Abstract
In prokaryotes, cognate toxin-antitoxin pairs have long been known, but no three-dimensional structure has been available for any given complex from Mycobacterium tuberculosis. Here we report the crystal structure and activity of a member of the VapBC family of complexes from M. tuberculosis. The toxin VapC-5 is a compact, 150 residues, two domain α/β protein. Bent around the toxin is the VapB-5 antitoxin, a 33-residue α-helix. Assays suggest that the toxin is an Mg-enabled endoribonuclease, inhibited by the antitoxin. The lack of DNase activity is consistent with earlier suggestions that the complex represses its own operon. Furthermore, analysis of the interactions in the binding of the antitoxin to the toxin suggest that exquisite control is required to protect the bacteria cell from toxic VapC-5.
Toxin-antitoxin (TA)2 loci were discovered 25 years ago on the mini-F plasmid of Escherichia coli as plasmid addiction modules responsible for the maintenance of extrachromosomal genetic elements (1). But the subsequent discovery of TA genes in the chromosomes of numerous diverse prokaryotes suggests alternative functions for the chromosomal TA systems (2). At least nine possible functions for chromosomal TA systems have been proposed, such as stabilization of genomic parasites, gene regulation, growth control, persistence, or programmed cell death (3) but the most probable hypothesis, supported by experimental evidence, is the cessation of growth under conditions of nutritional or environmental stress (4).
Indeed, a unifying feature of bacteria that contain TA operons is the propensity for dormancy or slow growth. For instance, Mycobacterium tuberculosis, which is able to arrest its growth during the intracellular phases of its lifecycle harbors 38 TA operons and Gloeobacter violaceus which grows slowly in the laboratory has 21 TA operons (5). The primary function of these TA systems could be to arrest growth, and thus enable survival in unfavorable or stressful environments (6).
Although the physiological role of TA loci is not clearly understood, their mechanisms of action have been thoroughly studied. TA loci consist of two genes organized in an operon, which encode an unstable antitoxin and a stable toxin (7). In favorable growth conditions, both proteins are expressed and the antitoxin sequesters the toxin in a complex allowing bacterial cell growth. In most cases, the complex and particularly the antitoxin act as a repressor of the operon transcription (8). Under stress, such as oxidative stress, elevated temperature, starvation, or addition of antibiotic, specific proteases are triggered which degrade the unstable antitoxin more rapidly than the toxin (4). This degradation leads to the release of the toxin in the cell. Most toxins are ribonucleases that cleave mRNA in a specific or nonspecific manner although some toxins have been determined to be inhibitors of gyrases and kinases (9).
The action of the toxin permits the bacterium to enter a reversible bacteriostatic state which can ultimately cause bacterial cell death (4, 6, 10–12). In most of these systems, toxins are homologues of the pilT N terminus domain (PIN domain), which are small proteins with structural homology to the T4 RNase H nuclease domain (13). Although sequence similarity is low within the PIN domains, multiple sequence alignments have shown that active site residues are highly conserved. These residues were first predicted in silico to have a nuclease activity which was then confirmed in vitro (14). These results support a ribonuclease function for the toxins.
Five TA families have been identified on the M. tuberculosis chromosome: one member from the higBA family, two from parDE (15), three from relBE (16), nine from mazEF (17), and 23 from vapBC (5, 14). The E. coli MazEF and the archaeal RelBE systems have been extensively studied. In the first system, the MazE antitoxin inhibits the toxic action of MazF, which cleaves mRNA specifically at ACA sequences, by mimicking ssRNA (17). The crystal structure of the complex shows that the antitoxin is organized as a long α-helix that binds a deep cleft formed by the oligomeric MazF (18). In the second system, RelBE adopts a different strategy: the toxin, RelE, binds to the A-site of the ribosome and cleaves nascent mRNA. RelB inhibits the action of RelE by wrapping around the toxin, enlarging it, and thus precluding it from penetrating the ribosomal A-site (19).
Here we present the first crystal structure of a prokaryotic VapBC complex, VapBC-5 from M. tuberculosis. The toxin, VapC-5, contains 126 residues (Mr = 14.0 kDa) and the antitoxin, VapB-5, contains 34 residues (Mr = 3.8 kDa). This structure provides insight into the modes of binding and inhibition of the toxin by the antitoxin and suggests that M. tuberculosis VapC-5 is an endoribonuclease.
EXPERIMENTAL PROCEDURES
Coexpression Vector—The cloning strategy for coexpression of Rv0626, which encodes for VapB-5 and Rv0627, which encodes for VapC-5 was designed for LIC using the pET46 Ek/LIC vector kit and LIC DUET minimal adaptor (both from Novagen). Gene-specific PCR primers contained 5′ extensions so that T4 polymerase treatment of PCR products in the presence of dATP would generate overhangs complementary to either the 5′-or3′-end of the pET46-LIC vector or the LIC DUET minimal adaptor. The PCR primers were designed so that Rv0627 would be in the upstream (5′) position separated from Rv0626 in the downstream (3′) position by the 138-base pair LIC DUET minimal adaptor. The construct resulted in Rv0627 having an N-terminal extension (MAHHHHHHVDDDDK) encoding a histidine tag and an enterokinase site for proteolytic removal of the tag, and the adaptor adds the amino acid sequence MQAGPAL to the N terminus of Rv0626.
Cloning of the Rv0626 and Rv0627 Genes—The Rv0626 and Rv0627 genes were PCR-amplified from M. tuberculosis H37Rv genomic DNA using Sure-Pol DNA polymerase (Denville Scientific). The following primers pairs were used: Rv0627Fwd (5′-GACGACGACAAGATGGTGAGCACGACGCCGGCCGCCGG), Rv0627Rev (5′-CGCGGGCGGCCGTATTAGACCCGAATGATCTCCACACTGGCCGCACCG), Rv0626Fwd (5′-GCGGGCCCGGCCTTGATGTCTGAGGTGGCCTCGCGTGAGCTGCG), and Rv0626Rev (5′-GAGGAGAAGCCCGGTATTACCGGATCGGCCCGAGATCCTCGGTCGTG). The PCR products were extracted from agarose gels using the QIAquick gel extraction kit (Qiagen) and eluted with TE buffer (10 mm Tris, pH 8.0, 1 mm EDTA). The products were then treated with T4 polymerase in the presence of dATP according to the protocol supplied with the pET46 Ek/LIC vector kit (Novagen). The LIC-treated PCR products were annealed to the pET46 Ek/LIC vector and LIC DUET minimal adaptor by following the LIC DUET Adaptor Kit protocol (Novagen). The annealed product was transformed into E. coli NovaBlue GigaSingles Competent Cells (Novagen); selection was for ampicillin resistance. Recombinant plasmids containing Rv0626/Rv0627 were isolated and sequenced (Davis Sequencing) to confirm the presence of the genes and adaptor.
Protein Preparation—The plasmid harboring the two genes was transformed into E. coli BL21 (DE3) pLysS and cells were grown to an A600 of 0.3 in LB containing 100 mg/liter ampicillin. Protein expression was then induced using 0.4 mm isopropyl-1-thio-β-d-galactopyranoside for 16 h at 25 °C. Cells were harvested and resuspended in 15 ml of 50 mm Tris, pH 7.0, 500 mm NaCl, and 10 mm β-mercaptoethanol (Buffer A) to which protease inhibitor mixture, RNase A, DNase I (Sigma), and phenylmethylsulphonyl fluoride were added. Cell lysis was performed by sonication on ice, and the lysate was centrifuged at 20,000 × g for 45 min at 4 °C. The filtered supernatant was then applied to a HisTrap Ni2+-chelating column (GE Healthcare) equilibrated in buffer A. The pure complex coeluted using a step gradient in 300 mm imidazole and was then concentrated to 6 mg/ml to screen for crystallization.
To prepare the selenomethionyl (SeMet) derivative of VapBC-5, E. coli BL21 (DE3) pLysS cells were grown in 2 ml of LB containing 100 mg/liter ampicillin for 6 h, then spun down, resuspended in 35 ml of M9 medium, and grown at 37 °C overnight. This overnight culture was then used to innoculate 2 liters of M9 medium. When the absorbance at 600 nm reached 0.15, solid amino acid supplements were added to the culture according to the protocol of Van Duyne et al. (20). Protein expression and purification of SeMet protein followed the same protocol as described earlier for the native protein.
Crystallization and Data Collection—Wild-type protein produced crystals in numerous conditions. The best diffracting crystal appeared in 30% MPD, 0.1 m sodium acetate pH 4.6, 0.2 m NaCl within 2 weeks. Crystals of the SeMet-substituted complex grew in 30% MPD, 0.1 m sodium acetate pH 4.2 after two months. Crystals were flash-frozen in liquid nitrogen, and native data were collected to 1.9-Å resolution at the Advanced Photon Source (24-ID-C, APS, IL) whereas selenium anomalous data were collected at Advanced Light Source Beamline (BL8-2-1, ALS, CA) (Table 1). All data were processed using DENZO and SCALEPACK (21).
TABLE 1.
Data collection statistics
| Data collection | Seleno-Met-substituted ALS BL8-2-1 | Native APS ID24C |
|---|---|---|
| Wavelength (Å) | 0.9795 | 1.5418 |
| Resolution (Å) | 2.5 | 1.86 |
| Space group | P6522 | P6522 |
| Unit cell | a = 64.7 Å b = 64.7 Å c = 165.1 Å γ = 120° | a = 64.8 Å b = 64.8 Å c = 164.7 Å γ = 120° |
| Completeness (%) | 99.9 (99.7)a | 93.0 (82.5) |
| Multiplicity | 21.9 (21.4) | 9.1 (4.5) |
| 〈I〉/〈σ(I)〉 | 22.7 (8.9) | 24.7 (3.8) |
| Rsym (%) | 11.3 (35.1) | 5.8 (36.1) |
| Number of selenium atoms/asymmetric unit | 2 observed of 6 expected |
Numbers in parentheses refer to a high-resolution bin of approximate width 0.08 Å
Structure Determination and Refinements—The structure of SeMet-VapBC-5 was solved by single wavelength anomalous diffraction (SAD) at 0.9795 Å. The program HKL2MAP (SHELXC, -D, and -E) (22, 23) was used to determine a substructure using data from 50.0 to 2.5 Å. SHELXE (23) was used to determine the correct enantiomorph of the substructure, and to calculate the phases to 1.9-Å resolution. Experimental phases were improved by density modification using the program DM (24). The resulting phases coupled with the observed structure factors were then input into ARP/wARP (25) for automated model building and side chain docking. The model was constructed by iterative rounds of refinement using Refmac5 (26) interspersed with map inspection in Coot (27). The refinement process was monitored using Rwork and Rfree (28) (Table 2).
TABLE 2.
Refinement statistics
| Refinement | Native APS ID24C |
|---|---|
| Toxin: antitoxin heterodimers/asymmetric unit | 1:1 |
| Protein residues | 164 (Toxin 130-antitoxin 34) |
| Water molecules | 56 |
| β-Mercaptoethanol | 1 |
| Acetate | 1 |
| Na+ ions | 3 |
| Rwork/Rfree (%) | 21.4 (24.5)/24.9 (35.0) |
| R.M.S.D. bond lengths ()/bond angles () | 0.012/1.282 |
| Wilson B, Å2 | 20.9 |
| Average isotropic thermal parameters, Å2 | |
| VapB-5, Main chain/side chain | 34.6/36.0 |
| VapC-5, Main chain/side chain | 15.9/17.6 |
| Solvents | 25.5 |
| Ramachandran analysis (%) | |
| Favored regions | 94.4 |
| Additionally allowed regions | 4.9 |
| Generously allowed regions | 0.7 |
| Disallowed regions | 0.0 |
Activity Assays—For fluorescence assays, we followed the protocol of the RNaseAlert™ kit (Ambion kit AM1964). This assay uses a RNA substrate containing a fluorescent reporter molecule on one end and a quencher on the other. In the intact substrate, the quencher is physically close to the reporter and dampens its fluorescence signal to extremely low levels. When the RNA substrate is cleaved, the quencher is separated from the reporter, allowing it to emit a bright green signal when excited. The cleavage of the RNA substrate is measured at 520 nm after excitation at 490 nm every minute for three hours using a fluorimeter. Each experiment was set up in triplicate for measurement accuracy. The positive control consisted of 3 × 10-5 units of RNase A with substrate. Negative controls consisted of substrate incubated with buffer and nuclease-free water or substrate incubated with 3 × 10-5 units of RNase A and 40 units of the RNase inhibitor RNaseOUT™ (Invitrogen). Purified and concentrated M. tuberculosis VapBC-5 was incubated at 25 μm in 50 mm Tris, pH 7.0, 150 mm NaCl and containing either 10 mm MgCl2 or 10 mm EDTA with 15 pmol of the fluorescent substrate resuspended in 5 μl of 10× buffer (Ambion kit AM1964). Although the composition of the substrate resuspension buffer is not provided in details, the presence of detergents is mentioned. These experiments were repeated in the presence of RNaseOUT™ to eliminate any RNase contamination.
For the in vitro ribonuclease assay, increasing amounts of the M. tuberculosis VapBC-5 complex (4, 8, 12, 16 μm) were incubated with 2 μm of a 150 nucleotide hairpin loop RNA in the presence of 10 mm EDTA, at 37 °C. Samples were also prepared in the same way using 10 mm MgCl2 instead of EDTA. Reactions were stopped after 5 h by the addition of gel-loading buffer (10 m urea, 10 mm EDTA, 0.5× TBE, 0.01% xylene cyanol) followed by heating at 90 °C for 3 min. Samples were run on a 15% polyacrylamide/urea denaturing gel and visualized using Sybr Green II RNA gel stain (Cambrex). Similar experiments using a 1:1 or 1:5 ratio of enzyme:dsDNA were set up using a 25-base pair DNA oligomer (data not shown).
RESULTS
Structure of the VapBC Heterocomplex—The structure of VapC-5 from M. tuberculosis in complex with a C-terminal fragment of VapB-5 was determined at 1.9-Å resolution by single wavelength anomalous dispersion (Fig. 1A). SHELXD was able to determine the positions of two selenium sites out of six putative sites in the asymmetric unit. ARP/WARP automatically built a total of 121 of 153 residues. The remaining residues were built in Coot (27) to provide the final model that was refined to Rwork and Rfree of 21.4 and 24.9%, respectively. The final model includes 56 water molecules, three Na+ ions, one molecule of β-mercaptoethanol, and one molecule of acetate. The geometry of the structure was checked using the Structure Analysis and VErification Server (SAVES), which integrates the programs PROCHECK, WHAT CHECK, ERRAT, VERIFY 3D, and PROVE. No residues were found in disallowed regions of the Ramachandran plot.
FIGURE 1.
Sequence and structure of M. tuberculosis VapBC-5. A, ribbon diagram of the VapBC-5 complex (β-strands are cyan, α-helices are magenta for VapC-5 and green for VapB-5). Dotted lines represent residues that are not visible in the density. B, surface representation of VapC-5 showing negative electrostatic potential in red and positive in blue. VapB-5 is shown as a ribbon diagram with amino acid side chains shown in stick representation. Black arrows designate the cavities that shelter the active site residues as well as residues that may be involved in the binding of Mg2+ ions. C, amino acid sequence of M. tuberculosis VapC-5 on top and VapB-5 at the bottom with their secondary structure elements assigned and colored according to the ribbon diagram. Residues in lowercase are not seen in the structure. Putative catalytic residues are marked with a red star and Arg-102 with a blue star.
The structure of M. tuberculosis VapC-5 toxin forms a compact α/β/α main domain and a protruding clip structure of two α-helices. These domains are built from 11 secondary structure elements (Fig. 1, A and C): β1 (residues 22–25), α1 (residues 28–31), α2 (residues 41–43), β2 (residues 47–51), α3 (residues 53–64), α4 (residues 68–84), α5 (residues 91–108), α6 (residues 113–126), β3 (residues 129–131), α7 (residues 137–141), β4 (residues 147–149). The compact core domain is organized as a four-stranded parallel β-sheet (β2-β1-β3-β4) surrounded by five α-helices. The clip structure protrudes from the core domain and consists of α-helices α3 and α4 linked by a short loop. These two domains are connected by two potentially flexible stretches of residues, which conceivably confer the necessary flexibility to bind the antitoxin. The possibility of flexibility is suggested by the coil structure of these segments. Electron density for the 19 N-terminal residues (15 from the histidine tag and four from the native N terminus) is not visible in the structure of VapC-5 nor are the five residues in the loop linking helices α1 and α2 (Fig. 1, A and C).
The structure of VapB-5 antitoxin shows clear electron density for 33 residues out of 93 (86 native and seven from the vector). The missing residues are part of the N-terminal region that is predicted to bind to DNA (29). VapB-5 has an extended geometry organized in two α-helices, α1 (residues 58–65) and α2 (residues 72–82) connected by a long and potentially flexible loop (Fig. 1, A–C). The antitoxin binds tightly to VapC-5 in a deep and wide groove formed between the core domain and the clip structure (Fig. 1, A and B). The N-terminal residues of VapB-5, and in particular the side chain atoms of VapB-5 Arg-53, are wrapped around VapC-5 to fortify the interactions. These interactions involve hydrophobic residues to stabilize the N terminus of helix α1 of the antitoxin as well as the C-terminal of helix α2 of the antitoxin. The remaining interactions are direct hydrogen bonds between charged residues of the antitoxin and the toxin. The groove narrows around the loop linking the two α-helices of VapB-5 by the alternative conformation of the side-chain atoms of VapC-5 Gln-73 and residue VapC-5 Ala-32 main chain (Fig. 1B). Because the antitoxin fills the large otherwise exposed groove, the interface between VapB-5 and VapC-5 buries 558 Å2 which represents 30% of the accessible surface area of the antitoxin. In the crystal, VapBC-5 forms a dimer through the 2-fold axis that involves helix α3, the loop linking β3 to α5, the helix α5, the loop linking α5 to α6 and the N-terminal part of helix α6 from VapC-5 as well as the C-terminal part of helix α2 and the extended coil region of VapB-5 (residues 82–86).
Geometry of the Putative Active Site Residues—As predicted from sequence alignment, the structure of M. tuberculosis VapC-5 belongs to the family of PIN domains (30). This family of proteins groups homologs of the N-terminal domain of the pili biogenesis protein that are organized as two α-helices, four parallel β-sheets, followed by two α-helices. The alignment of multiple PIN domains with M. tuberculosis VapC-5 reveals four highly conserved acidic residues, VapC-5 Asp-26, VapC-5 Glu-57, VapC-5 Asp-115, and VapC-5 Asp-135 consistent with exonuclease activity. In fact, experimental results have shown that these residues are involved in a Mg2+-dependent exonuclease activity (14). In the structure of VapBC-5, these residues cluster to form a negatively charged cavity structured by residues from both VapB-5 and VapC-5 (Figs. 1B and 2). The acidic cavity is shaped by side chains (VapB-5 Arg-75, VapB-5 Leu-78, and VapB-5 Ala-82) from VapB-5 helix α-2 which restrains accessibility to the putative catalytic residues on top of the cavity. Residues from VapC-5 that link strand β-1 to helix α-1, helices α-5 to α-6, and strand β-4 to helix α-7 as well as residues from helix α-6 and the N-terminal part of helix α-1 also participate in the formation of the acidic cavity.
FIGURE 2.
The putative active site of M. tuberculosis VapC-5. The main chain atoms are represented as a ribbon diagram and colored as follows: VapB-5 is shown in green, VapC-5 α-helices are shown in magenta, β-strands in cyan and loops in gray. Residues involved in the active site formation are shown as green sticks for VapB-5 and yellow for VapC-5. For both proteins, N and O atoms are blue and red, respectively. The four acidic catalytic residues are in bold font. Water molecules are shown as cyan balls. Potential hydrogen bonds are depicted as cyan-dashed lines. For the purpose of clarity, only selected water molecules and hydrogen bonds are shown.
The putative active site residues are tightly stabilized by a network of hydrogen bonds (Fig. 2). VapC-5 Glu-57 OE1 is stabilized by VapC-5 Thr-27 OG1 while VapC-5 Glu-57 OE2 accepts a hydrogen bond from the amide group NH1 of VapC-5 Arg-112 which also interacts with the main chain carbonyl of VapB-5 Ala-82. The VapC-5 Arg-112 NH2 donates a hydrogen bond to OD1 of VapC-5 Asp-115. A well-defined water interacts (31) with both VapC-5 Asp-115 OD1 and VapC-5 Asp-26 OD2. VapC-5 Asp-115 OD2 forms hydrogen bonds with a second water molecule. It is likely that VapC-5 Asp-115 OD2 is protonated given the short hydrogen bond (2.42 Å) formed with VapC-5 Asp135 OD2. VapC-5 Asp-26 OD1 interacts with both VapC-5 Ser-28 OG and its main chain amine group. VapC-5 Asp-135 OD2 is also stabilized by the main chain amide group of VapC-5 Arg-112 while its OD1 group forms a hydrogen bond with a third water molecule. A fourth water molecule stabilizes the main chain amide group of VapC-5 Asp-135, the main chain carbonyl of VapC-5 Asp-133 and VapC-5 Thr-131 OG1. This carbonyl atom also donates a hydrogen bond to VapC-5 Asp-26 OD2. OD1 and OD2 of VapC-5 Asp-133 are both stabilized by VapB-5 Arg-75, NH2 and NE, respectively.
Structural Homologs—The overall structure of VapC-5 in the structure of the M. tuberculosis VapBC-5 superimposes well on the structure of VapC from Pyrobaculum aerophilum (1V8P, R.M.S.D. = 3.2 Å on 113 Cα, % identity = 24%) (14) except for helix α-2 (Fig. 3, A and B). Compared with Pae VapC helix α-2 in M. tuberculosis VapC-5 is shifted by about 10 Å and is broken into two one-turn helices linked by a flexible loop for which no electronic density is observed in VapC-5 in our complex. The position occupied by helix α-2 in Pae VapC is occupied by residues of the C-terminal helix of the antitoxin in the structure of M. tuberculosis VapBC-5. It is thus unlikely that the difference around helix α-2 is due to the presence of VapB-5 bound to VapC-5. That difference could reflect that Pyrobaculum aerophilum is an archaea whereas M. tuberculosis is a prokaryote.
FIGURE 3.
Superposition of the structure of M. tuberculosis VapBC-5 with its structural homologues. In all figures, M. tuberculosis VapB-5 is shown in green, M. tuberculosis VapC-5 is in magenta, Ngo FitA (34) is in orange, Ngo FitB (34) is in red, and Pae VapC (14) is in blue. A, ribbon diagram of the superimposed M. tuberculosis VapBC-5 and Pae VapC, which shows that the structure between these two homologues is conserved except for helix α-2 in VapBC-5 that is shifted. B, top view of the zoomed region showing the large displacement of M. tuberculosis VapC-5 helix α-2 compared with the corresponding helix in Pae VapC-5. The disorganized loop linkingα-1 toα-2 is represented as red broken lines. C, ribbon diagram of the superimposed structures. The structure of the toxins in these different structural homologues is conserved whereas their respective cognates differ. D, zoom of the acidic cavity of the superimposed structures. It shows that Arg-112 from M. tuberculosis VapC-5 and Arg-68 from Ngo FitA both form hydrogen bonds with a residue that belongs to the active site and thus Arg-112 could play an indirect role in the mechanism of inhibition of the toxin. Residues from M. tuberculosis VapB-5 are shown as green sticks, those from M. tuberculosis VapC-5 are shown in yellow, Arg-68 from Ngo FitA is in orange, and active site residues from Ngo FitB are shown as red sticks. E, stereoview of the superposition of the putative active site residues of M. tuberculosis VapC-5 with the active site residues and magnesium ions of endo and exonuclease FEN-1 (31). Residues from M. tuberculosis VapC-5 are shown as yellow sticks, and active site residues and magnesium ions from endo and exonuclease FEN-1 are shown as gray sticks and green spheres, respectively. The conservation of most of the residues that bind the magnesium ions suggests that M. tuberculosis VapC-5 catalytic mechanism could involve the two metal ions as suggested by the mechanism of FEN-1 nuclease.
Searches with DALI (32) and DÉJÀ VU (33) using the VapC-5 structure as a probe found structural homologs of VapC-5 in PIN domain-containing proteins. The closest structural homolog is FitAB from Neisseria gonorrhoeae (% identityVapC-5/FitB = 21% and % identityVapB-5/FitA = 14%) for which the structure of the complex was determined with and without DNA bound to the N terminus of FitA (34). Superimposing VapC-5 on Ngo FitB shows the same deviation of helix α-2 compared with Pae VapC (Fig. 3C). Although the toxins superimpose well, Ngo FitA does not bind to the toxin in the same manner that M. tuberculosis VapB-5 binds VapC-5. Ngo FitA helix α-3 fills a large exposed hydrophobic groove on Ngo FitB while the C-terminal extended coil region of Ngo FitA binds to the positively charged part of the Ngo FitB groove. The extended coil region of Ngo FitA is organized as helix α-2 in the structure of M. tuberculosis VapBC-5 and the loop linking helices α-1 and α-2 is buried deeper in the VapC-5 groove. In their Ngo FitAB structure, the authors propose that Ngo FitA inhibits Ngo FitB by pointing the guanidinium group of Ngo FitA Arg-68, at the C-terminal extended coil region, to the carboxyl groups of the putative catalytic residues Ngo FitB Asp-5, Ngo FitB Glu-42 and Ngo FitB Asp-104. Although this Ngo FitA Arg-68 is not conserved in the primary amino acid sequence of M. tuberculosis VapB-5, structural superimposition shows that VapC-5 Arg-112 from M. tuberculosis VapC-5 itself points toward the carboxylic groups of the putative catalytic residues as seen in the structure of Ngo FitAB (Fig. 3D). This VapC-5 Arg-112 is tightly held in position by a hydrogen bond accepted by the main chain carbonyl of residue VapB-5 Ala-82 and thus may participate in the mechanism of inhibition of the toxin.
Structural comparisons also identified the endo and exonuclease FEN-1 as a member of the PIN domain superfamily (14, 31). Although the structural alignment gave poor statistics (R.M.S.D. of 2.25 Å on 102 equivalent positions with 6.5% sequence identity), the active site residues superimpose well. Moreover the structure of FEN-1 displays two Mg2+ ions bound to the active site residues that allow the identification of the putative Mg2+ binding sites in VapC-5 (Fig. 3E).
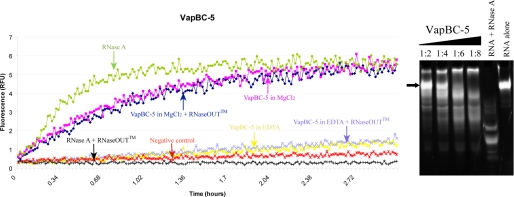
Activity Assays and Evidence for Endoribonuclease Activity—Assays were carried out to assess VapBC-5 activity on the fluorescent-labeled RNA substrate of unknown sequence in different buffers (Fig. 4A). In this assay, fluorescence is detected when the substrate is cleaved by nuclease activity. This assay was set-up using VapBC-5 as attempts to separate VapB-5 and VapC-5 in the complex were unfruitful. Although the fluorescence assays were set up with the purified VapBC-5 complex, native PAGE analysis showed dissociation of the two partners (data not shown), which may be due to the presence of detergents in the substrate resuspension buffer. Thus, M. tuberculosis VapC-5 shows Mg2+-dependent activity in Tris buffer, pH 7.0 containing 150 mm NaCl as shown by the increase of fluorescence over time (Fig. 4A, magenta and navy blue curves). In fact, the activity of VapC-5 in 150 mm NaCl and in the presence of 10 mm EDTA is abolished (Fig. 4A, gray and yellow curves) comparable to the levels of activity detected in the negative control and in RNaseA in the presence of RNase OUT™ (red and black curves, respectively). To check whether activity was due to the contribution of contamination by RNase A, B, or C, the activity of VapBC-5 was assayed in the presence and absence of the specific inhibitor, RNaseOUT™ (Invitrogen), for each buffer condition; no effect was seen on VapC-5 activity. However, low or no activity was observed in Tris buffer, pH 7.0 containing 500 mm NaCl in the presence or absence of divalent cations (Mg2+, Mn2+, and Zn2+). This suggests that the activity of M. tuberculosis VapC-5 is dependent on the presence of Mg2+.
FIGURE 4.
In vitro ribonuclease activity of M. tuberculosis VapBC-5. A, fluorescence measurements as a function of time. A fluorescent substrate is incubated with VapBC-5 in different conditions. Fluorescence is measured when the substrate is cleaved which indicates the presence of ribonuclease activity. It clearly shows that VapC-5 activity is dependent on the presence of magnesium as shown by the magenta and navy blue curves. B, nuclease assay: polyacrylamide/urea denaturing gel showing from left to right:2 μm RNA stained by Sybr Green II with increasing concentration of VapBC-5 in MgCl2. The RNA was incubated for 5 h at 37 °C with 4, 8, 12, 16 μm VapBC-5, respectively, and the subsequent lanes shows the RNA incubated with RNase A as a positive control and the RNA alone. The black arrow points to the intact RNA. Degradation products appear (smears) when the RNA is incubated with VapBC-5.
A nuclease assay was also carried out using a 150-nucleotide RNA with known sequence and an extensive secondary structure as well as single-stranded regions (Fig. 4B). RNA degradation products appear using increasing amounts of the complex (smears on Fig. 4B, lanes 1:2, 1:4, 1:6 and 1:8). The RNA alone is intact compared with RNA incubated with the complex. The positive control shows total degradation of the RNA substrate by RNase A. This assay shows that the complex VapBC-5 has a limited activity on that given RNA substrate.
DISCUSSION
Proposed Catalytic Mechanism—Our crystal structure of the M. tuberculosis VapBC-5 toxin-antitoxin suggests a possible mechanism for VapB-5 inhibition of the toxic activity of VapC-5 supported by the superposition of the endo and exonuclease FEN-1 in which the conserved acidic pockets each bind a Mg2+ ion (31). Superposition of the putative active site residues of M. tuberculosis VapC-5 and the residues that bind Mg2+ ions in the endo and exonuclease FEN-1 show conservation of the geometry and suggest a similar mechanism (Fig. 3E). Thus in M. tuberculosis VapC-5, the first Mg2+ ion could bind to VapC-5 Asp-26, VapC-5 Glu-57, and VapC-5 Asp115 to play a critical catalytic role as shown by FEN-1 mutants (31). The second Mg2+ ion seems to be required to bind the substrate and thus form an active complex. This second site could involve VapC-5 Asp133, VapC-5 Asp-135, and possibly VapC-5 Asp-134. Thus, the two Mg2+ ions could be involved in a two metal ion catalytic mechanism as suggested by the mechanism of FEN-1 nuclease.
It is noteworthy that three ions (Table 2) were identified in the structure of VapC-5. These are located away from the putative active site residues and on the surface of the protein. Because these ions only have two to three ligands and the distance with the coordination groups is comprised between 2.6 and 3.2 Å, they are more likely to be Na+ than Mg2+ for which coordination distances are shorter (2.07–2.26 Å). Moreover, the presence of low coordination number on protein surfaces can be explained by the crystallization procedure, which involved NaCl and sodium acetate but no magnesium (35).
Like Pae VapC for which it has been reported that magnesium is required for exonuclease activity (14), M. tuberculosis VapC-5 also requires magnesium for ribonuclease activity as shown by the fluorescence assay; this is also consistent with the two metal ion catalytic mechanism proposed (Fig. 4A). Furthermore, these results suggest that VapC-5 could be a 3′-endoribonuclease or an exoribonuclease or both, similar to the endo and exonuclease FEN-1.
The ribonuclease activity we detected on the 150-nucleotide RNA was weak considering the excess of enzyme relative to the amount of RNA substrate, regardless of whether magnesium was present (data not shown) (Fig. 4B). This may be the result of a non-optimal substrate for this enzyme and/or the presence of intact and dissociated complexes in the sample. Possibly like the archeal RelBE, the VapC family of enzyme may require association with the ribosome to be fully active (19). However, it has recently been shown that VapC-1 from Haemophilus influenzae is a ribonuclease that acts on free RNA in a concentration-dependent manner (36).
Similar nuclease experiments using dsDNA show that M. tuberculosis VapBC-5 also binds dsDNA (data not shown), which is not surprising as the antitoxin is predicted to bind to the operon promoter (6). Similar to H. influenzae VapC-1, VapC-5 shows no degradation of dsDNA.
Proposed Mechanism of Inhibition—Upon VapB-5 binding by VapC-5 the main chain carbonyl oxygen of VapB-5 Ala-82 reorients the side chain of VapC-5 Arg-112, which locks VapC-5 Glu-57 in an unfavorable conformation to bind a Mg2+ ion. Moreover, VapB-5 Arg-75 could abstract the side chain of VapC-5 Asp-133, pushing it out of the active site/catalytic cavity and could thus provide the binding of the second Mg2+ ion (Fig. 2). The inhibition of nuclease activity by the antitoxin would then be due to the direct or indirect seclusion of the active site residues in catalytically unfavorable conformations. VapC-5 could become active upon the release of VapB-5 triggered by a still unknown signal. This might allow the binding of the substrate to the groove formed between the core and clip domains on VapC-5.
While no nuclease activity could be detected for Ngo FitAB or Ngo FitB in vitro (34), VapBC-5 clearly shows low nuclease activity on dsRNA substrates as well as a magnesium dependence consistent with a two metal ion catalytic mechanism. Although no binding constant could be obtained, the difficulty to separate VapB-5 from VapC-5 as well as the high salt content in the buffer during purification let us hypothesize that the complex formed is tight. This tight binding of the toxin to the antitoxin, resulting in few molecules of the free toxin in the sample may explain the low activity on dsRNA substrate. In contrast, in fluorescence assays, the presence of detergents in the resuspension buffer of the fluorescent substrate result in the disruption of the complex and thus the activity detected may be attributed to the free VapC-5. These results are consistent with M. tuberculosis VapC-5 showing greater toxicity than Ngo FitB, given that a tighter control is necessary for cell viability. Indeed VapC-5 has been shown to be highly toxic to M. tuberculosis (data not shown).
CONCLUSIONS
The first structure of a VapBC complex has been determined for VapBC-5 from M. tuberculosis at 1.9 Å resolution, using single wavelength anomalous dispersion. The VapC-5 toxin structure and the C-terminal part of its cognate antitoxin, VapB-5 are well-ordered while the N-terminal DNA binding region of VapB-5 is completely disordered. It has been shown that toxins have deleterious activity on cell growth, and that this activity is inhibited by the binding of their antitoxins. An as yet unknown signal, which may be specific to each TA system, triggers the degradation of the antitoxin thus releasing the toxin to exert its effect on the cell. This leads to M. tuberculosis growth defect in the case of VapBC-5 as shown in preliminary tests by overexpression of the toxin in M. smegmatis mc24517 strain.
M. tuberculosis VapC-5 has a structural homolog, Ngo FitB, but interestingly their partners, VapB-5 and FitA, respectively, differ structurally (Fig. 3C). A structural comparison suggests that VapB-5 binds more tightly to VapC-5 in a deeper groove and makes more interactions with residues of VapC-5 that are involved in the structure of the acidic catalytic cavity. This tight interaction may be necessary for stringent control of the highly toxic VapC-5 in M. tuberculosis. Based on these results, we propose that M. tuberculosis VapC-5 is most likely both an endoribonuclease and an exoribonuclease that can act on free RNA in a similar manner to the endo and exonuclease FEN-1.
Acknowledgments
We thank Dr. Corie Ralston at the Advanced Light Source for data collection and the Advanced Photon Source (Drs. Malcom Capel, Kanagalaghatta Rajashankar and Igor Kourinov at NECAT 24-ID-E) for assistance in data collection. We also thank Dr. M. Sawaya for helpful discussions and Dr. S. Sievers for assistance with fluorescence assays.
The atomic coordinates and structure factors (code 3DBO) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
This work was supported, in whole or in part, by National Institutes of Health Grants 23616-002-06 F3:02 and TBSGC R01. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: TA, toxin-antitoxin; PIN, pilT N-terminus; LIC, ligation-independent cloning; SeMet, selenomethionine; MPD, 3-methyl-1,5-pentanediol; SAD, single wavelength anomalous diffraction; R.M.S.D., root mean square deviation; Pae, Pyrobaculum aerophilum; Ngo, Neisseria gonorrhoeae.
References
- 1.Ogura, T., and Hiraga, S. (1983) Proc. Natl. Acad. Sci. U. S. A. 80 4784-4788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gerdes, K., Rasmussen, P. B., and Molin, S. (1986) Proc. Natl. Acad. Sci. U. S. A. 83 3116-3120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Magnuson, R. D. (2007) J. Bacteriol. 189 6089-6092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buts, L., Lah, J., Dao-Thi, M. H., Wyns, L., and Loris, R. (2005) Trends Biochem. Sci. 30 672-679 [DOI] [PubMed] [Google Scholar]
- 5.Pandey, D. P., and Gerdes, K. (2005) Nucleic Acids Res. 33 966-976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gerdes, K., Christensen, S. K., and Lobner-Olesen, A. (2005) Nat. Rev. Microbiol. 3 371-382 [DOI] [PubMed] [Google Scholar]
- 7.Van Melderen, L., Bernard, P., and Couturier, M. (1994) Mol. Microbiol. 11 1151-1157 [DOI] [PubMed] [Google Scholar]
- 8.Christensen, S. K., Maenhaut-Michel, G., Mine, N., Gottesman, S., Gerdes, K., and Van Melderen, L. (2004) Mol. Microbiol. 51 1705-1717 [DOI] [PubMed] [Google Scholar]
- 9.Bahassi, E. M., O'Dea, M. H., Allali, N., Messens, J., Gellert, M., and Couturier, M. (1999) J. Biol. Chem. 274 10936-10944 [DOI] [PubMed] [Google Scholar]
- 10.Jensen, R. B., and Gerdes, K. (1995) Mol. Microbiol. 17 205-210 [DOI] [PubMed] [Google Scholar]
- 11.Engelberg-Kulka, H., and Glaser, G. (1999) Annu. Rev. Microbiol. 53 43-70 [DOI] [PubMed] [Google Scholar]
- 12.Gerdes, K. (2000) J. Bacteriol. 182 561-572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arcus, V. L., Rainey, P. B., and Turner, S. J. (2005) Trends Microbiol. 13 360-365 [DOI] [PubMed] [Google Scholar]
- 14.Arcus, V. L., Backbro, K., Roos, A., Daniel, E. L., and Baker, E. N. (2004) J. Biol. Chem. 279 16471-16478 [DOI] [PubMed] [Google Scholar]
- 15.Oberer, M., Zangger, K., Gruber, K., and Keller, W. (2007) Protein Sci. 16 1676-1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gotfredsen, M., and Gerdes, K. (1998) Mol. Microbiol. 29 1065-1076 [DOI] [PubMed] [Google Scholar]
- 17.Engelberg-Kulka, H., Hazan, R., and Amitai, S. (2005) J. Cell Sci. 118 4327-4332 [DOI] [PubMed] [Google Scholar]
- 18.Kamada, K., Hanaoka, F., and Burley, S. K. (2003) Mol. Cell. 11 875-884 [DOI] [PubMed] [Google Scholar]
- 19.Takagi, H., Kakuta, Y., Okada, T., Yao, M., Tanaka, I., and Kimura, M. (2005) Nat. Struct. Mol. Biol. 12 327-331 [DOI] [PubMed] [Google Scholar]
- 20.Van Duyne, G. D., Standaert, R. F., Karplus, P. A., Schreiber, S. L., and Clardy, J. (1993) J. Mol. Biol. 229 105-124 [DOI] [PubMed] [Google Scholar]
- 21.Otwinowski, Z., and Minor, W. (1997) Methods Enzymol. 276 307-326 [DOI] [PubMed] [Google Scholar]
- 22.Schneider, T. R., and Sheldrick, G. M. (2002) Acta Crystallogr. D Biol. Crystallogr. 58 1772-1779 [DOI] [PubMed] [Google Scholar]
- 23.Sheldrick, G. M. (2002) Z. Kristallogr. 217 644-650 [Google Scholar]
- 24.Cowtan, K. (1994) Joint CCP4 and ESF-EACBM Newsletter on Protein Crystallography 31 34-38 [Google Scholar]
- 25.Perrakis, A., Morris, R., and Lamzin, V. S. (1999) Nat. Struct. Biol. 6 458-463 [DOI] [PubMed] [Google Scholar]
- 26.Murshudov, G. N., Vagin, A. A., Lebedev, A., Wilson, K. S., and Dodson, E. J. (1999) Acta Crystallogr. D Biol. Crystallogr. 55 247-255 [DOI] [PubMed] [Google Scholar]
- 27.Emsley, P., and Cowtan, K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60 2126-2132 [DOI] [PubMed] [Google Scholar]
- 28.Kleywegt, G. J., and Brünger, A. T. (1996) Structure 4 897-904 [DOI] [PubMed] [Google Scholar]
- 29.Wilbur, J. S., Chivers, P. T., Mattison, K., Potter, L., Brennan, R. G., and So, M. (2005) Biochemistry 44 12515-12524 [DOI] [PubMed] [Google Scholar]
- 30.Clissold, P. M., and Ponting, C. P. (2000) Curr. Biol. 10 R888-890 [DOI] [PubMed] [Google Scholar]
- 31.Hosfield, D. J., Mol, C. D., Shen, B., and Tainer, J. A. (1998) Cell 95 135-146 [DOI] [PubMed] [Google Scholar]
- 32.Holm, L., and Sander, C. (1995) Trends Biochem. Sci. 20 478-480 [DOI] [PubMed] [Google Scholar]
- 33.Kleywegt, G. J., and Jones, T. A. (1997) Methods Enzymol. 277 525-545 [DOI] [PubMed] [Google Scholar]
- 34.Mattison, K., Wilbur, J. S., So, M., and Brennan, R. G. (2006) J. Biol. Chem. 281 37942-37951 [DOI] [PubMed] [Google Scholar]
- 35.Dokmanic, I., Sikic, M., and Tomic, S. (2008) Acta Crystallogr. D Biol. Crystallogr. 64 257-263 [DOI] [PubMed] [Google Scholar]
- 36.Daines, D. A., Wu, M. H., and Yuan, S. Y. (2007) J. Bacteriol. 189 5041-5048 [DOI] [PMC free article] [PubMed] [Google Scholar]