reaction were determined using polymerase κ and polymerase η (Fig. 5 and Table 2). The extension, as determined by Keff, was twice as efficient with the dA terminus when compared with the dT terminus in experiments using polymerase κ. This resulted mainly from the difference in Kcat values. Extension from the dA terminus was also more facile when polymerase η was used. In this case, Kcat and Km values both contributed to cause a 2-fold difference. Thus, we did not obtain any evidence indicating that the dA terminus was more resistant to extension by these polymerases than was the dT terminus. Therefore, the requirement of the dA terminus for polymerase ζ-REV1 does not seem to be due to a greater blocking effect on the subsequent extension compared with the dT terminus. This result suggests that the insertion of dA and dT is not catalyzed by the same polymerase. Rather, the two insertion events are catalyzed by two different polymerases: one polymerase (such as polymerase κ and polymerase ι) that inserts dT and can catalyze subsequent extension from this dT terminus to complete TLS, and another polymerase that inserts dA, but cannot catalyze the extension, and hence is replaced by polymerase ζ-REV1. Indeed, polymerase κ appears to catalyze a complete TLS (Fig. 4C) with dT insertion (Fig. 3).
FIGURE 4.
Translesion syntheses catalyzed by polymerase η or polymerase κ, starting from a dA terminus opposite H-εdC (A),adAandadT terminus pairing to template H-εdC (B), and a dT terminus one nucleotide 3′ to H-εdC (C). Amounts of polymerases in a 10-μl reaction mixture were 0.91, 2.73, 3.64, and 18.2 nm of polymerase η in A; 0.78, 2.33, 3.1, and 15.5 nm of polymerase κ in A and C; and 0.78, 2.33, 3.1, and 4.65 nm of polymerase κ in B. Reaction was conducted at 37 °C for 30 min.
FIGURE 5.
Steady-state kinetics of extension from a dA and a dT terminus opposite H-εdC catalyzed by polymerase η and polymerase κ.
TABLE 2.
Extension catalyzed by polymerase κ and polymerase η from dA and dT termini pairing to H-εdC template
| Enzyme | Terminus | Km | Vmax | Kcat | Keff (Kcat/Km) |
|---|---|---|---|---|---|
| μm | nm/min | min-1 | μm/min | ||
| Polymerase κ | A | 2.5 ± 0.71 | 2.6 ± 0.18 | 0.59 | 0.24 |
| T | 2.3 ± 0.55 | 1.2 ± 0.06 | 0.26 | 0.11 | |
| Polymerase η | A | 1.2 ± 0.36 | 0.70 ± 0.04 | 1.9 | 1.7 |
| T | 1.9 ± 0.77 | 0.50 ± 0.04 | 1.4 | 0.73 |
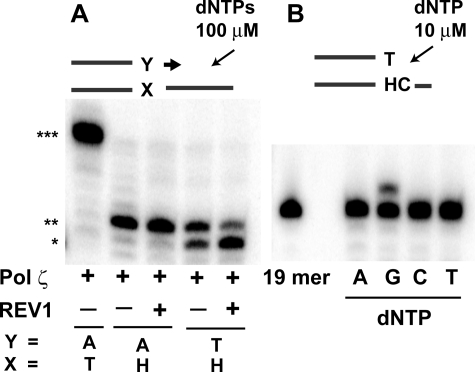
To obtain support for this concept, we studied the extension from the dA and dT termini, using yeast polymerase ζ and yeast REV1 (Fig. 6). In this experiment, we observed that majority of extension terminated following the incorporation of one nucleotide (Fig. 6A), which was the correct dG (Fig. 6B), opposite the next template dC. Very little full extension of the two primers to the end of the 28-mer template was observed. Because REV1 was reported to enhance the catalytic activity of polymerase ζ (42), we also conducted the reaction in the presence of REV1. However, there was no enhancing effect on the extension. Further addition of proliferating cell nuclear antigen (214 nm) to the reaction mixture did not result in any enhancing effect. Again, this one nucleotide extension was found to be more efficient with a dA terminus than with a dT terminus, as was observed in the experiments using polymerase κ and polymerase η.
FIGURE 6.
Translesion synthesis catalyzed by yeast polymerase ζ. A, annealed template (28-mer)-primer (19-mer) substrate (40 nm) was incubated with polymerase ζ (35.2 nm) in the presence (+) or absence (–) of REV1 (35.7 nm) in a 10-μl reaction mixture at 37 °C for 60 min. Asterisks indicate 19-mer (*), 20-mer (**), and 28-mer (***). B, nucleotide insertion opposite template dC located 5′ to H-εdC. Reaction was conducted at 37 °C for 60 min in the presence of polymerase ζ (35.2 nm).
DISCUSSION
To study the TLS mechanism in mammalian cells, we have reported here the development of an MEF-replicating vector system, which is complemented with in vitro TLS experiments to fortify findings in cells and to help to deduce a cellular mechanism. Because a DNA polymerase involved in TLS likely varies depending on lesions to be bypassed, our approach using various gene knockout MEFs will be useful in probing a TLS mechanism of each DNA adduct.
Here, we have applied it to a TLS study of H-εdC. This adduct blocks DNA synthesis and highly miscodes by directing mainly the incorporation of dT and dA in mammalian cells (10, and this study). Our study has revealed that TLS associated with dA, but not dT, insertion requires the functions of REV1 and polymerase ζ (Table 1). This indicates that two distinct TLS mechanisms operate on the H-εdC adduct in MEFs. What are these two pathways, and how are they chosen? Two scenarios are envisioned at the step of nucleotide insertion opposite H-εdC: the insertion of dA and dT is catalyzed by the same polymerase or by different polymerases.
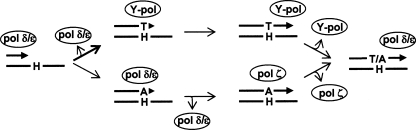
To address these issues, we conducted in vitro primer extension studies using purified polymerases. Although the activity of a polymerase is influenced, modified, and/or regulated by various factors in cells, the in vitro experiments may still be informative and help to deduce the cellular TLS mechanism. The in vitro primer extension studies have revealed that dA and dT are inserted opposite the adduct by pol η and by pol η, pol κ, and pol ι, respectively (Fig. 3). pol ζ did not insert any nucleotide opposite the H-εdC adduct. This result raises the possibility that pol η is responsible for the TLS with dA insertion. However, the inactivation of the Polh gene did not impair the frequency of dA insertion (Table 1). Therefore, the dA insertion is most likely catalyzed by a polymerase other than those from the Y-family. Considering that the Y-family TLS polymerases are very likely recruited first when a replicative polymerase is blocked by a DNA adduct, the polymerase that catalyzes this insertion could be a replicative polymerase (lower minor pathway of Fig. 7). However, the polymerase cannot extend a primer from this dA terminus and so it disengages from TLS. Under this situation, pol ζ-REV1, which is very adept at elongating DNA from a terminus opposite a DNA lesion and a mismatch (25, 29, 30), is recruited to the dA terminus to complete TLS. However, this TLS is a minor event when a replicative polymerase encounters H-εdC. A replicative polymerase largely disengages from DNA synthesis without inserting a nucleotide (dA) (upper major pathway). Upon this blockade, the Y-family polymerases (pol η, pol κ, and pol ι) are recruited to the site to catalyze nucleotide insertion. These three Y-family polymerases insert dT, which is almost exclusive with pol κ and pol ι (Fig. 3). Following the insertion of dT, possibly the same polymerase will catalyze extension from this dT terminus to complete TLS (Figs. 4 and 5). Inactivation of any one of these three polymerases caused no remarkable effect on the frequency of dT insertion (Table 1), which suggests that they complement this task each other. pol ι appears to contribute to this task more significantly than do the other two polymerases, because the frequency of dT insertion is lower in pol ι-deficient MEFs (Table 1). The possible contribution of the three polymerases to this TLS event will be clarified in future experiments using double and triple gene knockout MEFs.
FIGURE 7.
TLS model for H-εdC (H) adduct. Refer to text for explanation.
The idea that the incorporation of dA and dT is catalyzed by different polymerases is also supported by qualitative (Fig. 4) and quantitative (kinetic) (Fig. 5 and Table 2) in vitro studies. If the insertion is catalyzed by the same polymerase, extension from a dA:H-εdC pair should be more difficult than from a dT:H-εdC pair and hence the specialized extension ability of pol ζ-REV1 would be required to complete TLS. The experiments using pol η and pol κ have shown that extension from a dA:H-εdC pair is actually more facile than from a dT:H-εdC pair (Figs. 4 and 5 and Table 2). These findings suggest that the two incorporations are catalyzed by different polymerases.
Our results have also shown that polymerase κ and polymerase η cannot substitute for polymerase ζ-REV1 in the task of extension from a dA:H-εdC pair in cells (Table 1), although they can efficiently catalyze the extension in vitro (Fig. 4). This implies the existence of a cellular mechanism for the exclusive selection of pol ζ-REV1 for extension from a dA:H-εdC pair.
Although a complete understanding of the mechanism of TLS across the H-εdC adduct needs further studies, we have clearly shown the existence of two distinct pathways acting upon the same DNA adduct. Our results illustrate how cells can use different TLS pathways to overcome a synthesis block. This new strategy, in conjunction with the use of single and multiple gene knockout MEFs, will be informative for elucidating the mammalian TLS mechanisms operating on various DNA adducts.
Acknowledgments
We thank the following people for providing gene knockout MEFs, DNA polymerases, plasmid and technical advice: F. Hanaoka (Osaka University, polymerase η and MEF), H. Ohmori (Kyoto University, polymerase κ and MEF), R. Woodgate (NIH, polymerase ι and MEF), R. Wood (University of Pittsburg, REV3 MEF), M. Bemark (Medical Research Council, REV3 MEF), Y. Matsumoto (Fox Chase), M. Gassmann (Leiden University, mouse polyoma virus replication cassette), and V. Serenkov (SUNY at Stony Brook, advice in enzyme kinetic study).
This work was supported, in whole or in part, by National Institutes of Health Grants CA76163, CA47995, CA91016, ES11297, and ES013508. This work was also supported by Grant EU-IP FP6-51211. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: H-εdC, heptanone-etheno-dC; cREV1, cDNA of the human REV1 gene; MEFs, mouse embryonic fibroblasts; Py, polyoma; TLS, translesion DNA synthesis; pol, polymerase.
References
- 1.Davidson, J. F., Guo, H. H., and Loeb, L. A. (2002) Mutat. Res. 509 17–21 [DOI] [PubMed] [Google Scholar]
- 2.Klaunig, J. E., and Kamendulis, L. M. (2004) Annu. Rev. Pharmacol. Toxicol. 44 239–267 [DOI] [PubMed] [Google Scholar]
- 3.Kujoth, G. C., Hiona, A., Pugh, T. D., Someya, S., Panzer, K., Wohlgemuth, S. E., Hofer, T., Seo, A. Y., Sullivan, R., Jobling, W. A., Morrow, J. D., Van Remmen, H., Sedivy, J. M., Yamasoba, T., Tanokura, M., Weindruch, R., Leeuwenburgh, C., and Prolla, T. A. (2005) Science 309 481–484 [DOI] [PubMed] [Google Scholar]
- 4.Ragu, S., Faye, G., Iraqui, I., Masurel-Heneman, A., Kolodner, R. D., and Huang, M.-E. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 9747–9752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blair, I. A. (2008) J. Biol. Chem. 283 15545–15549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee, S. H., Rindgen, D., Bible, R. H., Hajdu, E., and Blair, I. A. (2000) Chem. Res. Toxicol. 13 565–574 [DOI] [PubMed] [Google Scholar]
- 7.Pollack, M., Oe, T., Lee, S. H., Elipe, M. V. S., Arison, B. H., and Blair, I. A. (2003) Chem. Res. Toxicol. 16 893–900 [DOI] [PubMed] [Google Scholar]
- 8.Rindgen, D., Nakajima, M., Wehrli, S., Xu, K., and Blair, I. A. (1999) Chem. Res. Toxicol. 12 1195–1204 [DOI] [PubMed] [Google Scholar]
- 9.Williams, M. V., Lee, S. H., Pollack, M., and Blair, I. A. (2006) J. Biol. Chem. 281 10127–10133 [DOI] [PubMed] [Google Scholar]
- 10.Pollack, M., Yang, I.-Y., Kim, H.-Y. H., Blair, I. A., and Moriya, M. (2006) Chem. Res. Toxicol. 19 1074–1079 [DOI] [PubMed] [Google Scholar]
- 11.Johnson, R. E., Washington, M. T., Prakash, S., and Prakash, L. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 12224–12226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Woodgate, R. (1999) Genes Dev. 13 2191–2195 [DOI] [PubMed] [Google Scholar]
- 13.Ling, H., Boudsocq, F., Woodgate, R., and Yang, W. (2001) Cell 107 91–102 [DOI] [PubMed] [Google Scholar]
- 14.Trincao, J., Johnson, R. E., Escalante, C. R., Prakash, S., Prakash, L., and Aggarwal, A. K. (2001) Mol. Cell. 8 417–426 [DOI] [PubMed] [Google Scholar]
- 15.Yang, W., and Woodgate, R. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 15591–15598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matsuda, T., Bebenek, K., Masutani, C., Hanaoka, F., and Kunkel, T. A., (2000) Nature 404 1011–1013 [DOI] [PubMed] [Google Scholar]
- 17.Tissier, A., McDonald, J. P., Frank, E. G., and Woodgate, R. (2000) Genes Dev. 14 1642–1650 [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang, Y., Yuan, F., Xin, H., Wu, X., Rajpal, D. K., Yang, D., and Wang, Z. (2000) Nucleic Acids Res. 28 4147–4156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhong, X., Garg, P., Stith, C. M., McElhinny, S. A. N., Kissling, G. E., Burgers, P. M. J., and Kunkel, T. A. (2006) Nucleic Acids Res. 34 4731–4742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ohmori, H., Friedberg, E. C., Fuchs, R. P. P., Goodman, M. F., Hanaoka, F., Hinkle, D., Kunkel, T. A., Lawrence, C. W., Livneh, Z., Nohmi, T., Prakash, L., Prakash, S., Todo, T., Walker, G. C., Wang, Z., and Woodgate, R. (2001) Mol. Cell. 8 7–8 [DOI] [PubMed] [Google Scholar]
- 21.Masutani, C., Kusumoto, R., Yamada, A., Dohmae, N., Yokio, M., Yuasa, M., Araki, M., Iwai, S., Takio, K., and Hanaoka, F. (1999) Nature 399 700–704 [DOI] [PubMed] [Google Scholar]
- 22.Lin, W., Xin, H., Zhang, Y., Wu, X., Yuan, F., and Wang, Z. (1999) Nucleic Acids Res. 27 4468–4475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jansen, J. G., Langerak, P., Tsaalbi-Shtylik, A., van den Berk, P., Jacobs, H., and de Wind, N. (2006) J. Exp. Med. 203 319–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ross, A.-L., and Sale, J. E. (2006) Mol. Immunol. 43 1587–1594 [DOI] [PubMed] [Google Scholar]
- 25.Haracska, L., Unk, I., Johnson, R. E., Johansson, E., Burgers, P. M. J., Prakash, S., and Prakash, L. (2001) Genes Dev. 15 945–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nelson, J. R., Gibbs, P. E. M., Nowicka, A. M., Hinkle, D. C., and Lawrence, C. W. (2000) Mol. Microbiol. 37 549–554 [DOI] [PubMed] [Google Scholar]
- 27.Lawrence, C. W. (2002) DNA Repair 1 425–435 [DOI] [PubMed] [Google Scholar]
- 28.Gibbs, P. E. M., McGregor, W. G., Maher, V. M., Nisson, P., and Lawrence, C. W. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 6876–6880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnson, R. E., Washington, M. T., Haracska, L., Prakash, S., and Prakash, L. (2000) Nature 406 1015–1019 [DOI] [PubMed] [Google Scholar]
- 30.Prakash, S., and Prakash, L. (2002) Genes Dev. 16 1872–1883 [DOI] [PubMed] [Google Scholar]
- 31.Bi, X., Slater, D. M., Ohmori, H., and Vaziri, C. (2005) J. Biol. Chem. 280 22343–22355 [DOI] [PubMed] [Google Scholar]
- 32.Ogi, T., Shinkai, Y., Tanaka, k., and Ohmori, H. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 15548–15553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu, X., Lao, Y., Yang, I.-Y., Hecht, S. S., and Moriya, M. (2006) Biochemistry 45 12898–12905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Camenisch, G., Gruber, M., Donoho, G., Sloun, P. V., Wenger, R. H., and Gassmann, M. (1996) Nucleic Acids Res. 24 3707–3713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ohkumo, T., Kondo, Y., Yokoi, M., Tsukamoto, T., Yamada, A., Sugimoto, T., Kanao, R., Higashi, Y., Kondoh, H., Tatematsu, M., Masutani, C., and Hanaoka, F. (2006) Mol. Cell. Biol. 26 7696–7706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McDonald, J. P., Frank, E. G., Plosky, B. S., Rogozin, I. B., Masutani, C., Hanaoka, F., Woodgate, R., and Gearhart, P. J. (2003) J. Exp. Med. 198 635–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wittschieben, J. P., Reshmi, S. C., Gollin, S. M., and Wood, R. D. (2006) Cancer Res. 66 134–142 [DOI] [PubMed] [Google Scholar]
- 38.Hirt, B. (1967) J. Mol. Biol. 26 365–369 [DOI] [PubMed] [Google Scholar]
- 39.Scott, S., Lao, Y., Yang, I.-Y., Hecht, S. S., and Moriya, M. (2006) Mutat. Res. 608 1–7 [DOI] [PubMed] [Google Scholar]
- 40.Yang, I.-Y., Miller, H., Wang, Z., Frank, E. G., Ohmori, H., Hanaoka, F., and Moriya, M. (2003) J. Biol. Chem. 278 13989–13994 [DOI] [PubMed] [Google Scholar]
- 41.Yang, I.-Y., Johnson, F., Grollman, A. P., and Moriya, M. (2002) Chem. Res. Toxicol. 15 160–164 [DOI] [PubMed] [Google Scholar]
- 42.Acharya, N., Johnson, R. E., Prakash, S., and Prakash, L. (2006) Mol. Cell. Biol. 26 9555–9563 [DOI] [PMC free article] [PubMed] [Google Scholar]