Abstract
The synthetic rexinoid bexarotene (Targretin, LGD1069) inhibits the formation of both estrogen receptor-negative and estrogen receptor-positive breast cancer in preclinical models and controls the expression of growth-regulatory biomarkers, such as IGFBP-6 (insulin-like growth factor-binding protein 6), RARβ, or cyclin D1. In this study, we identified a classical retinoic acid-responsive element in the first intron in the IGFBP-6 gene adjacent to a consensus AP-1 binding site, both elements essential for rexinoid-induced expression of IGFBP-6. In chromatin binding experiments, bexarotene increased the occupancy of the identified enhancer element by RXRα, RARβ, cJun, cFos, and p300. In normal mammary epithelial cells and T47D breast cancer cells, small interfering RNA-mediated knockdown of all RXR isoforms or RARβ, but not RARα or RARγ alone, blocked the induction of IGFBP-6 by bexarotene. Simultaneous knockdown of RARα and RARγ abrogated both the induction of RARβ and the up-regulation and secretion of IGFBP-6. The suppression of either RARβ or cJun by small interfering RNA blocked the recruitment of RXRα and cJun to the enhancer. These results demonstrate a novel cooperative interaction between retinoid receptors and AP-1 orchestrated by RARβ and highlight a novel mechanism by which RARβ can mediate the cancer-preventive effects of rexinoids.
Retinoids are lipid-soluble mediators that play essential roles in development and homeostatic regulation. Retinoic acid and its derivatives, such as 13-cis-retinoic acid, have been used for the treatment of various cancers, including certain forms of leukemia (1). In addition, retinoids and synthetic retinoid analogues, such as 9-cis-retinoic acid, 13-cis-retinoic acid, or N-(4-hydroxyphenyl) retinamide, have been shown to prevent cancer in animals (2, 3) and in humans (4, 5).
We and others have shown that the panagonist 9-cis-retinoic acid suppresses the development of breast cancer in mice and rats (6, 7). However, this agent has significant toxicity. In contrast, RXR-selective retinoids or rexinoids, such as bexarotene (LGD1069) and LG100268, were found to be very efficient in suppressing the incidence of estrogen receptor-negative mammary tumors in murine mammary tumor virus-ErbB2 mice (8, 9) without the typical toxicity of retinoids. In humans, bexarotene was found to induce remissions in lymphoma patients and thus has been approved by the Food and Drug Administration for the treatment of cutaneous T-cell lymphomas (10). Bexarotene is now being tested in a multicenter clinical trial for its chemopreventive activity against breast cancer.
In a previous study of gene expression profiles of rexinoid-induced biomarkers, we discovered that bexarotene modulated the expression of IGFBP-6 (insulin-like growth factor-binding protein-6), along with other growth-regulatory genes, such as COX-2, cyclin D1, and RARβ (retinoic acid receptor β) (11, 12). Insulin-like growth factors play a pertinent role in the growth regulation of mammary epithelial cells. Although IGF-I appears to be a critical factor in the regulation of breast cell growth, mice overexpressing IGF-II develop mammary tumors (13). IGF-II is a mitogen for many different cancer cells, and overexpression of IGF-II gives rise to mammary tumors (14, 15). As opposed to rodents, humans maintain high levels of IGF-II throughout life, and its stromal expression suggests that IGF-II may affect epithelial cell growth through paracrine pathways in the breast (16). One of the important mechanisms to modulate the effect of IGFs in the tissues is to regulate the amount of available IGFs2 through IGF-binding proteins (17), with IGFBP-1 and -3 being the major IGF-I binding factors. IGFBP-2 is frequently elevated in human prostate and brain tumors and was suggested to play a role in PTEN signaling (18). IGFBP-4, an early responsive estrogen-induced gene, and IGFBP-5 as an estrogen-repressed gene, have been investigated as potential predictive markers for endocrine therapy of breast cancer (19). IGFBP-3 is the most abundant IGF-binding protein in circulation carrying most of the circulating IGF, with intrinsic biologic activities independent of its potential to sequester IGF (20, 21). Although the biological effects of IGFBP-3 have been characterized best, less is known about IGFBP-6. IGFBP-6 has a high affinity for binding IGF-II and is able to inhibit the growth of various cancer cells and activate programmed cell death pathways (22, 23). In addition, patients with invasive breast cancer had significantly lower serum levels of IGFBP-6 than subjects with benign breast disease (24). IGFBP-6 belongs to the small group of genes recently identified as genes showing correlating expression profiles in human estrogen receptor-positive breast tumors and the estrogen receptor-positive cell lines T47D and MCF7 (25).
Here we have characterized the ability of the RXR-selective retinoid bexarotene to regulate the expression of IGFBP-6. Our results show that IGFBP-6 is up-regulated by the cooperative action of retinoid receptors and the AP-1 transcription factor through a complex response element in the first intron of the IGFBP-6 gene. In addition, the early induction of RARβ by bexarotene is essential for the formation of an active AP-1/RXR transcriptional complex, suggesting a novel mechanism by which RARβ can mediate the cancer preventive effects of rexinoids.
MATERIALS AND METHODS
Ligands—The synthetic RXR-ligand bexarotene (Targretin, LGD1069) was a kind gift of Dr. William Lamph (Ligand Pharmaceuticals) and was used at a 1 μm final concentration, unless otherwise indicated. siRNA SmartPools and Dharmafect 1 transfection reagent were purchased from Dharmacon (Lafayette, CO).
Cell Culture—Normal human mammary epithelial cells (HMECs) were derived from healthy women who had undergone reduction mammoplasties and were purchased from Clonetics (San Diego, CA). Cells between passages 4 and 12 were used. HMECs were maintained in mammary epithelial basal medium supplemented with 50 μg/ml bovine pituitary extract, 5 μg/ml insulin, 10 ng/ml human recombinant epidermal growth factor, 0.5 μg/ml hydrocortisone, 30 μg/ml gentamicin, and 15 ng/ml amphotericin-B (Clonetics). Cells were cultured in a humidified environment at 37 °C with 5% CO2 in the air. The human breast cancer cell lines T47D and MCF-7 were obtained from the ATTC (Manassas, VA) and maintained in Iscove's modified Eagle's medium containing 10% fetal bovine serum (Invitrogen).
RNA Extraction and Measurement of Transcript Levels—Total RNA was extracted using the RNeasy kit from Qiagen according to the manufacturer's instructions. Reverse transcription was performed in triplicate, with a “non-reverse transcription control” lacking reverse transcriptase in parallel, to control amplifications due to genomic DNA contamination. Transcript quantitation based on real time monitoring of amplification was carried out using an ABI 7700 sequence analyzer performing 40 cycles of 95 °C for 12 s and 60 °C for 30 s. Values of transcripts in unknown samples were obtained by interpolating their Ct (PCR cycles to threshold) values on a standard curve derived from known amounts of cognate, amplicon-specific synthetic oligonucleotides. Transcript levels were normalized to the level of cyclophilin mRNA.
Selective Gene Knockdown Experiments—To assess the effect of gene silencing on the consequent changes in transcript levels, 104 cells were seeded on 24-well tissue culture plates 24 h prior to transfection. Transfection conditions were optimized for highest level of knockdown at the lowest toxicity, and the active individual duplexes were identified for combination knockdown experiments. Suppression of transcript levels exceeding 70% was considered acceptable.
Generation of Reporter Constructs—Noncoding regions of the IGFBP-6 gene (accession number AJ006952) were amplified by polymerase chain reaction using the genomic clone RP11-158-I20 as a template. Fresh PCR products were TA-cloned into a TOPO2.1 TA-cloning vector (Invitrogen) and then transferred to the pGL3 basic luciferase reporter vector (Promega, Madison, WI). Site-directed mutagenesis was carried out using the QuikChange mutagenesis (Stratagene) protocol according to the manufacturer's recommendations. Three point mutations were introduced in the distal AP-1 binding site (AP1mut31; TGAGTCA → TTATTAA) and two into the RXRE (RXRmut23; AGGTCA → AGTTAA).
Reporter Assays—Reporter assays of luciferase expression vectors were carried out to assess the retinoid responsiveness of various regulatory regions of the IGFBP-6 gene promoter and introns. Transfections were conducted overnight in cells plated in 24-well tissue culture plates at ∼70% confluence, using 0.6 μl of FuGene6 transfection reagent (Roche Applied Science) and 0.2 μg of plasmid DNA in each well. Treatments were done in triplicate, and experiments were repeated three times. Measurements were performed using the Dual-Reporter Assay Kit (Promega, Madison, WI) on a Luminoskan apparatus. To control for variations in transfection efficiency, firefly luciferase readings were normalized to the activity of the anthozoan Renilla reniformis luciferase enzyme expressed from a constitutive promoter.
Western Blot—Cells were transfected with siRNAs for nuclear receptors or luciferase, as a control. Forty-eight hours later transfected cells were treated with bexarotene or vehicle. Another 24 h later, cell supernatants were harvested, and proteins from whole cell lysates were extracted using methods previously described (11). Equal amounts of total protein were electrophoresed on a 10% acrylamide denaturing gel and transferred onto a polyvinylidene difluoride membrane. Protein levels were detected by using the following antibodies: rabbit polyclonal antibody specific for IGFBP-6 at 1:200 (catalog number PB-383-9; Austral Biologicals, San Ramon, CA), RXRα (D-20, sc-553), RARα (C-20, sc-551), RARβ (C-19, sc-552X), RARγ (C-19, sc-550; Santa Cruz Biotechnology, Inc., Santa Cruz, CA), and mouse monoclonal antibody specific for β-actin (catalog number sc-7202; Santa Cruz Biotechnology; 1:200). The blots were visualized using the enhanced chemiluminescence Western blot detection system (Amersham Life Sciences).
Chromatin Immunoprecipitation (ChIP) Assay—Chromatin immunoprecipitation assays were performed as described before (26). Briefly, cells were grown for 4 days in 15-cm dishes to 60% confluence and kept in serum-free media for another 48 h before drugs were added. After washing in PBS, cells were fixed in 1% formalin for 15 min. Cellular DNA was fragmented by sonication with 8 times 5 pulses at power output 5 (Branson Sonifier 450). 500 μg of cell lysates were precleared with protein G-agarose, and the transcription factors in the supernatant were immunoprecipitated using antibodies against RXRα, RXRβ, RARα, RARβ, RARγ, cJun, cFos, ATF2, and p300. Normal rabbit IgG was used as negative control. After reversal of the cross-links, DNA was extracted, and the 2530–2700 section of intron 1 of IGFBP-6 was amplified by 32 cycles of PCR. To ensure reproducibility, experiments were repeated three times. DNA products were first analyzed on 1.5% agarose gels. For accuracy and increased sensitivity to detect chromatin precipitated by the RARβ antibody, extracted DNA was also measured by quantitative real time PCR. Optimal efficiency and linear nature of the reactions was determined by constructing a standard curve from serial dilutions of the BAC clone containing the IGFBP-6 gene.
Statistical Analysis—Statistical significance was determined using Student's t test or two-tailed analysis of variance. A p value of <0.05 was considered statistically significant. Values are presented as means ± S.D.
RESULTS
IGFBP-6 Is Induced by Bexarotene and Is Growth-suppressive—We previously identified IGFBP-6 as a marker gene of the chemopreventive rexinoid bexarotene (Targretin, LGD1069) in normal HMECs. In previous microarray experiments, IGFBP-6 was found to be up-regulated (mean change 2.8-fold at p < 0.05) by treatment with 1 μm bexarotene (among 12,384 genes present on the oligonucleotide chip) (11). This result stimulated us to investigate the molecular mechanism by which rexinoids induce IGFBP-6 expression. To understand what components of the IGF system may be affected by this rexinoid, we first measured the effect of bexarotene on the mRNA levels of the IGF-binding proteins 1–6 in HMECs after a 24-h exposure to the drug using real time quantitative reverse transcription-PCR. These studies showed that IGFBP-2 mRNA was most abundant in HMECs, whereas IGFBP-1 and -5 were barely detectable (Table 1). As shown in Table 1, bexarotene increased the expression of IGFBP-3 and IGFBP-6.
TABLE 1.
Relative mRNA levels of IGF-binding proteins in normal mammary epithelial cells are shown ± S.D. Relative mRNA levels were expressed per 106 of cyclophilin molecules.
| Vehicle | Bexarotene | |
|---|---|---|
| IGFBP-1a | 4 ± 1 | 3 ± 0 |
| IGFBP-2 | 74,244 ± 21,260 | 95,845 ± 28,967 |
| IGFBP-3a | 619 ± 85 | 1671 ± 191 |
| IGFBP-4 | 1293 ± 241 | 1134 ± 146 |
| IGFBP-5 | 9 ± 4 | 8 ± 2 |
| IGFBP-6a | 683 ± 73 | 1660 ± 460 |
Significant difference between vehicle and LGD1069 at p < 0.05
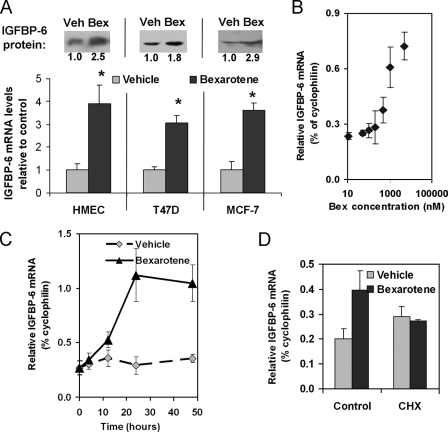
Nuclear Hormone Receptors Involved in Rexinoid-dependent Induction of IGFBP-6—We next confirmed that bexarotene induced IGFBP-6 in normal mammary epithelial cells and in multiple breast cancer cell lines. These studies showed an average 3–4-fold increase in the mRNA levels of IGFBP-6 upon the addition of 1 μm bexarotene to normal breast cells and breast cancer cells (T47D and MCF-7) (Fig. 1A). Increased levels of the mRNA were followed by the induction of the IGFBP-6 protein as well (Fig. 1A, upper inset). Based on the log-linear section of the dose-response curve, 1 μm concentration of bexarotene was chosen for treatment in further studies (Fig. 1B). As shown in Fig. 1C, the up-regulation of the IGFBP-6 mRNA occurred in a delayed fashion, reaching close to maximal change at ∼24 h, suggesting that the induction involves an indirect mechanism. The requirement of a retinoid-inducible intermediary factor for the up-regulation of IGFBP-6 was demonstrated by the lack of induction in the presence of cycloheximide prior to addition of bexarotene to the culture media (Fig. 1D).
FIGURE 1.
Characterization of the effect of the RXR-selective agonist bexarotene (LGD1069, Targretin) on the expression of IGFBP-6. A, comparison of the effects of bexarotene on the transcript levels of IGFBP-6 measured in normal and transformed mammary epithelial cells. Cells were treated with either DMSO/ethanol (50:50%, v/v) as vehicle (Veh) or 1 μm bexarotene (Bex) for 24 h. Relative molecule numbers of IGFBP-6 mRNA were measured by quantitative reverse transcription-PCR normalized to those of cyclophilin, and values in rexinoid-treated cells are expressed as -fold change over vehicle treatment. Statistical significance of the changes was determined by Student's t test. *, p < 0.05. Inset, IGFBP-6 protein levels after 48 h of vehicle or bexarotene treatment in the corresponding cell lines, as determined by Western blot. The numbers represent normalized expression levels relative to controls. B, dose-response curve. HMECs were treated with increasing doses of bexarotene (10 nm to 10 μm), and RNA was harvested 24 h later. IGFBP-6 RNA expression was measured using quantitative reverse transcription-PCR and expressed as a percentage of cyclophilin. C, time dependence of the induction of IGFBP-6 mRNA by 1 μm bexarotene in HME cells. D, comparison of the effect of bexarotene over vehicle on the expression of IGFBP-6 in cells with or without pretreatment with cycloheximide (CHX).
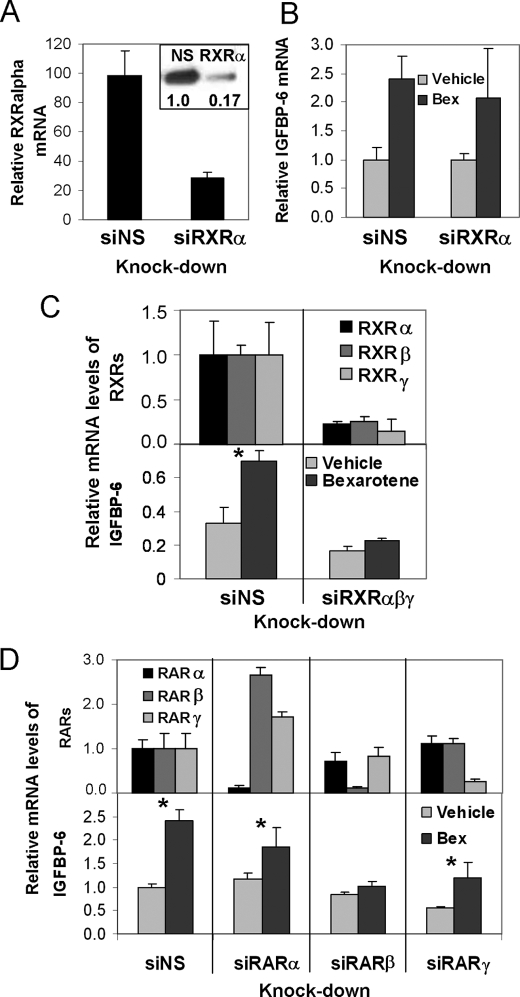
In order to confirm that the effect of bexarotene is RXR-dependent, we treated HMEC and T47D cells with the rexinoid after suppressing RXRs by specific siRNAs. Suppression of RXRα was seen as assessed at the level of mRNA (see Fig. 3A) and protein (Fig. 2A, inset) and by immunohistochemistry (data not shown). However, loss of RXRα resulted in no significant change in the induction of IGFBP-6 by bexarotene (Fig. 2B). Similarly, individual knockdown of RXRβ or RXRγ isoforms alone had no significant effect (data not shown). However, simultaneous knockdown of all three RXR isoforms diminished the induction of IGFBP-6 (Fig. 2C). Thus, RXR proteins are required for rexinoid induction of IGFBP-6; however, the different RXR isoforms are redundant in their ability to stimulate IGFBP-6 expression in response to rexinoid.
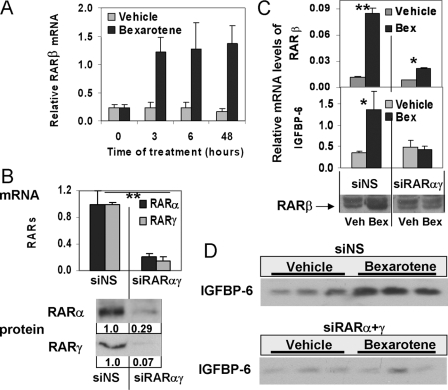
FIGURE 3.
The role of RARβ in the induction of IGFBP-6. A, time course of the mRNA levels of RARβ in HMECs treated with vehicle or 1 μm bexarotene. B, combined knockdown of RARα and RARγ in HMECs. Top, suppression of RARα and RARγ mRNAs upon transfection with gene-specific siRNAs. Bottom, changes in the protein levels of RARα and RARγ following 48-h knockdown by combined siRNAs. C, the effect of combined knockdown of RARα and RARγ on RARβ and IGFBP-6 expression. Top, RARβ mRNA levels upon Bex in HMECs transfected with nonspecific control siRNA or siRNAs against RARα and RARγ. Middle, IGFBP-6 mRNA levels after Bex in HMECs transfected with nonspecific control siRNA or siRNAs against RARα and RARγ. Bottom, Western blots showing the levels of cellular RARβ in response to Bex in HMECs transfected with nonspecific control siRNA or siRNAs against RARα and RARγ. D, Western blots demonstrating changes in the levels of secreted IGFBP-6 in response to bexarotene or vehicle in tissue culture media of HMECs following knockdown with a nonspecific siRNA (siNS) or siRNAs against RARα and RARγ.
FIGURE 2.
Characterization of the effects of individual and combined knockdown of retinoid X receptors and retinoic acid receptors on the response of IGFBP-6 to bexarotene. A, RXRα expression following siRNA-mediated knockdown in HMECs. mRNA levels were measured by quantitative reverse transcription-PCR. Inset, RXRα protein levels in cells transfected with nonspecific (NS) or RXRα-specific (RXRα) siRNA. The numbers indicate protein abundance normalized to glyceraldehyde-3-phosphate dehydrogenase, relative to control. B, IGFBP-6 mRNA levels following transfection of HMECs with RXRα siRNAs. C, comparison of the effects of nonspecific and RXR-specific siRNAs on the induction of IGFBP-6 by bexarotene. Top, RXR mRNA levels in cells transfected with control (siNS) or the combination of RXRα, -β, and -γ siRNAs. Bottom, mRNA levels of IGFBP-6 following knockdown of RXRα, RXRβ, and RXRγ. D, top, expression levels of RARs following siRNA knockdown of the individual receptors in HMECs. Bottom, induction of IGFBP-6 mRNA by bexarotene in HMECs following knockdown with a nonspecific siRNA (siNS) or siRNAs against RARα, RARβ, or RARγ.
To determine which potential RXR partner receptors may mediate the induction of IGFBP-6, we knocked down RARα, RARβ, or RARγ prior to the addition of bexarotene. All of these receptors had detectable transcript levels at base line, with RARβ mRNA levels being less than 2% of that of RARα and less than 0.75% of RARγ. siRNA treatment of retinoic acid receptors resulted in at least 70% suppression of their mRNAs, as determined by quantitative reverse transcription-PCR (Fig. 2D, top). Knockdown of RARβ was able to block the up-regulation of IGFBP-6 (Fig. 2D, bottom), whereas suppression of RARα or RARγ did not abrogate but only partially reduced the response of IGFBP-6 to bexarotene. Similar results were obtained when repeating the knockdown experiments in T47D cells (data not shown).
Induction of RARβ Is Critical for the Rexinoid-dependent Up-regulation of IGFBP-6 Expression—As shown in Fig. 2D, RARβ was the only retinoid receptor whose knockdown alone blocked the rexinoid response of IGFBP-6. Although RARβ is expressed at a low level in untreated cultured cells, the time course of RARβ expression shows that RARβ is greatly induced as early as 3 h following bexarotene treatment (Fig. 3A), and cells sustained elevated protein levels of RARβ 48 h after rexinoid exposure (Fig. 3C, bottom).
Simultaneous suppression of RARα and RARγ by siRNAs in HMECs reduced their mRNA levels by over 80% and protein levels correspondingly (Fig. 3B). The combined knockdown of RARα and RARγ greatly inhibited the induction of RARβ by bexarotene (Fig. 3C, top). Furthermore, the lack of induction of RARβ in RARα/γ-suppressed cells was associated with the abrogation of the up-regulation of IGFBP-6 transcript levels and protein by bexarotene (Fig. 3, C (middle) and D), suggesting that the induction of RARβ is critical for the up-regulation of IGFBP-6.
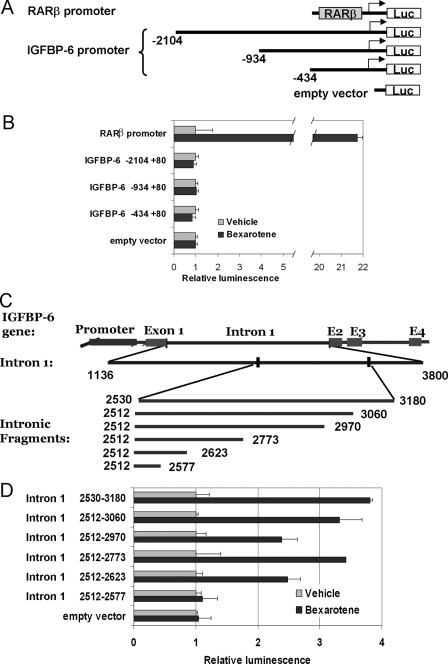
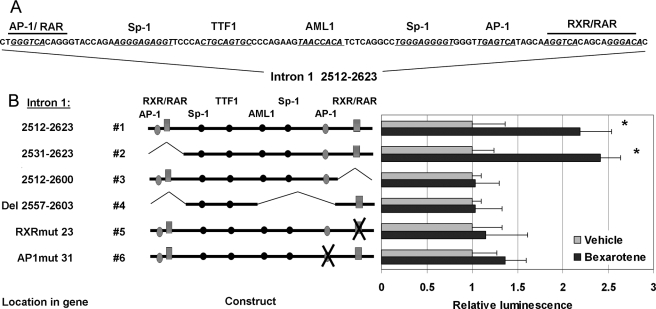
Identification of the Bexarotene-responsive Elements in the IGFBP-6 Gene—To further elucidate the mechanism by which retinoids regulate the expression of IGFBP-6, we isolated the 5′-regulatory sequences, including the 5′-untranslated region, of the IGFBP-6 gene upstream of the coding region and cloned a series of deletion constructs into the luciferase reporter vector pGL3 basic (Fig. 4A). The expression vectors containing these promoter fragments were transiently transfected into HMECs and T47D cells, and the bexarotene-dependent increase of luciferase activity was assayed. None of the reporter constructs spanning the proximal 2 kb of the putative IGFBP-6 promoter exhibited increased activity upon the addition of bexarotene (Fig. 4B). Therefore, the three introns and the 3′-untranslated region of the IGFBP-6 gene were also cloned and tested. All of the resulting reporter constructs proved unresponsive to bexarotene, with the exception of one containing the distal region of intron 1. Within this segment, spanning ∼650 bp (Fig. 4C, positions 2531–3180 in the IGFBP-6 gene), we identified an intronic region that showed identical response to bexarotene as the native IGFBP-6 gene. By creating a series of deletion mutants, we found that this rexinoid-responsive region is a complex regulatory element comprising 112 bp (2512–2623; Fig. 4D).
FIGURE 4.
Identification of rexinoid-responsive elements within the IGFBP-6 gene. A, promoter segments of the IGFBP-6 gene (-2104 to +80) inserted in the pGL3 basic luciferase vector. B, luciferase activity of the IGFBP-6 promoter constructs in bexarotene-treated HMECs expressed as -fold change over vehicle treatment. Reporter activity was measured using the dual luciferase assay (Promega), and enzyme activity was normalized to Renilla luciferase activity. C, schematic diagram of the reporter constructs containing serial deletions of the distal region of intron 1 from the IGFBP-6 gene. D, comparison of the luciferase activity in HMECs transfected with IGFBP-6 intronic reporter constructs and treated with vehicle or 1 μm bexarotene.
To identify relevant intron 1 response elements, this intron 1 region was studied in silico using the Transcription Element Search Software (available on the World Wide Web). The analysis suggested that the rexinoid-responsive region of intron 1 contains potential response elements for AP-1, RXR-RAR, Sp1, TTF1, and AP-3 between the locations 2512 and 2623 (Fig. 5A). Therefore, we next created and tested various 5′, 3′, and internal deletion constructs of intron 1 to determine which of these elements were functionally required for the activity of the construct (Fig. 5B). Induction of the luciferase reporter gene upon bexarotene treatment was moderate but was highly reproducible over multiple experiments (significant at p < 0.005 between groups by two-way analysis of variance). Removal of the proximal, overlapping AP-1/RXR-RAR response elements (see construct 2) did not reduce the activity of the reporter (compare construct 2 with construct 1; Fig. 5B). However, deletion of the DR5 consensus RXR-RAR response element (AGGTCA-5-GGGACA) at position 2605–2622 (construct 3) completely eliminated the bexarotene response of the reporter (Fig. 5B). When the AP-1 binding site (TGAGTCA) upstream of the RXRE was deleted (Del 2557–2603) while the distal RXR·retinoic acid response element was left intact, again all response to retinoids was lost. To establish whether the distal RXR and AP-1 binding sites are indeed critical, these putative response elements were individually altered by site-directed mutagenesis. Inactivation of either the putative distal RXRE (construct 5, RXRmut23, 2605–2610) or the AP-1 response element (construct 6, APmut31, 2593–2599) resulted in significant reduction or the abrogation of the bexarotene response. Similar results were obtained in both T47D and HMECs, demonstrating that both the distal RXRE and the adjacent AP-1 elements are required in rexinoid-induced transactivation of the IGFBP-6 gene.
FIGURE 5.
Analysis of reporter constructs to identify responsive elements to bexarotene in intron 1 of the IGFBP-6 gene. A, sequence and putative response elements (underlined) of the bexarotene-responsive fragment (positions 2512–2623) of intron 1 of the IGFBP-6 gene. Nucleotides replaced by site-directed mutagenesis of the putative AP-1 and RXR-response elements are highlighted. B, comparison of internal deletion reporter constructs (left) of the 2512–2623 fragment of intron 1 of IGFBP-6 created to identify critical response elements to bexarotene. Luciferase activity was measured (right) in HMECs treated with vehicle or 1 μm bexarotene after transfection with reporter constructs harboring deletions of individual response elements within the 2512–2623 fragment of intron 1 of the IGFBP-6 gene. Data represent the means of normalized light units ± S.D. from five different experiments (performed in triplicate) graphed as -fold induction over vehicle-treated controls. Statistical significance of the changes was determined by two-way analysis of variance tests. *, p < 0.005.
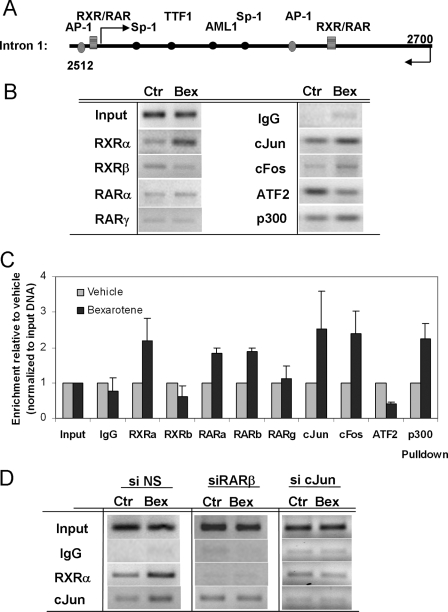
To determine which transcription factors in vivo bind the regulatory region shown to be bexarotene-responsive in reporter assays, we performed ChIP assays in T47D cells targeting the rexinoid-responsive region of intron 1 (Fig. 6A). In the absence of bexarotene, this segment was occupied by cJun and ATF2 and showed weak binding of RXRα, RXRβ, and the coactivator p300 (Fig. 6B), suggesting that in the inactive state, a cJun·ATF2 heterodimer binds the element. Serial ChIP assays indicated that upon the addition of bexarotene, increased binding of RXRα to the active intronic segment occurred no earlier than 12 h (data not shown). Further ChIP analyses showed that recruitment of RXRα and cJun were the dominant changes on the response element, but increased binding of cFos, RARα, and p300 were also observed (Fig. 6, B and C). At the same time, bexarotene treatment markedly reduced its occupancy by ATF2 and RXRβ (Fig. 6, B and C). Enhanced recruitment of RARβ upon bexarotene could be detected by quantitative real time PCR but not conventional PCR analyzed on gel (Fig. 6C). The concurrent binding of AP-1 and retinoid receptors is consistent with our reporter assay data showing that selective deletion of either the RXR-RAR response element or the AP-1 site abrogates the activation of the bexarotene-responsive construct.
FIGURE 6.
ChIP assay in T47D cells to assess recruitment of transcription factors to the rexinoid-responsive fragment of the IGFBP-6 gene after 12 h of bexarotene treatment. A, design of the PCR assay encompassing the bexarotene-responsive element in intron 1 of IGFBP-6. B, PCR amplification of the DNA segment 2524–2697 following chromatin immunoprecipitation with antibodies against RXRα, RXRβ, RARα, RARγ, cJun, cFos, ATF2, and p300. Normal rabbit IgG was used as negative control. C, determination of immunoprecipitated chromatin upon bexarotene treatment using quantitative real time PCR. Samples were normalized to their respective input, and values are expressed relative to vehicle-treated counterparts. D, binding of RXRα in the presence or absence of RARβ or cJun to the IGFBP-6 gene. Chromatin IP detecting recruitment of RXRα and cJun to the bexarotene-responsive site of IGFBP-6 in controls (si NS) and cells after depletion of RARβ or cJun (siRARβ, si cJun).
Both RARβ and cJun Are Required for the Recruitment of RXRα to the IGFBP-6 Gene—To determine whether RARβ is essential for the recruitment of transcription factors to the rexinoid-responsive IGFBP-6 enhancer, we performed chromatin immunoprecipitation experiments in bexarotene-treated cells following the knockdown of RARβ. The suppression of RARβ was associated with the loss of recruitment of RXRα and cJun to the enhancer in response to bexarotene (Fig. 6D). Conversely, ATF2 binding markedly increased upon bexarotene treatment, whereas binding of cFos diminished (data not shown). In a parallel experiment, the siRNA-mediated suppression of cJun removed cJun and decreased RXRα occupancy to the response element. This demonstrates that the cooperative action of AP-1 and induced RARβ are necessary for the recruitment of RXRα to the IGFBP-6 gene.
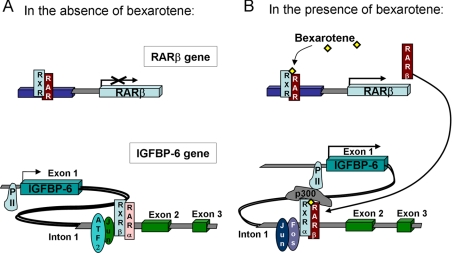
Based on these observations, we propose the model shown in Fig. 7 to explain how bexarotene induces the expression of IGFBP-6. In unstimulated cells or in the absence of elevated levels of RARβ, the identified response element in intron 1 of the IGFBP-6 gene may be occupied by ATF2·cJun and RXRβ· RARα or -γ dimers and remains inactive (Fig. 7, left). Treatment of the cells by the cancer-preventive rexinoid bexarotene results in the early induction of RARβ, which binds RXR proteins, in particular RXRα (Fig. 7, right). This dimeric RXR· RARβ complex then binds to the IGFBP-6 intron, displaces RXRβ and ATF2, and causes recruitment of cJun and cFos, which then facilitates binding of p300 and RNA polymerase II, ultimately inducing IGFBP-6 mRNA expression, through a unique collaboration between complexes of RXR·RAR heterodimers and Jun·Fos heterodimers.
FIGURE 7.
The role of RARβ induction and RXR-AP1 cooperation in the regulation of IGFBP-6. Shown is a schematic diagram showing the state of the rexinoid-responsive gene IGFBP-6 in the absence (A) or presence (B) of bexarotene.
DISCUSSION
In the present study, we characterized the ability of the RXR-selective retinoid bexarotene to up-regulate the expression of IGFBP-6. We demonstrated that bexarotene regulates IGFBP-6 through a complex enhancer element in intron 1 of the gene, and this regulation requires permissive interaction between the AP-1 transcription factor complex and RXR·RARβ heterodimers. Although the involvement of RARβ is essential for the bexarotene-induced up-regulation of IGFBP-6, its own induction requires RARα or RARγ. As shown in Fig. 7, induction of RARβ leads to the recruitment of RXRα, the displacement of ATF2 and RXRβ, and the binding of a cJun·cFos heterodimer, ultimately recruiting the cofactor p300 and RNA polymerase II to drive the induction of IGFBP-6.
In previous studies of rexinoid-induced biomarkers, we discovered that bexarotene modulated the expression of IGFBP-6, along with other growth-regulatory genes, such as COX-2, cyclin D1, and RARβ (11). Because the effect of IGFs in the tissues is modulated by the abundance of IGF-binding proteins (17), the regulation of IGF-binding proteins by bexarotene may contribute to the cancer-preventive activity of the rexinoid. IGFBP-6 has a high affinity for binding IGF-II and is able to inhibit the growth of various cancer cells and activate programmed cell death pathways (22, 23). The pleiotropic effects of RXR-selective retinoids make it likely that the cancer-preventive effect of this class of agents is due to regulation of many genes. However, understanding how bexarotene regulates IGFBP-6 helps create the next set of biomarkers that can be used as surrogates of breast cancer as an end point of clinical studies. Furthermore, the mechanism and exact composition of the transcriptional complexes occupying promoters or enhancers in biomarker genes reveal novel targets for new, improved chemopreventive agents with better efficacy and less toxicity.
IGFBP-6 has a high affinity for binding IGF-II and is able to inhibit the growth of various cancer cells (22, 23) and low serum levels of IGFBP-6 in patients correlated with invasive breast cancer (24). Recombinant IGFBP-6 in physiologic concentrations is able to suppress growth of HMECs.3 However, blocking IGFBP-6 expression using siRNAs did not reverse the antiproliferative effect of bexarotene in breast epithelial cells, indicating that the induction of IGFBP-6 is not solely responsible for the growth-suppressive effects and possibly the cancer-preventive activity of this rexinoid. Indeed, we have previously shown that rexinoids modulate many growth-regulatory genes, such as cyclin D1, RARβ, or COX-2, in addition to IGFBP-6. These results strongly suggest that rexinoids act through a network of pathways to suppress cancer formation.
RARβ has been shown to distinctly mediate the growth-inhibitory effect of retinoids and as such is a potential target for cancer-preventive treatment (27–29). Silencing of the RARβ gene and repressing methylation of the promoter were shown to be causative reasons in the genesis of various cancers (30, 31). Studies in RARβ (-/-) teratocarcinoma cells have identified genes that were specifically regulated by RARβ(2) (32). Similarly, our data show that RARβ plays a critical role in the induction of IGFBP-6 and cannot be substituted by other RARs. However, in breast epithelial cells, the abundance of RARβ is low, and the up-regulation of IGFBP-6 is preceded by the induction of RARβ. The RARβ gene is directly regulated by RXR·RAR heterodimers through a classical retinoic acid response element, and its expression can occur in a tissue-specific manner. The knockdown and chromatin binding experiments furthermore indicate that although RARα or RARγ is required for the up-regulation of RARβ, they are not sufficient to activate the intronic bexarotene-responsive enhancer of IGFBP-6.
A high level of redundancy was observed on the IGFBP-6 gene with retinoid X receptors. Although ChIP results indicate that RXRβ binding to the response element is not needed for gene induction, individual knock-down of RXRα was not sufficient to abrogate it. This suggests that RXRβ can substitute for RXRα when the latter is depleted. Because RXRγ is not detectable in these cells (data not shown), this isoform is unlikely to participate in the regulation of IGFBP-6.
Our results demonstrate that the AP-1 transcription factor also has an integral role in the regulation of IGFBP-6 expression. Our data showed a reciprocal relationship between cFos and ATF2, where cFos and cJun replaced ATF2 on the response element during activation of IGFBP-6. ATF2 (activating transcription factor 2, CREBP, HB16, CREB2) is a member of the ATF/CREB family of transcription factors. ATF2 binds the promoters of various viral and cellular genes, many of which are important in cell growth and differentiation. Loss of ATF2 can promote formation of breast cancer (33). The ATF2 protein can form either homodimers or heterodimers with cJun and other members of the ATF/CREB and Jun/Fos families. Thus, increased binding of ATF2 to the enhancer may compete off cFos from active cFos-cJun heterodimers. Our data suggest that an ATF2-cJun dimer is either inhibitory or at best an inactive complex on the IGFBP-6 enhancer element. Replacement of the ATF2-cJun dimers by cJun·cFos heterodimers greatly increased transactivation of the enhancer and suggests that cJun·cFos heterodimers can enter a permissive interaction with RXR·RAR heterodimers. ATF2 and p300 can cooperate in the control of transcription by forming a protein complex that is responsive to differentiation-inducing signals, such as retinoic acid or E1A (34). However, on the IGFBP-6 enhancer element, greater p300 occupancy was seen upon formation of an AP-1/retinoid receptor complex and decreased ATF2 binding.
In neuroblastoma cells, the AP-1 activator TPA up-regulates IGFBP-6 (35). In our experiments, suppression of cFos levels by siRNA causes decreased base-line expression and blunts the induction of IGFBP-6 in HMECs (data not shown). Previous studies showed that growth suppression by retinoids is associated with AP-1 antagonism (36), and retinoid-induced inhibition of AP-1 was dependent on the orphan receptor COUP-TF (37). In particular, RARβ-selective ligands were shown to inhibit AP-1 and induce apoptosis in breast cells (38). Gel shift assays demonstrated that RXRα inhibits Jun and Fos DNA binding and that 9-cis-retinoic acid enhances this inhibition, suggesting that a mechanism involving direct protein-protein interaction between RXR and AP-1 components mediates the inhibitory effect observed in vivo (39). Furthermore, domains of RAR responsible for the inhibitory action of retinoids on AP-1 activity were identified (40). These studies and the natural functional antagonism demonstrated by retinoids and mitogenic pathways ending on AP-1 suggest that genes with antioncogenic properties would be regulated in an opposing manner by retinoid receptors and AP-1. This discovery indicates that, depending on the cellular context and the mechanism of action of the drug used, AP-1 may be involved in regulatory mechanisms that counteract cell proliferation. This notion opens a new path to the development of pharmacological agents that aim to modulate AP-1 activity.
The studies presented here demonstrate a novel mechanism of cooperative action of the AP-1 transcription factor and retinoid receptors through a complex response element in the first intron of the IGFBP-6 gene. Furthermore, we demonstrate that bexarotene-induced RARβ is essential for the formation of an active AP-1/RXR transcriptional complex, revealing a novel mechanism by which RARβ can mediate the tumor-preventing effects of RXR-selective retinoids.
This work was supported, in whole or in part, by National Institutes of Health, NCI, Grants RO1 CA10121, RO1 CA78480, and U19 CA086809 (to P. H. B.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: IGF, insulin-like growth factor; siRNA, small interfering RNA; HMEC, human mammary epithelial cell; ChIP, chromatin immunoprecipitation; RXR, retinoid X receptor; RXRE, RXR element.
I. P. Uray, Q. Shen, H.-S. Seo, H. Kim, W. W. Lamph, R. P. Bissonnette, and P. H. Brown, unpublished observation.
References
- 1.Warrell, R. P., Jr., Maslak, P., Eardley, A., Heller, G., Miller, W. H., Jr., and Frankel, S. R. (1994) Leukemia 8 929-933 [PubMed] [Google Scholar]
- 2.Moon, R. C., and Constantinou, A. I. (1997) Breast Cancer Res. Treat. 46 181-189 [DOI] [PubMed] [Google Scholar]
- 3.Moon, R. C., Thompson, H. J., Becci, P. J., Grubbs, C. J., Gander, R. J., Newton, D. L., Smith, J. M., Phillips, S. L., Henderson, W. R., Mullen, L. T., Brown, C. C., and Sporn, M. B. (1979) Cancer Res. 39 1339-1346 [PubMed] [Google Scholar]
- 4.Hong, W. K., Lippman, S. M., Itri, L. M., Karp, D. D., Lee, J. S., Byers, R. M., Schantz, S. P., Kramer, A. M., Lotan, R., and Peters, L. J. (1990) N. Engl. J. Med. 323 795-801 [DOI] [PubMed] [Google Scholar]
- 5.Lippman, S. M., and Hong, W. K. (1992) J. Natl. Cancer Inst. Monogr. 13 111-115 [PubMed] [Google Scholar]
- 6.Wu, K., Kim, H. T., Rodriquez, J. L., Munoz-Medellin, D., Mohsin, S. K., Hilsenbeck, S. G., Lamph, W. W., Gottardis, M. M., Shirley, M. A., Kuhn, J. G., Green, J. E., and Brown, P. H. (2000) Clin. Cancer Res. 6 3696-3704 [PubMed] [Google Scholar]
- 7.Teplitzky, S. R., Kiefer, T. L., Cheng, Q., Dwivedi, P. D., Moroz, K., Myers, L., Anderson, M. B., Collins, A., Dai, J., Yuan, L., Spriggs, L. L., Blask, D. E., and Hill, S. M. (2001) Cancer Lett 168 155-163 [DOI] [PubMed] [Google Scholar]
- 8.Wu, K., Kim, H. T., Rodriquez, J. L., Hilsenbeck, S. G., Mohsin, S. K., Xu, X. C., Lamph, W. W., Kuhn, J. G., Green, J. E., and Brown, P. H. (2002) Cancer Epidemiol. Biomarkers Prev. 11 467-474 [PubMed] [Google Scholar]
- 9.Liby, K., Rendi, M., Suh, N., Royce, D. B., Risingsong, R., Williams, C. R., Lamph, W., Labrie, F., Krajewski, S., Xu, X., Kim, H., Brown, P., and Sporn, M. B. (2006) Clin. Cancer Res. 12 5902-5909 [DOI] [PubMed] [Google Scholar]
- 10.Cohen, M. H., Hirschfeld, S., Flamm Honig, S., Ibrahim, A., Johnson, J. R., O'Leary, J. J., White, R. M., Williams, G. A., and Pazdur, R. (2001) Oncologist 6 4-11 [DOI] [PubMed] [Google Scholar]
- 11.Kim, H. T., Kong, G., Denardo, D., Li, Y., Uray, I., Pal, S., Mohsin, S., Hilsenbeck, S. G., Bissonnette, R., Lamph, W. W., Johnson, K., and Brown, P. H. (2006) Cancer Res. 66 12009-12018 [DOI] [PubMed] [Google Scholar]
- 12.Kong, G., Kim, H. T., Wu, K., DeNardo, D., Hilsenbeck, S. G., Xu, X. C., Lamph, W. W., Bissonnette, R., Dannenberg, A. J., and Brown, P. H. (2005) Cancer Res. 65 3462-3469 [DOI] [PubMed] [Google Scholar]
- 13.Sachdev, D., and Yee, D. (2001) Endocr. Relat. Cancer 8 197-209 [DOI] [PubMed] [Google Scholar]
- 14.Osborne, C. K., Coronado, E. B., Kitten, L. J., Arteaga, C. I., Fuqua, S. A., Ramasharma, K., Marshall, M., and Li, C. H. (1989) Mol. Endocrinol. 3 1701-1709 [DOI] [PubMed] [Google Scholar]
- 15.Bates, P., Fisher, R., Ward, A., Richardson, L., Hill, D. J., and Graham, C. F. (1995) Br. J. Cancer 72 1189-1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Giani, C., Cullen, K. J., Campani, D., and Rasmussen, A. (1996) Breast Cancer Res. Treat 41 43-50 [DOI] [PubMed] [Google Scholar]
- 17.Sachdev, D., and Yee, D. (2006) J. Mammary Gland Biol. Neoplasia 11 27-39 [DOI] [PubMed] [Google Scholar]
- 18.Mehrian-Shai, R., Chen, C. D., Shi, T., Horvath, S., Nelson, S. F., Reichardt, J. K., and Sawyers, C. L. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 5563-5568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mita, K., Zhang, Z., Ando, Y., Toyama, T., Hamaguchi, M., Kobayashi, S., Hayashi, S., Fujii, Y., Iwase, H., and Yamashita, H. (2007) Jpn. J. Clin. Oncol. 37 575-582 [DOI] [PubMed] [Google Scholar]
- 20.Kelley, K. M., Oh, Y., Gargosky, S. E., Gucev, Z., Matsumoto, T., Hwa, V., Ng, L., Simpson, D. M., and Rosenfeld, R. G. (1996) Int. J. Biochem. Cell Biol. 28 619-637 [DOI] [PubMed] [Google Scholar]
- 21.Imai, Y., Moralez, A., Andag, U., Clarke, J. B., Busby, W. H., Jr., and Clemmons, D. R. (2000) J. Biol. Chem. 275 18188-18194 [DOI] [PubMed] [Google Scholar]
- 22.Bach, L. A., Hsieh, S., Brown, A. L., and Rechler, M. M. (1994) Endocrinology 135 2168-2176 [DOI] [PubMed] [Google Scholar]
- 23.Sueoka, N., Lee, H. Y., Walsh, G. L., Fang, B., Ji, L., Roth, J. A., LaPushin, R., Hong, W. K., Cohen, P., and Kurie, J. M. (2000) Am. J. Respir. Cell Mol. Biol. 23 297-303 [DOI] [PubMed] [Google Scholar]
- 24.Kaulsay, K. K., Ng, E. H., Ji, C. Y., Ho, G. H., Aw, T. C., and Lee, K. O. (1999) Eur. J. Endocrinol. 140 164-168 [DOI] [PubMed] [Google Scholar]
- 25.Zhu, Y., Wang, A., Liu, M. C., Zwart, A., Lee, R. Y., Gallagher, A., Wang, Y., Miller, W. R., Dixon, J. M., and Clarke, R. (2006) Int. J. Oncol. 29 1581-1589 [PubMed] [Google Scholar]
- 26.DeNardo, D. G., Kim, H. T., Hilsenbeck, S., Cuba, V., Tsimelzon, A., and Brown, P. H. (2005) Mol. Endocrinol. 19 362-378 [DOI] [PubMed] [Google Scholar]
- 27.Li, X. S., Shao, Z. M., Sheikh, M. S., Eiseman, J. L., Sentz, D., Jetten, A. M., Chen, J. C., Dawson, M. I., Aisner, S., Rishi, A. K., Gutien, P., Schnap, L., and Fontai, F. A. (1995) J. Cell. Physiol. 165 449-458 [DOI] [PubMed] [Google Scholar]
- 28.Liu, Y., Lee, M. O., Wang, H. G., Li, Y., Hashimoto, Y., Klaus, M., Reed, J. C., and Zhang, X. (1996) Mol. Cell. Biol. 16 1138-1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wan, H., Oridate, N., Lotan, D., Hong, W. K., and Lotan, R. (1999) Cancer Res. 59 3518-3526 [PubMed] [Google Scholar]
- 30.Virmani, A. K., Rathi, A., Zochbauer-Muller, S., Sacchi, N., Fukuyama, Y., Bryant, D., Maitra, A., Heda, S., Fong, K. M., Thunnissen, F., Minna, J. D., and Gazdar, A. F. (2000) J. Natl. Cancer Inst. 92 1303-1307 [DOI] [PubMed] [Google Scholar]
- 31.Sirchia, S. M., Ren, M., Pili, R., Sironi, E., Somenzi, G., Ghidoni, R., Toma, S., Nicolo, G., and Sacchi, N. (2002) Cancer Res. 62 2455-2461 [PubMed] [Google Scholar]
- 32.Zhuang, Y., Faria, T. N., Chambon, P., and Gudas, L. J. (2003) Mol. Cancer Res. 1 619-630 [PubMed] [Google Scholar]
- 33.Maekawa, T., Shinagawa, T., Sano, Y., Sakuma, T., Nomura, S., Nagasaki, K., Miki, Y., Saito-Ohara, F., Inazawa, J., Kohno, T., Yokota, J., and Ishii, S. (2007) Mol. Cell. Biol. 27 1730-1744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kawasaki, H., Song, J., Eckner, R., Ugai, H., Chiu, R., Taira, K., Shi, Y., Jones, N., and Yokoyama, K. K. (1998) Genes Dev. 12 233-245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chambery, D., De Galle, B., Ehrenborg, E., and Babajko, S. (2000) Growth Horm. IGF Res. 10 349-359 [DOI] [PubMed] [Google Scholar]
- 36.Wan, H., Hong, W. K., and Lotan, R. (2001) Cancer Res. 61 556-564 [PubMed] [Google Scholar]
- 37.Lin, F., Kolluri, S. K., Chen, G. Q., and Zhang, X. K. (2002) J. Biol. Chem. 277 21414-21422 [DOI] [PubMed] [Google Scholar]
- 38.Li, Y., Hashimoto, Y., Agadir, A., Kagechika, H., and Zhang, X. (1999) J. Biol. Chem. 274 15360-15366 [DOI] [PubMed] [Google Scholar]
- 39.Salbert, G., Fanjul, A., Piedrafita, F. J., Lu, X. P., Kim, S. J., Tran, P., and Pfahl, M. (1993) Mol. Endocrinol. 7 1347-1356 [DOI] [PubMed] [Google Scholar]
- 40.DiSepio, D., Sutter, M., Johnson, A. T., Chandraratna, R. A., and Nagpal, S. (1999) Mol. Cell. Biol. Res. Commun. 1 7-13 [DOI] [PubMed] [Google Scholar]