Abstract
Low volatility, lipid-like cuticular hydrocarbon pheromones produced by Drosophila melanogaster females play an essential role in triggering and modulating mating behavior, but the chemosensory mechanisms involved remain poorly understood. Recently, we showed that the CheB42a protein, which is expressed in only 10 pheromone-sensing taste hairs on the front legs of males, modulates progression to late stages of male courtship behavior in response to female-specific cuticular hydrocarbons. Here we report that expression of all 12 genes in the CheB gene family is predominantly or exclusively gustatory-specific, and occurs in many different, often non-overlapping patterns. Only the Gr family of gustatory receptor genes displays a comparable variety of gustatory-specific expression patterns. Unlike Grs, however, expression of all but one CheB gene is sexually dimorphic. Like CheB42a, other CheBs may therefore function specifically in gustatory perception of pheromones. We also show that CheBs belong to the ML superfamily of lipid-binding proteins, and are most similar to human GM2-activator protein (GM2-AP). In particular, GM2-AP residues involved in ligand binding are conserved in CheBs but not in other ML proteins. Finally, CheB42a is specifically secreted into the inner lumen of pheromone-sensing taste hairs, where pheromones interact with membrane-bound receptors. We propose that CheB proteins interact directly with lipid-like Drosophila pheromones and modulate their detection by the gustatory signal transduction machinery. Furthermore, as loss of GM2-AP in Tay-Sachs disease prevents degradation of GM2 gangliosides and results in neurodegeneration, the function of CheBs in pheromone response may involve biochemical mechanisms critical for lipid metabolism in human neurons.
Detection of pheromones in animals involves specialized chemosensory organs of two distinct types. Volatile pheromones are detected, often at great distances from the source, by highly sensitive olfactory organs (1). However, many pheromones that trigger specific sexual behaviors are poorly volatile and act through direct contact with chemosensory organs (2, 3). In Drosophila melanogaster, pheromones modulate multiple aspects of mating behavior (4, 5). The past few years have brought remarkable progress in our understanding of the mechanisms underlying pheromone perception by the olfactory system. In particular, cis-vaccenyl acetate (cVA),4 a volatile pheromone produced in the male ejaculatory bulb, is detected by one or perhaps two olfactory receptor proteins expressed in specific subsets of olfactory hairs on the antennae of both sexes (6-8). Despite the apparently identical responses it engenders in the peripheral olfactory organs of males and females (8), cVA inhibits male courtship behavior, but accelerates female mating (7). Another olfactory receptor is tuned to male-specific odors distinct from cVA, and two others respond indistinguishably to male and female odors (8).
Less is known about the molecular mechanisms involved in detecting a number of pheromones that have very low volatility, which also play critical roles in modulating Drosophila mating behavior (2, 4). These long-chain hydrocarbons (C23-C27) are produced by specialized epidermal cells in the abdomen. In particular, female-specific cuticular hydrocarbons are required for normal stimulation of male courtship behavior (9, 10), and even trigger homosexual male courtship when ectopically produced by males (11). Remarkably, whereas these compounds have very low volatility and are only effective over a radius of less than 1 cm (12), their detection may involve both olfactory (13, 14) and gustatory organs (15-17). Although there have been recent breakthroughs in the characterization of the olfactory perception of these cuticular hydrocarbon pheromones (8, 18), more is known about the gustatory organs and molecules involved. Differences in the number and innervation patterns of taste sensilla between the sexes (19, 20), as well as amputation and masking experiments (21, 22), provided early evidence that taste sensilla on male forelegs are involved in detection of pheromones. We and others therefore identified genes that are specifically expressed in subsets of gustatory sensilla on male front legs, at least two of which are required for normal male courtship response to female-specific cuticular hydrocarbons (16, 17, 23). CheA29a and CheB42a, two genes expressed specifically in subsets of gustatory sensilla on male front legs, defined two novel and unrelated families of small secreted proteins (23). More recently, we have shown that CheB42a is expressed in the same taste hairs as Gr68a (17), a putative gustatory pheromone receptor (16). Furthermore, loss of CheB42a results in a remarkably specific effect on the elaborate courtship behavior that males direct at females (17). Although unaffected in several other behaviors, including the early steps in the courtship sequence, CheB42a mutant males attempt to copulate earlier and more frequently than controls with other individuals of the same species, whether male or female, as long as they express female-specific hydrocarbons. These results indicate that CheB42a functions in gustatory perception of female cuticular hydrocarbon pheromones that modulate male courtship. By what mechanism does CheB42a modulate pheromone perception? What is the function of the other 11 CheBs encoded by the D. melanogaster genome? Based on the evidence in this report, we propose that CheB42a and other CheBs are gustatory-specific pheromone-binding proteins that modulate detection of specific contact pheromones.
EXPERIMENTAL PROCEDURES
Analysis of the Expression of Drosophila CheB Genes—RNA preparation, Northern blots, semi-quantitative PCR, and in situ hybridization were described previously (23). For Northern blots, RNA was extracted from appendages (antennae, legs, and wings), or heads (without antennae) separated in bulk by sieving (24). To allow for a rough comparison of the expression levels from different genes, all probes were of ∼500 nucleotides, labeled at similar specific activities with [32P]dCTP, and hybridized to identical filters that were exposed for the same amount of time (except for the exposure of the rp49 filter for 1/10th the time) to x-ray film. For semi-quantitative RT-PCR analysis, body parts of each type were hand-dissected from male or females, total nucleic acids were extracted, and cDNA generated using oligo(dT) primers. Amplification primers were designed to flank an intron so that the ratio of the short product resulting from amplification of the cDNA, to the long product resulting from amplification of genomic DNA, provides a relative measure of the mRNA levels in different samples (25). Expression of CheB38c was further analyzed by generating a CheB38c-Gal4 fusion, in which 5.2 kb of genomic DNA upstream of the CheB38c initiation codon were amplified by PCR from genomic DNA and inserted 5′ of the hsp70 TATA box in the pGATB vector (26). Transgenic flies were generated using standard methods (27) and GFP expression was analyzed for several independent insertions in crosses to UAS-GFP lines obtained from the Drosophila stock center (Bloomington, IN). For quantitative real-time PCR, RNA was extracted from whole flies with TRIzol (Invitrogen), treated with RNase-free DNase (Qiagen), and further purified on RNeasy/QIAamp columns (Qiagen). First-strand cDNA was synthesized using oligo(dT)20 and SuperScript III reverse transcriptase (Invitrogen). Real time PCR were performed in 96-well thin-wall plates (Applied Biosystems) using an Applied Biosystems 7300/7500 Real Time PCR System according to the manufacturer's suggested procedure, with the following modifications. One primer in each pair was designed to span an exon-exon junction, resulting in specific amplification of cDNA as confirmed in pilot experiments (data not shown). For every sample, specificity of amplifications was further confirmed by analysis of the dissociation curve. In every experiment, the relative concentration of each mRNA was obtained by comparison to a standard curve generated by amplification of serial dilutions of the corresponding cDNA product. To account for possible differences in total RNA concentration between samples, all values were normalized to the relative concentration of a ubiquitously expressed ribosomal protein mRNA (rp49). Independent replications of these measurements were performed at least once for seven of the 12 CheB genes, in four cases with different primers, with similar results (not shown).
Sequence Analysis—PSI-BLAST searches (28, 29) were performed for multiple iterations until no new significant similarities were uncovered, on the National Center for Biotechnology Information web server (www.ncbi.nlm.nih.gov). The sequences of over 100 ML domain proteins were obtained from the SMART server (30). Sequences were aligned using ClustalW (31) at the European Bioinformatics Institute server, and alignments edited and displayed using BOXSHADE. PSI-BLAST significance scores depend on a number of parameters, including the number of sequences in the data base at the time of the search, but in all cases discussed here, p < 1 × 10-10. Several PSI-BLAST searches using one of the CheBs as query indicate significant similarities with CheBrs and vice versa, and searches with either type of insect protein indicate significant similarities with human GM2-AP and related proteins from other organisms. In the same searches, similarities with NPC2, Der f2, and many other ML proteins are either not detected, or associated with much poorer significance scores than GM2-APs. In addition, PSI-BLAST searches using human GM2-AP as query yield significant scores with CheBrs but not with NPC2, Der f2, or many other MLs. An identical search restricted to arthropod sequences also identifies significant similarities with D. melanogaster CheBs but not to Drosophila NPC2 (32).
Immunochemistry—Our immunohistochemistry protocol was modified from Ref. 33. Briefly, front legs of Canton S males were dissected by hand and fixed overnight at 4 °C in phosphate-buffered saline (PBS) containing 4% paraformaldehyde. After three rinses in PBS solution, the legs were dehydrated in a series of ethanol solutions (25, 50, 75, and 90% and three times at 100%), and embedded in Paraplast Plus (Fisher Scientific) tissue embedding media at 60 °C. Samples were sectioned on a microtome (Olympus Cut 4060), collected on Superfrost plus microslides (Fisher Scientific), and dried at 39 °C overnight. The slides were then dewaxed in xylenes, rehydrated in 100, 75, 50, and 25% ethanol, rinsed in PBS, and blocked in PBT (PBS with 1% bovine serum albumin and 0.1% Triton X-100) containing 5% normal goat serum (Roche) for 1 h at room temperature. Slides were incubated for 4 h at room temperature with guinea pig anti-CheB42a antibody (23) at a 1:200 dilution, and when applicable, rat anti-PBPRP2 (34) at a 1:300 dilution for 4 h at room temperature, followed by three washes in PBT. The sections were then incubated for 2 h at room temperature with goat anti-guinea pig antibodies conjugated to Alexa Fluor 488 (Invitrogen), and when appropriate, with goat anti-rat antibodies conjugated to Alexa Fluor 568 (Molecular Probes), at dilutions of 1:200. After three washes in PBT, the stained sections were mounted in Vectashield and analyzed under a Leica TCS confocal microscope.
RESULTS
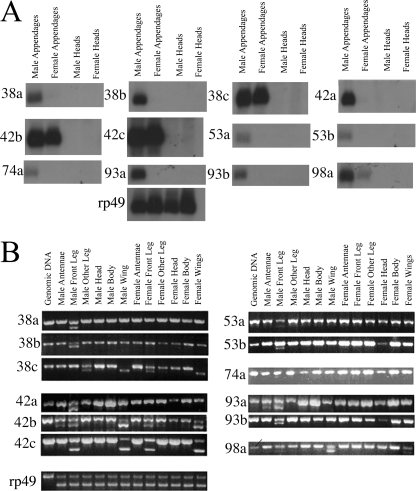
Genes of the Drosophila CheB Family Are Expressed in a Variety of Sexually Dimorphic, Gustatory-specific Patterns in Adult Flies—We have previously shown that, in addition to CheB42a, the mRNAs for three other D. melanogaster CheB genes are enriched and present at sexually dimorphic levels in a preparation of mixed fly appendages with chemosensory function (legs, wings, and third antennal segments) (23). The same mRNAs are not detected in heads lacking the olfactory third antennal segment. We have now confirmed and extended this observation using Northern blots to analyze the distribution of all 12 CheB mRNAs (Fig. 1A). The mRNAs for 8 CheB genes (CheB38a, -38b,-42a,-53a,-53b,-74a,-93a, and -93b) are detected only in male appendages, whereas CheB98a mRNA is found at higher levels in the appendages of males than those of females. In the above experiments, RNA was extracted from a pool of appendages with different chemosensory functions: legs and wings carry large numbers of gustatory sensilla, whereas the third antennal segment is the main olfactory organ of the fly (35). We therefore further analyzed expression of all 12 CheB genes in specific appendages (Fig. 1B) using a semi-quantitative RT-PCR method that allows comparison of relative concentrations of an mRNA among small samples of different tissues (23, 25). RT-PCR was performed on total nucleic acids extracted from distinct body parts dissected from either males or females: front legs, second and third pairs of legs (pooled), wings, third antennal segment, heads (without third antennal segment, but retaining many taste hairs on the labelum as well as inside the pharynx), and bodies (decapitated and with no legs or wings). As primers were designed to hybridize to either side of an intron, the relative concentrations of the short product resulting from amplification of the cDNA, and the long product, resulting from amplification of genomic DNA, provides a measure of the relative levels of mRNA in different samples (Fig. 1B). This analysis confirms that expression of all 12 CheB genes is specific to appendages and undetectable in heads and bodies. Furthermore, expression of all 12 CheB genes is found almost exclusively in the legs and wings, the two gustatory appendages. Only CheB42b and CheB93a are expressed detectably, albeit at levels lower than in wings and legs, in an olfactory appendage: the third antennal segment. The coincidence of the expression of CheB family genes with gustatory function is further reinforced by their preferential expression in the front legs relative to the other two pairs of legs, mirroring the higher concentrations of taste sensilla found on the front legs (19, 20). The only exceptions to this rule are CheB98a, which is not detectably expressed in the legs of animals of either gender, and the remarkable absence of CheB38c expression in the front legs of males, see below.
FIGURE 1.
CheB gene expression is almost exclusively sexually dimorphic and restricted to legs and wings, appendages enriched in gustatory hairs. A, Northern blot analysis of RNA extracted from appendages (antennae, legs, and wings) or heads (without antennae) from males or females. Radiolabeled probes are of similar size and specific activity, allowing for the approximate comparison of expression levels of different genes. B, semi-quantitative RT-PCR analysis of CheB gene expression. Expression of all 12 CheB genes in each body part was assessed semi-quantitatively (25). Relative expression levels in different tissues can be evaluated by comparing the ratio of the larger DNA product, amplified from intron-containing genomic DNA, to the smaller DNA product, resulting from amplification of the cDNA (23).
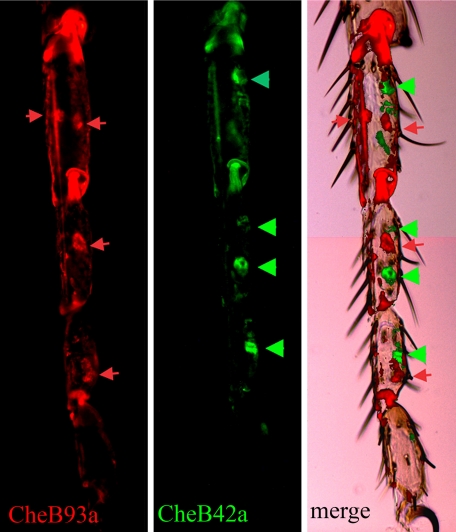
Although expression of the 12 CheBs is almost exclusively gustatory-specific, it occurs in a variety of patterns. CheBs can be assigned to one of two groups based on their expression (Table 1). CheB42a and seven other CheBs in Group I are expressed exclusively or at highest levels in the front legs of males. As expression of CheB42a is restricted to a subset of hairs on the front legs of males (17, 23), we compared it to that of CheB93a, another Group I gene, by double label in situ hybridization (Fig. 2). As previously reported for CheB42a (17, 23), both genes are expressed in punctate patterns that correspond to cells or groups of cells associated with gustatory sensilla. However, the mRNAs for CheB93a and CheB42a are never found in the same cell, or even in association with the same sensillum. Therefore, expression of CheB42a and CheB93a is associated with two distinct subsets of taste hairs on the front legs of males.
TABLE 1.
D. melanogaster CheBs can be classified into two groups based on their expression patterns
| Group I: specific expression in male front legs | Group II: specific or preferential expression on wings of either sex |
|---|---|
| CheB38a, CheB38b, CheB42a, CheB53a, CheB53b, CheB74a, and CheB93b are only expressed in male front legs | CheB38c is also expressed in legs, except male front legs |
| CheB93a is expressed in a distinct subset of hairs from CheB42a and, at lower levels, in the third antennal segment | CheB42b is also expressed in the third antennal segment of males, and front legs of both males and females |
| CheB42c is also expressed in the front legs of both males and females | |
| CheB98a is wing-specific, higher in males |
FIGURE 2.
CheB42a and CheB93a are expressed in two non-overlapping subsets of taste sensilla on male front legs. Frozen sections of male front legs were analyzed by in situ hybridization using fluorescently labeled oligonucleotides. Signal from probes for CheB42a and CheB93a are shown in green (arrowheads, right panel) and red (arrows, center panel), respectively, and superimposed on a Nomarsky image of the leg (left panel). Both genes are expressed at the base of gustatory sensilla recognizable by their curved shafts (23).
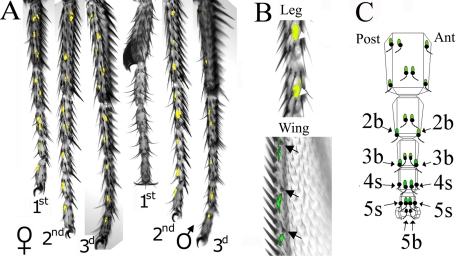
In contrast to Group I genes, all four CheB genes in Group II are expressed in both males and females. Furthermore, whereas three Group II genes are detectably expressed in legs, all are also expressed at high levels in the wings, another appendage with large numbers of gustatory sensilla. We therefore investigated in more detail the expression pattern of the Group II gene CheB38c by generating a transgenic construct in which 5.2 kb of sequences upstream of the CheB38c gene are fused to the yeast transcriptional activator Gal4 (26) (Fig. 3). In the presence of a UAS-GFP reporter construct, this transgene results in specific, punctate production of GFP on the legs and wings of males and females but, conspicuously, not on the front legs of males (Fig. 3A). This pattern is consistent with the distribution of CheB38c mRNA (Fig. 1B), suggesting that it faithfully replicates expression of the endogenous CheB38c gene. Higher resolution imaging shows that all GFP-expressing cells or groups of cells on the wings and legs are associated specifically with gustatory sensilla (Fig. 3B and data not shown). Therefore, the three CheB genes whose expression we have analyzed at the cellular level, CheB42a, CheB93a, and CheB38c, are expressed specifically in three non-overlapping subsets of taste hairs.
FIGURE 3.
CheB38c expression is associated with most gustatory sensilla on legs and wings, but is absent from male front legs and from most of the taste bristles on the legs that respond to sugar, salt, or bitter compounds. A and B, fluorescence and Nomarsky images from the appendages of animals expressing GFP under indirect control of CheB38c upstream sequences using the Gal4/UAS system (26) are superimposed. A, punctate GFP expression is visible in all three pairs of legs in females, but only in the second and third pairs of male legs. B, higher magnification images showing that GFP expression is found at the base of gustatory sensilla (arrows) on the legs, recognizable by their characteristic curvature and the lack of associated bracts (19) (upper panel), and on the wing margins by their smaller size, curved shaft and offset position relative to the wing margin (67) (lower panel). C, schematic map indicating the position (in black) of all 25 gustatory sensilla present on the five tarsal segments of female front legs. A green oval at the base of the sensillum indicates GFP expression under control of CheB38c-Gal4. The 10 sensilla on the tarsi of female front legs that respond to sugars, salts, or bitter compounds are indicated by arrows (36, 37). CheB38c expression is not found in the two 5b sensilla at the very tip of the leg that respond to both sugars and salts, or in the four 4s/5s, which respond to sugars, salts, and several bitter compounds. The only CheB38c-expressing sensilla known to respond to any food-related compound are the four 2b/3b sensilla, which respond to sugars but not to salts or bitter compounds.
Intriguingly, the combined expression of the 12 CheB genes is also restricted to a subset of all taste hairs of Drosophila, and is almost completely excluded from sensilla involved in detection of food stimuli. In particular, we have not detected any CheB expression in the many gustatory hairs of the head. Those include a high concentration of taste hairs on the surface of the labelum, many of which have been shown to function in gustatory detection of food components, as well as a number of taste hairs located inside the pharynx where they may allow the last sampling of any food before it is transferred to the digestive organs (35). Furthermore, whereas almost all sensilla on the front legs of males and females express CheB42a, CheB93a, or CheB38c, none of these genes is expressed in the two terminal sensilla that respond to sugars and salts (36) (Fig. 3C and data not shown). In addition, expression of CheB42a, CheB93a, or CheB38c is also undetectable in the four shorter sensilla on segments 4 and 5 that respond to bitter compounds (37). The only food-tasting sensilla in which we have found CheB expression are four sugar-sensing sensilla on the second and third tarsal segments of female legs (36) (Fig. 3C). Gustatory-specific expression of the 12 CheB genes therefore occurs largely in taste hairs that are unresponsive to food components such as sugars, salts, and bitter compounds.
In addition to displaying a variety of gustatory-specific patterns of expression, at least 11 of the 12 CheB genes are expressed in a sexually dimorphic manner. Expression of the 8 genes in Group I is detected only in males (Table 1). Although all four genes in Group II are expressed in both males and females, expression in the two sexes is either qualitatively or quantitatively different in at least three cases. In a pattern strikingly complementary to that of Group I genes, CheB38c is expressed in all legs and wings of both sexes, with the exception of the front legs of males (Figs. 1 and 3). CheB42b mRNA, whereas present at comparable levels in the legs and wings of both sexes, is expressed in the third antennal segments of males but not females (Fig. 1). Finally, whereas CheB98a expression is only detected in the wings, it is higher in males than females (Fig. 1). CheB42c is the only member of the CheB gene family for which we have not detected a difference in expression between the sexes (Fig. 1).
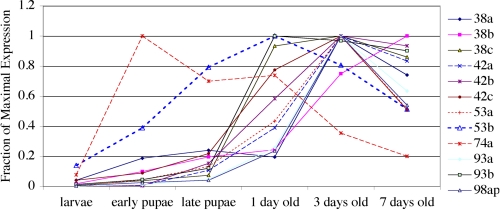
To test whether expression of CheBs is consistent with a role in the development rather than the function of adult gustatory organs, we measured the concentration of all 12 CheB mRNAs using quantitative PCR in larvae, early and late pupae, and in adult flies 1, 3, or 7 days after eclosion (Fig. 4). For all 12 CheBs, expression in larvae is at least 10 times lower than at later stages. In all but one case expression remains low in early pupae, and peaks either 1 or 3 days after eclosion. Although expression of CheB74a presents an exception, peaking in early pupae, it remains high after eclosion. As previously shown for CheB42a protein (23), expression of 10 of the other 11 CheB genes therefore peaks after completion of gustatory sensilla differentiation during late pupal stages (38). In summary, the 12 genes in the CheB family are expressed primarily in adult flies, in a variety of gustatory-specific patterns that are sexually dimorphic in all but one case.
FIGURE 4.
Expression of CheB genes initiates during late pupation and, in all but one case, peaks after eclosion. Quantitative PCR was performed on RNA extracted from animals at each developmental stage. The concentration of each mRNA is normalized relative to the concentration of a ubiquitous ribosomal protein mRNA (rp49) in the same sample, with the maximal concentration for that mRNA set at 1.
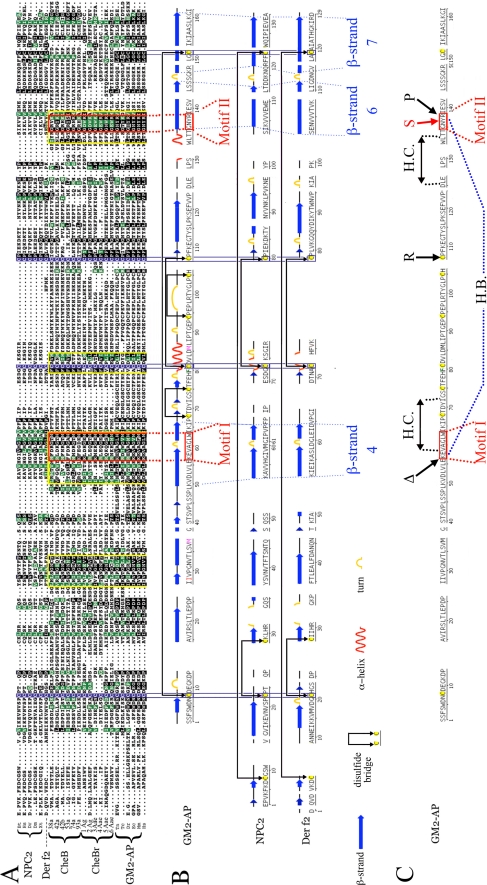
Drosophila CheB Proteins Are Related to GM2-activating Protein (GM2-AP), a Human Lipid-binding Protein Whose Absence Results in Neurodegeneration in Tay-Sachs Disease—Standard BLAST searches do not reveal significant similarities between CheBs and known proteins (17, 23). To test for the existence of proteins with relatively low levels of sequence similarity to CheBs, we used PSI-BLAST, a reiterative algorithm that detects conserved sequences present in multiple proteins (28, 29). Indeed, this approach revealed the existence of significant similarities between CheBs and a number of known proteins (see “Experimental Procedures”). In particular, several proteins encoded by the genomes of the mosquitoes Anopheles gambiae and Aedes aegypti are related to Drosophila CheBs, and we will therefore refer to them as CheBrs (for CheB-related proteins, Fig. 5 and data not shown). Furthermore, PSI-BLAST searches also indicate the existence of significant sequence similarities between Drosophila CheBs, mosquito CheBrs, and human GM2-AP. GM2-AP is a soluble protein that binds to the GM2 glycolipid, and whose absence in humans results in the Tay-Sachs neurodegenerative disease (39). GM2-AP belongs to the ML (MD-like, where MD stands for myeloid differentiation protein) superfamily of lipid-binding proteins found in all eukaryotes (40). Like CheBs and CheBrs, GM2-APs and other MLs are either known or predicted soluble proteins of 150-200 amino acids in length, with two or more disulfide bonds and signal peptides at their amino termini. Primary sequence identity among ML proteins is generally low (40), even among proteins that are functionally interchangeable (41). However, several ML proteins have been shown to fold into similar β-sandwiches, consisting largely of seven β-strands organized into two β-sheets; these include GM2-AP (42-45), Niemann-Pick type C2 protein (NPC2), a cholesterol-binding protein named for its role in the Niemann-Pick neurodegenerative disease (46), and two related dust mite antigens that play a major role in dust allergies, Der p2 (47) and Der f2 (48-50) (Fig. 5).
FIGURE 5.
CheB42a and other Drosophila CheBs are related to GM2-AP and other members of the ML family of lipid-binding proteins. A, alignment of six sequences from each of four groups: proteins related to NPC2, including the mite antigen Der f2, CheBs, CheBrs, and proteins related to GM2-AP. Three of the four cysteines involved in disulfide bonds in GM2-AP, NPC2, and Der f2 and the perfectly aligned cysteines in all other proteins in this alignment are boxed in purple. Residues present in human GM2-AP and conserved in at least 10 of the other 24 proteins in the alignment are shaded in black when identical, and green when similar. Sequence blocks found in CheBs, CheBrs, and GM2-APs but not in NPC2 and Der f2 are boxed in yellow, and Motifs I and II, discussed in the text, are boxed in red. B, the sequence alignment above was used to align the known secondary structures of Der f2 (50), bovine NPC2 (46), and human GM2-AP (42-45), obtained from the Protein Data Bank server. Dotted lines indicate several secondary structure elements that are precisely aligned in the three proteins (the fourth, six, and seventh β-strands, and two turns, one interrupting β-strand 4, the other between β-strands 6 and 7). C, three known point mutations that inactivate human GM2-AP and result in Tay-Sachs disease, ΔK88, C138R, and R169P (45), are indicated by black arrows. The synthetic Y137S mutation resulting in closure of the hydrophobic cleft and loss of ligand binding (44) is shown by a red arrow. Two double-headed arrows indicate sequences that are nearby in space (45), forming a hydrophobic cleft (H.C.) that interacts with ligands at the surface of the protein (42-44). A hydrogen bond (H.B.) between Lys-88 and Tyr-137 is shown by a dotted line. The sequences of CheBs were reported previously (23). CheBr2 Ag is A. gambiae hypothetical protein AGAP006698. CheBr3Aa, CheBr5Aa, and CheBr6Aa are AAEL004735, AAEL009978, and AAEL004712 from A. aegyptii. CheBr1Ag and CheB4Aa are two hypothetical proteins encoded by the two mosquito genomes that are not currently annotated. The six GM2-AP sequences are Q60648, 1G13, NP_001075381, CAG04534, CAG04534, LOC550529, and XP_973964.1 from the genomes of Mus musculus, (Mm), Homo sapiens (Hs), Equus caballus (Ec), Tetraodon negroviridis (Tn), Danio rerio (Dr), and Tribolium castaneum (Tc). The five NPC2 sequences are 1NEP, P61916, Q9DGJ3, Q9VQ62, and UPI00006A080B from Bos taurus (Bt), H. sapiens (Hs), D. rerio (Dr), D. melanogaster (Dm), and Xenopus tropicalis (Xt), and Der f2 is Q9BIX2 from Dermatophagoides farinae.
Interestingly, systematic PSI-BLAST searches suggest that Drosophila CheBs, mosquito CheBrs, and GM2-APs from a wide range of organisms are significantly more similar to each other than any of them is to NPC2, Der f2, or many other ML proteins (see “Experimental Procedures”). Because GM2-AP, NPC2, and Der f2 are all established members of the ML protein superfamily, this result suggests that CheBs and CheBrs represent previously unidentified members of the ML superfamily that are most closely related to GM2-AP. To further analyze sequence conservation between CheBs, CheBrs, and GM2-APs, we used ClustalW (31) to align the sequences of the 12 D. melanogaster CheBs, six mosquito CheBrs, and over 100 other ML family proteins, including proteins related to human GM2-AP and NPC2 from a variety of organisms. A subset of the aligned sequences is shown in Fig. 5A, with color shading that highlights residues of human GM2-AP that are conserved in several other aligned proteins. This alignment reveals that the majority of residues that are conserved between GM2-AP, NPC2, and Der f2 are also present in CheBs and CheBrs (Fig. 5A). In particular, three of the four ML cysteines known to be involved in disulfide bond formation are in perfect alignment with highly conserved cysteines in CheBs and CheBrs, whereas the fourth cysteine involved in a disulfide bond is aligned within a few residues of cysteines that are less conserved in the insect proteins. The presence in CheBs and CheBrs of most residues conserved between GM2-AP, NPC2, and Der f2 suggests that both types of insect proteins represent previously unrecognized subgroups of the ML protein superfamily.
Furthermore, consistent with PSI-BLAST analysis, this alignment also reveals several blocks of sequences that are conserved between GM2-APs, CheBs, and CheBrs, but are not found in NPC2, Der f2, or other ML proteins (boxed in yellow in Fig. 5A, and data not shown). Two short motifs are particularly well conserved; Motif I, (KR)-X-X-X-G-X-W, is found 12 to 30 residues before the second cysteine, and Motif II, G-X-(YWH)-(KR), occurs 12 to 20 residues before the fourth cysteine (Fig. 5A, boxed in red). The existence of sequences that are conserved between GM2-AP, CheBs, and CheBrs, but are not found in NPC2 or Der f2 confirms the inclusion of CheBs and CheBrs into the ML protein family and further suggests that GM2-AP, CheBs, and CheBrs belong to a common subgroup within that superfamily.
As the above conclusions depend on the accuracy of the alignment in Fig. 5A, we determined whether this alignment is consistent with the known crystal structures of GM2-AP (45), NPC2 (46), and Der f2 (50) (Fig. 5B). Indeed, corresponding primary sequences with similar secondary structures in the three proteins (seven β-strands, one α-helix and several turns) are accurately matched, or within a few residues of each other in the ClustalW alignment. In particular, the position of the fourth, six, and seventh β-strands, and the placement of two turns (one interrupting β-strand 4, the other between β-strands 6 and 7), are precisely conserved in all three proteins. In addition, four cysteines are found at identical positions, and similarly paired to form two disulfide bonds in GM2-AP, NPC2, and Der f2. Finally, in the known structure of GM2-AP, Motif I overlaps the turn that interrupts β-strand 4, and Motif II occurs at the beginning of β-strand 7; thus both motifs are placed at the center of the two structural elements that are best matched with the corresponding structures in NPC2 and Der f2. The accurate alignment of the known structural elements of GM2-AP, NPC2, and Der f2 therefore validates the accuracy of the alignment in Fig. 5A, and supports our conclusions that CheBs and CheBrs not only contain most sequences conserved between GM2-AP, NPC2, and Der f2, but also share sequence motifs with GM2-AP that are not found in NPC2, or Der f2.
The significance of Motifs I and II is independently supported by the key roles these sequences play in human GM2-AP (42-45) (Fig. 5C). Of the three mutations in GM2-AP known to result in Tay-Sachs disease (45), one occurs in Motif I (ΔK88), and another in Motif II (R169P). In addition, mutation of the tyrosine in Motif II (Tyr-137) prevents ligand binding (44). Furthermore, the two motifs are located in close proximity in the three-dimensional structure of GM2-AP, and a hydrogen bond between Lys-88 in Motif I and Tyr-137 in Motif II (H.B. in Fig. 5C) is important for “open” and “closed” configurations, both of which are required for ligand binding (44). Finally, residues in Motif I contribute directly to a ligand-binding hydrophobic cleft (H.C. in Fig. 5C) on the surface of the protein, whereas residues in Motif II are located near the cleft entrance, and may control its access to ligand molecules (45).
In summary, the presence of most residues conserved between three established ML proteins indicates that both CheBs and CheBrs represent previously unrecognized members of the ML protein superfamily. Furthermore, the presence in both types of insect proteins, of sequence motifs that play critical roles in the structure and function of human GM2-AP, but are absent in other ML proteins, suggests that GM2-AP, CheBs, and CheBrs represent a distinct subgroup within the ML superfamily.
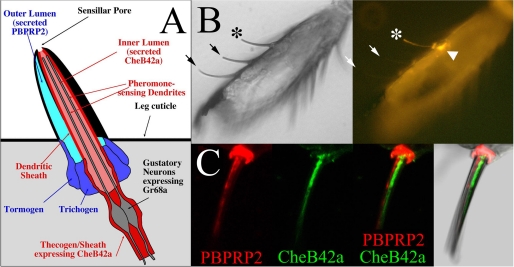
CheB42a Is Present at the Site of Pheromone Detection: The Inner Lumen of Pheromone-sensing Hairs—The inclusion of CheBs in a superfamily of lipid-binding proteins suggests that CheBs may represent a family of gustatory-specific pheromone-binding proteins, accounting for the role of CheB42a in gustatory perception of female-specific pheromones (17). This hypothesis predicts that CheB42a must be present at a precise site inside pheromone-sensing taste hairs. Taste perception in insects occurs within cuticle-covered taste hairs that house three to five sensory neurons and several types of specialized non-neuronal cells (35). An internal fluid-filled cavity or lumen within each hair contains the dendrites of gustatory neurons and communicates with the outside environment through a single cavity at the hair tip. Environmental chemicals and pheromones diffusing into the inner lumen of taste hairs are detected by specific receptors on the surface of gustatory dendrites (Fig. 6A). In addition to this inner lumen, the taste hairs on the legs of Drosophila contain an outer lumen whose function is unclear as it lacks any neuronal dendrite, and is separated both from the inner lumen and from the environment by impermeable cuticular walls (19, 51). Thus, for CheB42a to bind directly to pheromones, it must be secreted into the inner lumen of pheromone-sensing hairs. Consistent with this possibility, all 12 CheBs contain predicted signal peptides at their amino termini (23), and CheB42a is expressed specifically in non-neuronal sheath or thecogen cells: secretory cells that line the inner lumen of pheromone-sensing hairs (17).
FIGURE 6.
CheB42a is secreted into the inner lumen of pheromone-sensing taste hairs where pheromones bind to their receptors. A, schematic showing the complex organization of a pheromone-sensing taste hair, see text. B, a paraffin section of a male front leg was immunolabeled with anti-CheB42a (17, 23). One or more cells (arrowhead) at the base of a taste hair and the corresponding hair shaft (asterisk) are specifically labeled, whereas no labeling is associated with two other taste hairs visible in this field (arrows). C, double-labeling with anti-CheB42a (green) and anti-PBPRP2 (red) (34) shows that both antibodies decorate the shaft of this taste hair through its entire length, but the two signals are adjacent and do not overlap.
To determine whether CheB42a is present in the inner lumen of pheromone-sensing hairs, we adopted a modified immunofluorescence procedure that affords improved sensitivity and preservation of the tissue in the legs of Drosophila (Fig. 6, B and C). As reported previously (17), anti-CheB42a antibody labels the cell bodies of thecogen cells (arrowhead) at the base of some taste hairs (asterisk), but not others (arrows), consistent with the expression of CheB42a in a subset of taste hairs (17, 23) (Fig. 2). Furthermore, using this modified protocol, we find that anti-CheB42a antibody labels the shafts of the taste hairs that express CheB42a (asterisk). Labeling is specific as the shaft of neighboring taste or mechanosensory hairs where CheB42a is not expressed are not labeled (arrows in Fig. 6B and data not shown). To determine whether CheB42a is found in the inner and/or outer hair lumen, we compared its distribution with that of PBPRP2, a protein of unknown function expressed in most if not all taste hairs of the legs, including the pheromone-sensing hairs that express CheB42a (34). PBPRP2 is expressed specifically in the tormogen and trichogen types of non-neuronal cells, and subsequently secreted into the outer lumen, but not the inner lumen of leg taste hairs. Double labeling of male leg sections using our modified protocol, with antibodies against both PBPRP2 and CheB42a, confirms that these proteins are expressed in adjacent cells within the same taste hairs (Ref. 17 and data not shown). Furthermore, anti-PBPRP2 antibody also specifically labels the shaft of taste hairs (Fig. 6C), but not of neighboring mechanosensory hairs (not shown). Comparison of the staining obtained with the two antibodies shows that the distributions of CheB42a and PBPRP2, whereas adjacent through the length of the taste hair shafts, do not overlap (Fig. 6C). These data indicate that CheB42a is secreted specifically into the inner lumen of pheromone-sensing taste hairs.
DISCUSSION
The Diversity of Gustatory-specific and Sexually Dimorphic Expression Patterns among Drosophila CheBs Suggests That, Like CheB42a, Other Proteins in this Family Modulate Gustatory Detection of Specific Pheromones—CheB42a, a protein expressed specifically in pheromone-sensing taste hairs on the front legs of males modulates behavioral response to low-volatility female cuticular hydrocarbon pheromones that trigger male courtship (17). In this study we show that, whereas quite diverse, expression of all 12 D. melanogaster CheBs is almost exclusively gustatory-specific and sexually dimorphic, suggesting that their function is similar to that of CheB42a. Expression of genes of the CheB family is largely restricted to legs and wings, two types of appendages that are highly enriched in taste hairs. Furthermore, expression of at least three CheB genes is associated with specific subsets of taste hairs. In the only cases of non-gustatory expression, we find that two CheBs, whereas detectably expressed in the third antennal segment, the main olfactory organ, are still expressed at highest levels in legs and wings. Although almost exclusively restricted to gustatory organs, expression of the 12 CheB genes is remarkably varied. CheB42a and CheB93a are both expressed predominantly or exclusively in the front legs of males, but in two non-overlapping subsets of taste sensilla. Expression of CheB38c defines a third non-overlapping set of gustatory sensilla distributed on all legs and wings of both males and females, with the striking exception of male front legs. In all, the 12 CheBs are expressed in at least 6 distinct subsets of taste hairs, some of which may overlap partially (Table 1). The variety of gustatory-specific expression patterns observed for CheB genes is only matched by that of the Gr taste receptors, some of which are also expressed in olfactory organs (52, 53). Like Grs, individual CheBs may therefore be involved in gustatory perception of specific chemicals or groups of chemicals. Furthermore, expression of the 12 CheB genes is largely excluded from the many taste sensilla on the labelum and legs that detect food stimuli (35-37). This observation suggests that taste hairs fall into two functional categories, similar to the functional division between olfactory hairs that respond to a relatively wide spectrum of odors and those narrowly tuned to detect one or more pheromones (5, 54). Indeed, the 10 taste hairs on the front legs of males that express CheB42a and Gr68a detect courtship-stimulating female-specific cuticular hydrocarbon pheromones (16, 17), but do not respond to sugar, salt, or bitter compounds (36, 55). By extension, the subsets of sensilla that express CheB93a, CheB38c, or other CheBs may also be narrowly tuned to other low-volatility cuticular hydrocarbon pheromones that modulate Drosophila behavior (2, 4).
Finally, a role for CheBs other than CheB42a in gustatory response to pheromones is further supported by the sexually dimorphic expression of at least 11 of the 12 CheB genes. To our knowledge, no other gene family in any organism displays a comparable level of sexually dimorphic expression outside of the reproductive system. Of particular relevance is the paucity of sexual dimorphism in other families of chemosensory proteins, in both insects and vertebrates. Some insect gustatory and olfactory pheromone receptors (35, 56), as well as several olfactory pheromone-binding proteins (PBPs) (57) are expressed differentially between the sexes. However, each of those proteins represent exceptions in the large families of olfactory receptors, taste receptors, or odorant-binding proteins, the majority of which are expressed indistinguishably in the two sexes and likely function in response to general environmental stimuli. The highly unusual prevalence of sexually dimorphic expression patterns among members of the CheB gene family therefore suggests that CheBs play a specific role in gustatory detection of sexually dimorphic chemical cues, most likely pheromones. In summary, the gustatory-specific, diverse and sexually dimorphic expression patterns of the CheB genes suggest that, as shown for CheB42a (17), they modulate gustatory response to pheromones.
CheBs May Bind to the Lipid-like Pheromones of Drosophila—By what mechanism does CheB42a, and probably other CheBs, modulate gustatory response to pheromones? A likely answer to this question is suggested by our finding that CheBs belong to the ML superfamily of single domain, lipid-binding proteins. Importantly, the highest amino acid conservation between GM2-AP, Drosophila CheBs, and mosquito CheBrs, is found in two motifs that interact closely in the three-dimensional structure of GM2-AP and contribute to its ligand-binding site (42-45), suggesting that all three types of proteins bind chemically similar ligands. Furthermore, the ganglioside ligand of GM2-AP, GM2, contains two long aliphatic chains that resemble the long-chain hydrocarbon pheromones that are key modulators of sexual behaviors in Drosophila and other flies (2, 4). We therefore propose that CheBs bind directly to these pheromones and modulate their interactions with transmembrane gustatory pheromone receptors. In support of this hypothesis, we show here that CheB42a is secreted specifically into the inner lumen of pheromone-sensing hairs, the precise compartment where the dendrites of pheromone-sensing neurons detect the presence of pheromone molecules.
How might binding to CheBs modify the gustatory response to pheromones? Although CheB42a is expressed in the same subset of taste hairs as Gr68a, inactivation of synaptic transmission from Gr68a-expressing neurons or knock-down of Gr68a mRNA inhibits male response to female-specific cuticular hydrocarbons (16), whereas mutant males lacking the CheB42a protein display an increased response to the same pheromones (17). Binding to CheB42a may therefore inhibit interactions between courtship-stimulating female pheromones and their receptor. Given the role of GM2-AP as a cofactor in the degradation of GM2 by β-hexosaminidase A, an attractive possibility is that CheBs act as cofactors for pheromone-degrading enzymes (58). Alternatively, CheB42a may enhance the interaction of inhibitory pheromones with yet unknown receptors expressed in the same hairs. In this scenario, the function of CheBs in the gustatory system may be analogous to that of LUSH, an olfactory pheromone-binding protein, in olfactory detection of cVA (59-61). Indeed, MD-2, another human protein in the ML superfamily, acts as a co-receptor for the Toll-like receptor TLR4, binding to lipopolysaccharides of Gram-negative bacteria as part of the innate immune response (62).
Finally, the conservation in CheBs of amino acid motifs with key roles in the function of GM2-AP suggests that CheB function in gustatory perception of the lipid-like pheromones of Drosophila is related to critical steps of lipid metabolism disrupted in the degenerating neurons of Tay-Sachs patients (63). Interestingly, pheromone detection by olfactory organs has also been linked to fundamental aspects of human lipid metabolism: SNMP1, a protein required for olfactory perception of cVA, is related to CD36, a co-receptor for TLR2, another Toll-like receptor involved in innate immune response to bacteria (64-66). Future insights into the function of CheB42a and other CheBs, and more generally into the gustatory and olfactory detection of the lipid-like pheromones of Drosophila, may therefore shed light on aspects of lipid metabolism involved in a variety of biological processes.
Acknowledgments
We are grateful to Yashi Ahmed, Stephen Goodwin, and Frederic Marion-Poll for critical reading of the manuscript and Larry Zwiebel for advice on immunochemistry.
This work was supported, in whole or in part, by National Institutes of Health NIDCD Grants RO1DC04284 and R01DC007911 (to C. W. P.). This work was also supported by Award 76200-560801 from the Biomedical Research Support Program for Medical Schools from the Howard Hughes Medical Institute to Dartmouth Medical School. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: cVA, cis-vaccenyl acetate; CheB, chemosensory B; CheBr, CheB-related; AP, activator protein; Gr, gustatory receptors; ML, MD-like; MD, myeloid differentiation protein; NPC2, Niemann-Pick gene 2 protein; PBP, pheromone-binding protein; RT, reverse transcriptase; GFP, green fluorescent protein; PBS, phosphate-buffered saline.
References
- 1.Wyatt, T. D. (2003) Pheromones and Animal Behaviour: Communication by Smell and Taste, Oxford University Press, Oxford
- 2.Wicker-Thomas, C. (2007) J. Insect Physiol. 53 1089-1100 [DOI] [PubMed] [Google Scholar]
- 3.Zufall, F., and Leinders-Zufall, T. (2007) Curr. Opin. Neurobiol. 17 483-489 [DOI] [PubMed] [Google Scholar]
- 4.Ferveur, J. F. (2005) Behav. Genet. 35 279-295 [DOI] [PubMed] [Google Scholar]
- 5.Vosshall, L. B. (2008) Neuron 59 685-689 [DOI] [PubMed] [Google Scholar]
- 6.Ha, T. S., and Smith, D. P. (2006) J. Neurosci. 26 8727-8733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kurtovic, A., Widmer, A., and Dickson, B. J. (2007) Nature 446 542-546 [DOI] [PubMed] [Google Scholar]
- 8.van der Goes van Naters, W., and Carlson, J. R. (2007) Curr. Biol. 17 606-612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marcillac, F., and Ferveur, J. F. (2004) J. Exp. Biol. 207 3927-3933 [DOI] [PubMed] [Google Scholar]
- 10.Savarit, F., Sureau, G., Cobb, M., and Ferveur, J.-F. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 9015-9020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ferveur, J., Savarit, F., O'Kane, C., Sureau, G., Greenspan, R., and Jallon, J. (1997) Science 276 1555-1558 [DOI] [PubMed] [Google Scholar]
- 12.Gailey, D. A., Lacaillade, R. C., and Hall, J. C. (1986) Behav. Genet. 16 375-405 [DOI] [PubMed] [Google Scholar]
- 13.Stockinger, P., Kvitsiani, D., Rotkopf, S., Tirian, L., and Dickson, B. J. (2005) Cell 121 795-807 [DOI] [PubMed] [Google Scholar]
- 14.Ejima, A., Smith, B. P., Lucas, C., Levine, J. D., and Griffith, L. C. (2005) Curr. Biol. 15 194-206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boll, W., and Noll, M. (2002) Development 129 5667-5681 [DOI] [PubMed] [Google Scholar]
- 16.Bray, S., and Amrein, H. (2003) Neuron 39 1019-1029 [DOI] [PubMed] [Google Scholar]
- 17.Park, S. K., Mann, K. J., Lin, H., Starostina, E., Kolski-Andreaco, A., and Pikielny, C. W. (2006) Curr. Biol. 16 1154-1159 [DOI] [PubMed] [Google Scholar]
- 18.Ejima, A., Smith, B. P., Lucas, C., van der Goes van Naters, W., Miller, C. J., Carlson, J. R., Levine, J. D., and Griffith, L. C. (2007) Curr. Biol. 17 599-605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nayak, S. V., and Singh, R. N. (1983) Int. J. Insect Morphol. Embryol. 12 273-291 [Google Scholar]
- 20.Possidente, D. R., and Murphey, R. K. (1989) Dev. Biol. 132 448-457 [DOI] [PubMed] [Google Scholar]
- 21.Robertson, H. M. (1983) Experientia (Basel) 39 333-335 [Google Scholar]
- 22.Venard, R., Anthony, C., and Jallon, J.-M. (1989) in Neurobiology of Sensory Systems (Singh, R. N., and Strausfeld, N. J., eds) pp. 377-385, Plenum Press, New York
- 23.Xu, A., Park, S. K., D'Mello, S., Kim, E., Wang, Q., and Pikielny, C. W. (2002) Cell Tissue Res. 307 381-392 [DOI] [PubMed] [Google Scholar]
- 24.Pikielny, C. W., Hasan, G., Rouyer, F., and Rosbash, M. (1994) Neuron 12 35-49 [DOI] [PubMed] [Google Scholar]
- 25.Diviacco, S., Norio, P., Zentilin, L., Menzo, S., Clementi, M., Biamonti, G., Riva, S., Falaschi, A., and Giacca, M. (1992) Gene (Amst.) 122 313-320 [DOI] [PubMed] [Google Scholar]
- 26.Brand, A. H., and Perrimon, N. (1993) Development 118 401-415 [DOI] [PubMed] [Google Scholar]
- 27.Rubin, G. M., and Spradling, A. C. (1982) Science 218 348-353 [DOI] [PubMed] [Google Scholar]
- 28.Altschul, S. F., Madden, T. L., Schaffer, A. A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D. J. (1997) Nucleic Acids Res. 25 3389-3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schaffer, A. A., Aravind, L., Madden, T. L., Shavirin, S., Spouge, J. L., Wolf, Y. I., Koonin, E. V., and Altschul, S. F. (2001) Nucleic Acids Res. 29 2994-3005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schultz, J., Milpetz, F., Bork, P., and Ponting, C. P. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 5857-5864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thompson, J. D., Higgins, D. G., and Gibson, T. J. (1994) Nucleic Acids Res. 22 4673-4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang, X., Warren, J. T., Buchanan, J., Gilbert, L. I., and Scott, M. P. (2007) Development 134 3733-3742 [DOI] [PubMed] [Google Scholar]
- 33.Pitts, R. J., Fox, A. N., and Zwiebel, L. J. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 5058-5063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Park, S. K., Shanbhag, S. R., Wang, Q., Hasan, G., Steinbrecht, R. A., and Pikielny, C. W. (2000) Cell Tissue Res. 300 181-192 [DOI] [PubMed] [Google Scholar]
- 35.Vosshall, L. B., and Stocker, R. F. (2007) Annu. Rev. Neurosci. 30 505-533 [DOI] [PubMed] [Google Scholar]
- 36.Meunier, N., Ferveur, J. F., and Marion-Poll, F. (2000) Curr. Biol. 10 1583-1586 [DOI] [PubMed] [Google Scholar]
- 37.Meunier, N., Marion-Poll, F., Rospars, J. P., and Tanimura, T. (2003) J. Neurobiol. 56 139-152 [DOI] [PubMed] [Google Scholar]
- 38.Ray, K., Hartenstein, V., and Rodrigues, V. (1993) Dev. Biol. 155 26-37 [DOI] [PubMed] [Google Scholar]
- 39.Mahuran, D. J. (1998) Biochim. Biophys. Acta 1393 1-18 [DOI] [PubMed] [Google Scholar]
- 40.Inohara, N., and Nunez, G. (2002) Trends Biochem. Sci. 27 219-221 [DOI] [PubMed] [Google Scholar]
- 41.Berger, A. C., Vanderford, T. H., Gernert, K. M., Nichols, J. W., Faundez, V., and Corbett, A. H. (2005) Eukaryot. Cell 4 1851-1862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wright, C. S., Zhao, Q., and Rastinejad, F. (2003) J. Mol. Biol. 331 951-964 [DOI] [PubMed] [Google Scholar]
- 43.Wright, C. S., Mi, L. Z., and Rastinejad, F. (2004) J. Mol. Biol. 342 585-592 [DOI] [PubMed] [Google Scholar]
- 44.Wright, C. S., Mi, L. Z., Lee, S., and Rastinejad, F. (2005) Biochemistry 44 13510-13521 [DOI] [PubMed] [Google Scholar]
- 45.Wright, C. S., Li, S. C., and Rastinejad, F. (2000) J. Mol. Biol. 304 411-422 [DOI] [PubMed] [Google Scholar]
- 46.Friedland, N., Liou, H. L., Lobel, P., and Stock, A. M. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 2512-2517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Derewenda, U., Li, J., Derewenda, Z., Dauter, Z., Mueller, G. A., Rule, G. S., and Benjamin, D. C. (2002) J. Mol. Biol. 318 189-197 [DOI] [PubMed] [Google Scholar]
- 48.Johannessen, B. R., Skov, L. K., Kastrup, J. S., Kristensen, O., Bolwig, C., Larsen, J. N., Spangfort, M., Lund, K., and Gajhede, M. (2005) FEBS Lett. 579 1208-1212 [DOI] [PubMed] [Google Scholar]
- 49.Ichikawa, S., Takai, T., Inoue, T., Yuuki, T., Okumura, Y., Ogura, K., Inagaki, F., and Hatanaka, H. (2005) J. Biochem. (Tokyo) 137 255-263 [DOI] [PubMed] [Google Scholar]
- 50.Suzuki, M., Tanaka, Y., Korematsu, S., Mikami, B., and Minato, N. (2005) Biochem. Biophys. Res. Commun. 339 679-686 [DOI] [PubMed] [Google Scholar]
- 51.Shanbhag, S. R., Park, S. K., Pikielny, C. W., and Steinbrecht, R. A. (2001) Cell Tissue Res. 304 423-437 [DOI] [PubMed] [Google Scholar]
- 52.Scott, K., Brady, R., Jr., Cravchik, A., Morozov, P., Rzhetsky, A., Zuker, C., and Axel, R. (2001) Cell 104 661-673 [DOI] [PubMed] [Google Scholar]
- 53.Dunipace, L., Meister, S., McNealy, C., and Amrein, H. (2001) Curr. Biol. 11 822-835 [DOI] [PubMed] [Google Scholar]
- 54.de Bruyne, M., and Warr, C. G. (2006) Bioessays 28 23-34 [DOI] [PubMed] [Google Scholar]
- 55.Meunier, N., Marion-Poll, F., Lansky, P., and Rospars, J. P. (2003) Chem. Senses 28 671-679 [DOI] [PubMed] [Google Scholar]
- 56.Hallem, E. A., Dahanukar, A., and Carlson, J. R. (2006) Annu. Rev. Entomol. 51 113-135 [DOI] [PubMed] [Google Scholar]
- 57.Pelosi, P., Zhou, J. J., Ban, L. P., and Calvello, M. (2006) Cell Mol. Life Sci. 63 1658-1676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Leal, W. S. (2005) Chemistry of Pheromones and Other Semiochemicals Ii 240 1-36 [Google Scholar]
- 59.Xu, P., Atkinson, R., Jones, D. N., and Smith, D. P. (2005) Neuron 45 193-200 [DOI] [PubMed] [Google Scholar]
- 60.Pophof, B. (2002) Naturwissenschaften 89 515-518 [DOI] [PubMed] [Google Scholar]
- 61.Laughlin, J. D., Ha, T. S., Jones, D. N., and Smith, D. P. (2008) Cell 133 1255-1265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Visintin, A., Iliev, D. B., Monks, B. G., Halmen, K. A., and Golenbock, D. T. (2006) Immunobiology 211 437-447 [DOI] [PubMed] [Google Scholar]
- 63.Mahuran, D. J. (1999) Biochim. Biophys. Acta 1455 105-138 [DOI] [PubMed] [Google Scholar]
- 64.Rogers, M. E., Sun, M., Lerner, M. R., and Vogt, R. G. (1997) J. Biol. Chem. 272 14792-14799 [DOI] [PubMed] [Google Scholar]
- 65.Benton, R., Vannice, K. S., and Vosshall, L. B. (2007) Nature 450 289-293 [DOI] [PubMed] [Google Scholar]
- 66.Jin, X., Ha, T. S., and Smith, D. P. (2008) Proc. Natl. Acad. Sci. U. S. A. 105 10996-11001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Singh, R. N. (1997) Microscopy Res. Tech. 39 547-563 [DOI] [PubMed] [Google Scholar]