Abstract
Transforming growth factor-β (TGF-β) family members, including TGF-βs, activins, and bone morphogenetic proteins, exert diverse biological activities in cell proliferation, differentiation, apoptosis, embryonic development, and many other processes. These effects are largely mediated by Smad proteins. Smad7 is a negative regulator for the signaling of TGF-β family members. Dysregulation of Smad7 is associated with pathogenesis of a variety of human diseases. However, the in vivo physiological roles of Smad7 have not been elucidated due to the lack of a mouse model with significant loss of Smad7 function. Here we report generation and initial characterization of Smad7 mutant mice with targeted deletion of the indispensable MH2 domain. The majority of Smad7 mutant mice died in utero due to multiple defects in cardiovascular development, including ventricular septal defect and non-compaction, as well as outflow tract malformation. The surviving adult Smad7 mutant mice had impaired cardiac functions and severe arrhythmia. Further analyses suggest that Smad2/3 phosphorylation was elevated in atrioventricular cushion in the heart of Smad7 mutant mice, accompanied by increased apoptosis in this region. Taken together, these observations pinpoint an important role of Smad7 in the development and function of the mouse heart in vivo.
TGF-β3 superfamily members exert their biological functions via binding to serine/threonine kinase receptors at the cell surface (1), followed by signal transduction by a group of intracellular transducers called Smad (2, 3). Smad proteins can be classified into three functional subclasses: receptor-regulated Smad (R-Smad, including Smad1, -2, -3, -5, and -8), common partner Smad (Co-Smad or Smad4), and inhibitory Smad (I-Smad, including Smad6 and -7). Upon ligand binding, R-Smads are phosphorylated by the serine kinase of the receptors and form a heteromeric complex with Smad4. The R-Smad/Smad4 complex is subsequently translocated into the nucleus in which they function as transcriptional regulators that modulate expression of target genes. Smad6 and Smad7 comprise the group of inhibitory Smads that antagonize signaling of TGF-β family members (2, 3). Smad6 preferentially inhibits bone morphogenetic protein (BMP) signaling, whereas Smad7 is inhibitory to both TGF-β/activin- and BMP-mediated signaling by a series of in vitro biochemical analyses (2, 3).
The physiological functions of Smads have been revealed by gene-targeting analyses in mouse. Mice deficient in Smad1, Smad2, and Smad5 are dead in utero (4–8), indicating critical functions of these Smads in early embryonic development. Mice with deletion of Smad8 are viable and fertile (9), likely caused by functional compensation by Smad1 and Smad5. Deletion of Smad3 leads to abnormalities in the immune system (10, 11). Mice devoid of Smad4 also die in utero (12, 13). Deficiency of Smad6 results in multiple cardiovascular defects, including outflow tract defects and hyperplasia of the cardiac valves in mutant embryos, as well as development of aortic ossification and increased blood pressure in viable mutant mice (14).
The physiological importance of Smad7 is exemplified by recent findings that dysregulation of Smad7 is associated with a variety of pathological conditions in humans, including development of malignancy (15, 16), scleroderma formation (17, 18), and chronic inflammatory bowel diseases (19). To further elucidate the physiological functions of Smad7, it is urgently needed to have an animal model with functional loss of Smad7. Previously, Li et al. (20) reported a hypomorphic Smad7 mouse model in which exon 1 of the mouse Smad7 gene was deleted. These mutant mice appeared to have normal phenotype, except for some alterations in B-cell responses (20). The function of Smad7 was not completely lost in this mutant mouse model, because deletion of exon 1 of the Smad7 gene resulted in a truncated Smad7 protein containing intact MH2 domain due to alternative splicing. The retaining of Smad7 function in these mutant mice is consistent with a previous study demonstrating that the inhibitory effect of Smad7 is mainly mediated by the MH2 domain of the protein (21).
Studies of the expression pattern of Smad7 in early embryos suggest that it may play a role in cardiovascular development. Using in situ hybridization, Smad7 was found highly expressed in endothelial cells throughout aorta and various arteries in the embryo and adult tissues in the mouse (22). In the developing heart (E11.5 to E12.5), Smad7 is predominantly expressed in atrioventricular cushion (22). Recently, using a transgenic approach, we demonstrated that a 4.3-kb Smad7 promoter is able to direct reporter expression in various endothelial cells, including atrioventricular cushion (23).
To generate a mouse model with complete functional loss of Smad7, we targeted the MH2 domain of Smad7. As a result, we found that the majority of Smad7 mutant mice died in utero due to multiple development defects in the heart. On the other hand, the surviving adult mutant mice had declined cardiac function and arrhythmia. These findings, therefore, have uncovered an important physiological function of Smad7 in the development of mouse heart in vivo.
EXPERIMENTAL PROCEDURES
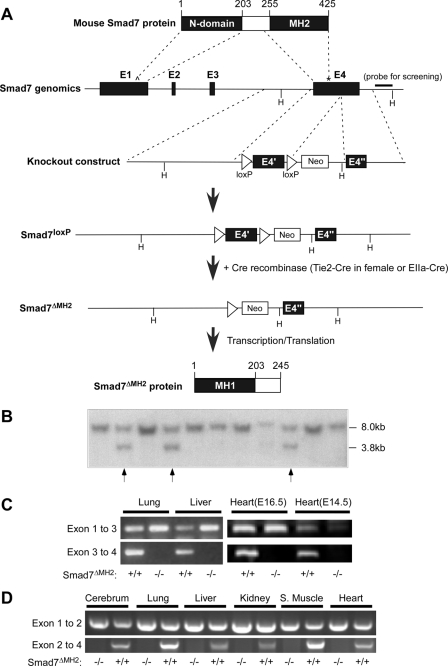
Generation of Smad7 Mutant Mice—The mouse Smad7 gene contains four exons in which exon 4 harbors the entire MH2 domain. In the knockout construct, the first loxP element was inserted immediately before exon 4 and the second loxP was inserted after the stop codon within exon 4 (Fig. 1A). A neomycin selection cassette was placed after the second loxP. The linearized construct was electroporated into embryonic stem cells that were subjected to G418 selection. Chimeric mice were generated by blastocyst injection at the Indiana University School of Medicine Trangenic/Knockout facility, and they were used to intercross with C57BL/6 mice to obtain heterozygous animals (Smad7loxP) that were characterized by Southern blotting analysis (Fig. 1B). Systemic deletion of Smad7 MH2 domain was achieved by crossing Smad7loxP male mice with Tie2-Cre female mice, or crossing Smad7loxP mice with EIIa-Cre mice.
FIGURE 1.
Generation of Smad7 mutant mice. A, diagrams of the Smad7 protein, the exon-intron structure of mouse Smad7 gene, the knockout construct, and the outcomes of homologous recombination. The numbering of amino acid is illustrated at the top. The position of the genomic region used for Southern blotting analysis is labeled with a bold bar. E stands for exons and H for HindIII restriction sites. The translation initiation site is marked as Λ, and the stop codon is marked as *. B, Southern blotting analysis of Smad7loxP alleles. Genomic DNA was isolated from mouse tails and used in Southern blotting analysis after HindIII digestion. The wild-type Smad7 allele is shown as a single 8.0-kb band, and the targeted allele was at 3.8 kb. The arrows indicate heterozygous Smad7loxP mice. C, RT-PCR analyses of lung, liver, and heart from adult or embryonic wild-type and Smad7ΔMH2-/- mice (created via Tie2-Cre) with specific primers that amplify mRNA corresponding to exons 1–3 or exons 3–4, respectively. D, RT-PCR analysis of total RNA isolated from multiple tissues in adult wild-type and Smad7ΔMH2-/- mice (created via EIIa-Cre) with specific primers that amplify mRNA corresponding to exons 1–2 or exons 2–4, respectively.
RNA Isolation and RT-PCR—Total RNA was isolated using TRIzol reagent (Invitrogen) from various tissues of adult mice or mouse embryos. The RNA was treated with RNase-free DNase I and reverse-transcribed with oligo(dT) primer using the SuperScript First-Strand Synthesis System for RT-PCR (Invitrogen). Oligonucleotide primers used for RT-PCR to detect Smad7 mRNA were: 5′-AAGTGTTCAGGTGGCCGGATCTCAG-3′ and 5′-ACAGCATCTGGACAGCCTGCAGTTG-3′ for the region of exons 1–3, 5′-CAACTGCAGGCTGTCCAGATGCTGTAC-3′ and 5′-GTAAACCCACACGCCATCCACTTCC-3′ for exons 3–4, 5′-GTTCAGGACCAAACGATCTG-3′ and 5′-GGGGAGACTCTAGTTCACAG-3′ for exons 1–2, and 5′-CCTCCTCCTTACTCCAGATACCC-3′ and 5′-CTGGGAGAAACCTGATGAAA-3′ for exons 2–4.
Histological Analysis, BrdUrd Labeling, TUNEL Assay, and Immunohistochemistry—Mouse embryos were harvested by cesarean section. Embryos and isolated hearts were fixed in 10% neutral buffered formalin, paraffin-embedded, sectioned (at 7 μm), and stained with hematoxylin and eosin. For BrdUrd staining, pregnant mice were injected with BrdUrd 1 h prior to dissection (0.1 mg/g body weight). The embryos were collected and fixed, paraffin-embedded, and sectioned (at 7 μm). Antigen retrieval was performed by citrate buffer methods, followed by treatment with 1 n HCl for 90 min at room temperature. Primary anti-BrdUrd antibody was incubated at 4 °C overnight, followed by incubation with secondary antibody and nuclear staining with Hoechst 33342. The images were examined using a Leica DM500 fluorescence microscope. The cell proliferation rate was calculated as the ratio of BrdUrd-positive cells versus Hoechst-positives cells. TUNEL assays were performed on paraffin sections using the Apoptag kit (Chemicon, Temecula, CA). Positive controls of TUNEL assay were performed via treatment with 0.1 μg/ml of DNase I (Sigma-Aldrich) for 10 min at room temperature for generation of nicks on double-stranded DNA. Immunohistochemical staining was performed on paraffin sections using a Vectastain ABC kit according to manufacturer's instructions (Vector Laboratories, Burlingame, CA). The following primary antibodies were used: rabbit anti-phosphorylated-Smad2/3 antibody (Chemicon) and rabbit anti-phosphorylated-Smad1/5/8 antibody (Cell Signaling Technology).
Echocardiography and ECG—Transthoracic echocardiograms were performed under 1.5–2.0% isoflurane. Two-dimensional short-axis images were obtained using high resolution Vevo 660 and 770 Imaging Systems (Visualsonics Inc., Toronto, Canada) equipped with a 35-MHz scan probe. Left ventricular chamber dimensions at systolic and diastolic phases were measured from M-mode recording. Heart rate was calculated from simultaneous electrocardiogram recording. Left ventricular volume, fractional shortening (FS%), and ejection fraction (EF%) were calculated using the Vevo Analysis program. ECG was performed using a protocol as previously described (24). A representative 2-s segment of each mouse was selected for measuring the P wave durations, P-R intervals, QRS durations, and RR intervals, as well the baselines and the morphology of QRS complexes.
Animal Protocols—All protocols involving the use of animals were in compliance with the National Institutes of Health's and the Indiana University Laboratory Animals Research Committee's Guidelines for the Care and Use of laboratory Animals. C57BL/6 mouse strain was purchased from Harlan (Indianapolis, IN). For timing of the pregnant mice and embryos, the noon of the day that the vaginal plug was observed was designated embryonic day 0.5 (E0.5).
Statistical Analysis—Analysis of variance was used for the analysis of R-R variation of ECG. A chi-square test was used for the analysis of the number of mice survived to weaning stage. Student's t test was performed with all other comparisons, and p < 0.05 was considered statistically significant.
RESULTS
Generation of Smad7 Mutant Mice—To generate mutant mice with functional loss of Smad7, we targeted exon 4 of Smad7 gene with two loxP sites (Fig. 1A). This region of Smad7 gene harbors the entire MH2 domain (Fig. 1A). Correctly targeted ES cell clones and heterozygous animals were identified by Southern blot analysis (Fig. 1B). Heterozygous and homozygous Smad7loxP mice are viable, fertile, and phenotypically normal, indicating that the neomycin selection cassette had no detrimental effect on Smad7 gene expression. In the presence of Cre recombinase, the 5′-half of exon 4 would be deleted, resulting in complete loss of the C-terminal half of Smad7 protein (amino acids 247–426) that covers the entire MH2 domain (amino acids 260–426). We created a systemic Smad7 MH2 deletion mouse (Smad7ΔMH2) by taking advantage of maternal germ line expression of Cre in oocytes in the Tie2-Cre transgenic female mouse, as the oocytes have a transient expression of Cre protein from maternally derived Cre mRNA (25). By crossing Smad7loxP male mice with Tie2-Cre female mice, we were able to generate systemic deletion of Smad7 MH2 domain (Fig. 1C). We also used EIIa-Cre mice to generate systemic Smad7ΔMH2-/- mice (26) (Fig. 1D). Both the Tie2-Cre- and EIIa-Cre-generated Smad7ΔMH2-/- mice had similar phenotypes. Systemic deletion of Smad7 MH2 domain was confirmed by RT-PCR in different tissues (Fig. 1, C and D). In these experiments, the cDNA region corresponding to exon 4 could not be detected in all the homozygous Smad7-defficient mice, although the region corresponding to exon 1–3 was still detected in these animals.
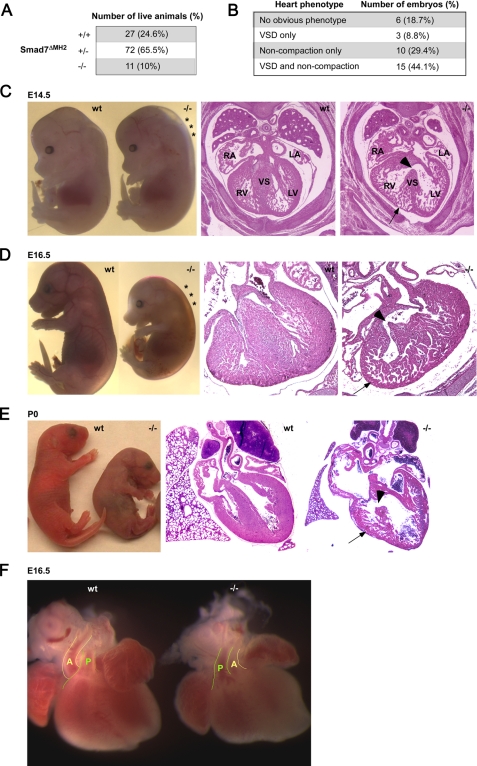
Impaired Cardiovascular Development in Smad7 Mutant Mice—Heterozygous Smad7 mutant mice (Smad7ΔMH2+/-) were fertile and apparently normal, and were intercrossed to generate homozygous mutant mice (Smad7ΔMH2-/-). We found pre-weaning loss of the homozygous mutant mice (Fig. 2A). By analyzing the gross morphology of the developing embryos, we observed signs of defects in cardiovascular development in Smad7ΔMH2-/- mice. A large number of Smad7ΔMH2-/- embryos from E14.5 to E16.5 appeared pale and displayed prominent subcutaneous edema (Fig. 2, C and D), characteristic of failure in the circulatory system. The majorities of neonatal Smad7ΔMH2-/- mice were smaller than wild-type and heterozygous littermates. Histological analysis of these Smad7ΔMH2-/- mice revealed abnormal ventricular development, including ventricular septal defect (VSD), thin ventricular wall, and ventricular non-compaction (Fig. 2, C–E). The alignment of aortic and pulmonary artery was perturbed in some of the Smad7ΔMH2-/- mice (Fig. 2F). Although histological analysis revealed various developmental defects in Smad7ΔMH2-/- embryos, the abnormal phenotype was not fully penetrated (Fig. 2B).
FIGURE 2.
Defects of cardiac development in Smad7ΔMH2-/- mice. A, under-presentation of Smad7ΔMH2-/- mice. The table summarizes the number of mice that survived to weaning stage. B, summary of cardiac phenotypes of Smad7ΔMH2-/- embryos at E14.5 and E16.5. C and D, embryo images and histological analyses of Smad7ΔMH2-/- mice at E14.5 and E16.5. Note that the Smad7ΔMH2-/- mice had edema in the dorsal region (marked by *). Transverse heart sections (right panels) at atrioventricular valve level showed VSD (arrowheads) and ventricular non-compaction (arrows). RA, right atrium; LA, left atrium; RV, right ventricle; LV, left ventricle; and VS, ventricular septum. E, neonatal Smad7ΔMH2-/- mouse was smaller and pale in comparison with wild-type littermate (left panel). Transverse heart sections (right panel) revealed VSD (arrowhead) and ventricular non-compaction (arrow). F, outflow tract defects in a Smad7ΔMH2-/- mouse. The positioning of aorta (A) relative to pulmonary artery (P) was altered in Smad7 mutant embryos at E16.5.
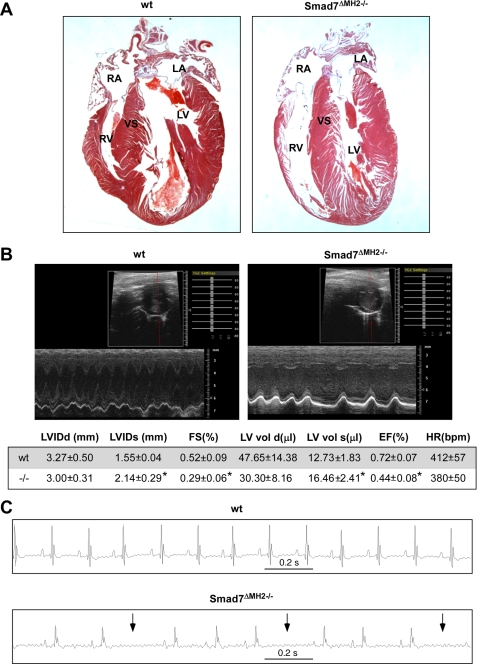
Altered Cardiac Function and Arrhythmia in Viable Smad7 Mutant Mice—A small percentage of homozygous mutant mice could survive to adulthood. However, the viable homozygous mutant mice were significantly smaller in size when compared with wild-type and heterozygous littermates (23.78 ± 2.72 g versus 28.11 ± 2.16 g, at 6 months old, p < 0.01). Histological analysis showed a thinner right ventricular wall in adult Smad7ΔMH2-/- mice than that of the wild-type animals (Fig. 3A). Using echocardiography, we analyzed cardiac function in the viable Smad7 mutant mice (Fig. 3B). Echocardiography showed a dramatic increase in the systolic ventricular chamber volumes associated with declined cardiac functions in viable Smad7ΔMH2-/- mice, including significantly decreased fractional shortening (FS) and ejection fraction (EF) as compared with the wild-type controls (Fig. 3B). In addition, ECG was recorded in four pairs of Smad7ΔMH2-/- mice and wild-type littermate controls. Three of the four Smad7 mutant mice had abnormalities in ECG recordings, whereas all control mice had normal sinus rhythm and normal morphology of the QRS complex (Fig. 3C). The R-R intervals of representative 2-s recordings of the mice were measured, and statistic analysis indicated that Smad7ΔMH2-/- mice had significantly higher incidence of irregular heart beat than the wild-type control animals. Smad7ΔMH2-/- mice also appeared to have abnormal morphologies of the QRS complexes, although the QRS duration was not altered (Fig. 3C). The R-R interval was significant increased in Smad7 mutant mice (219 ± 58 ms in mutant mice versus 154 ± 8 ms in controls, p < 0.01).
FIGURE 3.
Altered cardiac function and arrhythmia in viable adult Smad7ΔMH2-/- mice. A, histological analysis showed a very thin wall in the right ventricle in adult Smad7ΔMH2-/- mice (at 9 months old). RA, right atrium; LA, left atrium; RV, right ventricle; LV, left ventricle; and VS, ventricular septum. B, echocardiograph analysis of wild-type and viable Smad7ΔMH2-/- mice (at 3 months old, n = 5). Representative two-dimensional and M-mode images are shown. Measurement of various parameters of cardiac functions is shown in the lower panel, and * stands for p < 0.01. C, ECG analysis of wild-type and viable Smad7ΔMH2-/- mice (at 3 months old). Representative ECG images reveal arrhythmia (arrows) in Smad7 mutant mice.
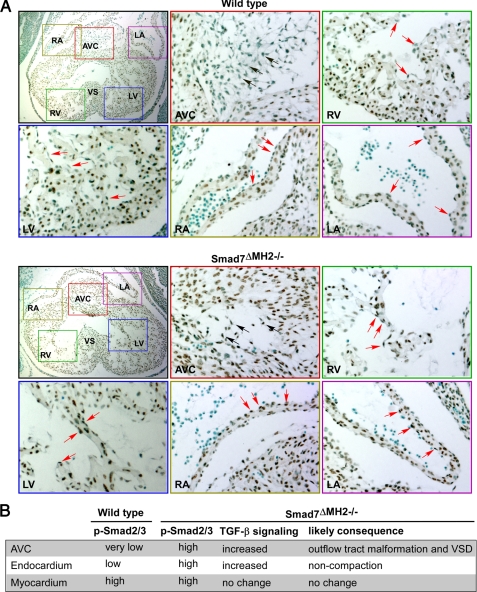
Increased Phosphorylation of Smad2/3 in Atrioventricular Cushion and Ventricular Endocardium in Smad7 Mutant Mice—One of the major signaling events subsequent to TGF-β receptor activation is Smad2/3 phosphorylation (2, 3). We hypothesized that functional loss of Smad7 in Smad7ΔMH2-/- mice could activate the TGF-β signaling through an increase in Smad2/3 phosphorylation. To address this issue, we performed an immunohistochemical staining to determine the level of Smad2/3 phosphorylation (p-Smad2/3) in Smad7-deficient hearts. In wild-type controls, we found that nuclear p-Smad2/3 staining was present in atrial and ventricular myocardia, while largely absent in atrioventricular cushion and at low level in atrial and ventricular endocardia (Fig. 4A). In Smad7ΔMH2-/- mice, p-Smad2/3 staining was significantly increased in the endocardial cells in atrioventricular cushion (Fig. 4A). Surprisingly, a large number of p-Smad2/3-positive cells were also found in atrial and ventricular endocardial cells (Fig. 4A). The change of Smad2/3 phosphorylation in endocardium may partly underlie the non-compaction phenotype observed in Smad7ΔMH2-/- mice (Fig. 4B), because recent studies have demonstrated that ventricular endocardium plays a pivotal role in regulating ventricular trabeculation and compaction (27–31). We also analyzed phosphorylation of Smad1/5/8 (p-Smad1/5/8), the Smads downstream of BMP receptors, in the Smad7-deficient hearts. Interestingly, p-Samd1/5/8 level was not altered in Smad7-deficient hearts when compared with the littermate controls (supplemental Fig. S1), suggesting that Smad7 is more relevant to TGF-β-mediated signaling than to the BMP pathway under physiological conditions.
FIGURE 4.
Increased phosphorylation of Smad2/3 in atrioventricular cushion and endocardium in Smad7 mutant embryos. A, immunohistochemical staining of heart sections at E11. 5 with an antibody against phosphorylated Smad2/3 and counterstained with methyl green. Amplified pictures are shown for the areas of atrioventricular cushion, atrium, and ventricle. The arrows in black indicate representative cells in cardiac cushion region. The arrows in red indicate representative cells in ventricular or atrial endocardium. Note that Smad2/3 phosphorylation is increased in atrioventricular cushion and endocardium of Smad7ΔMH2-/- mouse in comparison with the wild-type control. RA, right atrium; LA, left atrium; AVC, atrioventricular cushion; RV, right ventricle; LV, left ventricle; and VS, ventricular septum. B, summary of the Smad2/3 phosphorylation levels in various areas of mouse heart and the effect of Smad7 deletion on TGF-β signaling, as well as the potential physiological consequences.
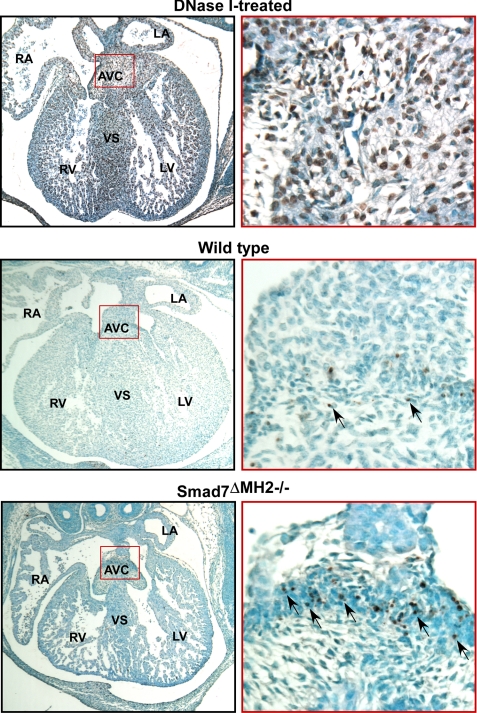
Increased Apoptosis in Atrioventricular Cushion in Smad7 Mutant Embryos—Previously, studies have shown that TGF-βs are key cytokines in regulating developmental apoptotic events during cardiac development (32). Apoptosis in endocardial cushion is a tightly regulated event (33). Lack of apoptosis would lead to malformed valvular tissues (34). Abnormally enhanced apoptosis activity would lead to various other cardiac defects, including VSD (35–38). We analyzed cellular apoptosis at various heart development stages using TUNEL assay. These experiments revealed that Smad7 mutant mice had elevated apoptosis in localized regions of the atrioventricular cushion at E14.5 (Fig. 5). DNase I-treated heart sections at E14.5 were used as positive controls for TUNEL staining (Fig. 5). The increase of apoptosis in atrioventricular cushion in the mutant mice is consistent with the observations that Smad2/3 phosphorylation is elevated in endocardial cushion (Fig. 4), and that Smad7 is predominantly expressed in this region (22, 23).
FIGURE 5.
Increased apoptosis in atrioventricular cushion in Smad7 mutant embryos. TUNEL assay of heart sections at E14.5 with methyl green as a counterstain. DNase I treatment was used as a positive control for TUNEL assay. Representative apoptotic cells are marked by arrows. Note that the Smad7ΔMH2-/- mouse displayed an increase of apoptosis-positive cells in localized regions of atrioventricular cushion in comparison with the wild-type littermate controls. RA, right atrium; LA, left atrium; AVC, atrioventricular cushion; RV, right ventricle; LV, left ventricle; and VS, ventricular septum.
Analysis of Cell Proliferation in the Heart of Smad7 Mutant Mouse—Ventricular trabeculation is commonly associated with cellular proliferative activity in the myocardium (39), whereas little is known for the molecular mechanism involved in the process of compaction. It was assumed that increasing hemodynamic load during mid-gestation stage plays a role in initiating the compaction process (39). Ventricular endocardium could play an important role in regulating this activity (27–31). To understand the potential mechanisms underlying the unique non-compaction phenotype in Smad7ΔMH2-/- mice, we analyzed cell proliferative activity in the developing hearts. Using BrdUrd incorporation and immunostaining, we revealed a slight increase of cell proliferation rate in the trabecular myocardium and endocardium in the right ventricle of Smad7ΔMH2-/- mice when compared with wild-type controls (supplemental Fig. S2, p < 0.05). We did not find significant alteration in cell proliferation profile in other areas of the heart (supplemental Fig. S2B), including the left ventricle. The lack of overall alteration in cellular proliferative activity in Smad7-deficient hearts suggests that the primary defect in the myocardium is not in the cellular proliferation-associated trabeculation, but rather in the process of ventricular compaction. This observation is consistent with our histological finding that the size and morphology of ventricular trabecula were not altered in Smad7-defficient mice when compared with wild-type controls (Fig. 2), whereas the compaction is defective in the mutant mice.
DISCUSSION
Although numerous studies have suggested that Smad7 plays a role in human diseases (17, 19), a direct relationship between a loss of Smad7 function and dysregulated physiology in animal models has not been established. The hypomorphic mouse model with exon 1 deletion of the Smad7 gene does not have a cardiovascular phenotype (20), likely due to the fact that the MH2 domain is still expressed and the inhibitory function of Smad7 is not completely lost in these mutant mice. Smad7 is involved in the inhibition of TGF-β superfamily signaling through interaction of its MH2 domain with type I receptors. In vitro studies have indicated that mutations in the MH2 domain are able to abolish the inhibitory effect of Smad7 on transcriptional responses induced by TGF-β (21). The deletion of the entire MH2 domain of Smad7 in our mouse model gave rise to profound defects in cardiac development. These findings indicate that the MH2 domain is indispensable for the in vivo function of Smad7.
Our study reveals for the first time that Smad7 is essential for cardiogenesis, consistent with the observation that Smad7 is profoundly expressed in the cardiovascular system in mouse embryos (22, 23). A majority of Smad7ΔMH2-/- embryos from E14.5 to E16.5 had subcutaneous edema, a sign of failure in the circulatory system due to cardiac defects. Serial histological sections of Smad7ΔMH2-/- embryos revealed ventricular non-compaction as well as ventricular septal defects and defect in the alignment of aorta and pulmonary artery. Interestingly, we did not find major defects in heart valves in Smad7ΔMH2-/- embryos examined, indicating that Smad7 may not play a vital role in the development of heart valves, but has an effect in the development of other structures derived from the cushion tissue, such as outflow tract and ventricular septum.
Ventricular non-compaction is a unique type of congenital cardiac defect. Usually, hypertrabeculation is associated with ventricular non-compaction. In Smad7-deficient hearts, the size and morphology of ventricular trabeculation appears to be normal. The thickness of trabecular myocardium is not significantly altered, consistent with the observation that the cellular proliferative activity in Smad7-deficient ventricles is largely normal, except in right ventricular trabeculea (supplemental Fig. S2). The change of cell proliferation rate in right ventricular trabecula could have resulted from a compensatory mechanism subsequent to either cardiac non-compaction or altered cardiac function. Nevertheless, the cardiac compaction process is clearly compromised in the Smad7-deficient mice.
The molecular mechanism for cardiac compaction is still not known. Interestingly, mice deficient in FKBP12, another negative regulator for TGF-β signaling, also develop cardiac non-compaction (40), suggesting that a common TGF-β-mediated pathway contributes in cardiac ventricular compaction. FKBP12 has been shown to interact with TGF-β type I receptor and inhibit the signaling of TGF-β superfamily members (41). Because FKBP12 can function as an adaptor protein for the efficient recruitment of Smad7-Smurf1 complex to activin type I receptor (42), functional loss of Smad7 may share a similar mechanism with that of the FKBP12 deficiency underlying the formation of cardiac defects. However, further analyses are needed to elucidate the functional interaction between Smad7 and FKBP12 in vivo.
Intriguingly, a small percentage of Smad7ΔMH2-/- mice could survive to adulthood. However, these viable Smad7 mutant mice had a reduction in body size. Statistical analysis of the body weight of the 3-month-old mice indicated that homozygous Smad7ΔMH2 mice were significantly smaller than the littermate wild-type or heterozygous control animals (p < 0.01). The functional assessment using echocardiography revealed a decline of cardiac functions in Smad7ΔMH2-/- adult mice, manifested as a significant increase in the systolic ventricular chamber as well as significant decreases in fraction shortening (FS) and ejection fraction (EF). Decreases in FS and EF are indicative of a decrease in contractile activity in the ventricular wall of the Smad7 mutant animals. In addition, three out of four Smad7ΔMH2-/- adult mice had severe cardiac arrhythmia by ECG analysis. The abnormal morphologies of the QRS complexes of the Smad7 mutant mice could be caused by cardiac non-compaction. In humans, clinical manifestations of ventricular non-compaction include decreased systolic function in the left ventricle and arrhythmia (43). As Smad7ΔMH2-/- mice demonstrated declined cardiac functions and arrhythmia in addition to non-compaction, these mice mimic the clinical features of cardiac non-compaction in humans.
The incomplete penetrance of the cardiac defects in Smad7 mutant mice could be due to potential functional redundancy with another inhibitory Smad, Smad6. Both Smad6 and Smad7 are inhibitory Smads that are expressed in the cardiovascular system in mouse embryos (44). Smad6 predominantly antagonizes BMP signaling. Although our analysis of Smad7 mutant mice strongly indicates that Smad7 is more relevant to TGF-β-mediated signaling than BMP signaling in vivo, previous in vitro biochemical analysis demonstrated that Smad7 is capable of inhibiting both TGF-β/activin and BMP signaling. Mouse embryos with deletion of the MH2 domain of Smad6 have defects in outflow tract septation and hyperplasia in cardiac valves (14). Meanwhile, aortic ossification and elevated blood pressure are found in viable adult Smad6 mutant mice (14). However, the non-overlapping phenotypes in Smad7ΔMH2-/- mice in comparison with the Smad6 mutant mice indicate a high degree of functional specificity within the inhibitory Smad subfamily under normal physiological condition in vivo. In addition, we could not rule out the possibility that genetic background may contribute to the variation of penetrance in the Smad7-deficient mice.
Our study indicates that phosphorylation of Smad2/3 is at a high level in atrial and ventricular myocardium but is not prominent in the atrioventricular cushion and endocardium in wild-type heart at E11.5 (Fig. 4A), largely consistent with the observation as previously reported (45). However, phosphorylation of Smad2/3 was markedly increased in atrioventricular cushion and endocardium in Smad7ΔMH2-/- mouse (Fig. 4A). From E11.5 to E12.5, Smad7 is predominantly expressed in atrioventricular cushion tissues (22, 23). Whether or not Smad7 is expressed in endocardium has not been carefully examined. However, because Smad7 is strongly expressed in the endothelium of major arteries (22), it is possible that the negative staining in in situ data is due to a lower abundance of Smad7 in this tissue. The increased Smad2/3 phosphorylation levels in cardiac cushion tissues and endocardium in Smad7-mutant mouse are highly likely due to disrupted expression of Smad7 in these areas. The consequent changes of TGF-β signaling in these tissues may contribute to various heart defects such as outflow tract malformation, VSD, and non-compaction (summarized in Fig. 4B). Consistent with previous reports that demonstrated absence of Smad7 in atrial and ventricular myocardium (22), we did not find changes of Smad2/3 phosphorylation in these areas (Fig. 4A). Considering that TGF-β signaling is able to trigger apoptosis (46), the elevated phosphorylation of Smad2/3 found in the atrioventricular cushion due to functional loss of Smad7 could lead to apoptosis in this region. Consistently, we did find an increase of apoptosis in endocardial cushion in the Smad7ΔMH2-/- mouse (Fig. 5), partially explaining the defects of cardiac development observed in these animals.
In summary, our data demonstrate that Smad7 is required for cardiac development in embryonic stage and cardiac function in adult. Considering such an important role of Smad7 in cardiac development, the next challenge would be to determine whether Smad7 is associated with congenital heart defects in humans.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants R21-CA122764 and R03-HD049556, by NIH Grants R01-HL81092 and R01-HL70259 (to W. S.), and by NIH Grant P01-HL85098 (to L. J. F. and W. S.). This work was also supported by the Indiana Genomics Initiative (INGEN), the National Natural Science Foundation of China (Grant 30588002), the Ministry of Science and Technology of China (Grants 2006CB943902 and 2007CB947100), the Science & Technology Commission of Shanghai Municipality (Grant 07DJ14005) (to Y. C.), and the Riley Children Foundation (to W. S. and L. J. F.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
Footnotes
The abbreviations used are: TGF-β, transforming growth factor-β; BMP, bone morphogenetic protein; ECG, electrocardiogram; MH2, Mad homology 2; Smad, Sma- and Mad-related protein; VSD, ventricular septal defect; RT, reverse transcription; BrdUrd, bromodeoxyuridine; TUNEL, terminal deoxynucleotidyl transferase-mediated dUTP nick end labeling; E, embryonic day; FS, fractional shortening; EF, ejection fraction.
References
- 1.Massague, J. (1998) Annu. Rev. Biochem. 67 753-791 [DOI] [PubMed] [Google Scholar]
- 2.Heldin, C. H., Miyazono, K., and ten Dijke, P. (1997) Nature 390 465-471 [DOI] [PubMed] [Google Scholar]
- 3.Shi, Y., and Massague, J. (2003) Cell 113 685-700 [DOI] [PubMed] [Google Scholar]
- 4.Lechleider, R. J., Ryan, J. L., Garrett, L., Eng, C., Deng, C., Wynshaw-Boris, A., and Roberts, A. B. (2001) Dev. Biol. 240 157-167 [DOI] [PubMed] [Google Scholar]
- 5.Nomura, M., and Li, E. (1998) Nature 393 786-790 [DOI] [PubMed] [Google Scholar]
- 6.Waldrip, W. R., Bikoff, E. K., Hoodless, P. A., Wrana, J. L., and Robertson, E. J. (1998) Cell 92 797-808 [DOI] [PubMed] [Google Scholar]
- 7.Weinstein, M., Yang, X., Li, C., Xu, X., Gotay, J., and Deng, C. X. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 9378-9383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang, H., Huylebroeck, D., Verschueren, K., Guo, Q., Matzuk, M. M., and Zwijsen, A. (1999) Development 126 1631-1642 [DOI] [PubMed] [Google Scholar]
- 9.Arnold, S. J., Maretto, S., Islam, A., Bikoff, E. K., and Robertson, E. J. (2006) Dev. Biol. 296 104-118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhu, Y., Richardson, J. A., Parada, L. F., and Graff, J. M. (1998) Cell 94 703-714 [DOI] [PubMed] [Google Scholar]
- 11.Yang, X., Letterio, J. J., Lechleider, R. J., Chen, L., Hayman, R., Gu, H., Roberts, A. B., and Deng, C. (1999) EMBO J. 18 1280-1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sirard, C., de la Pompa, J. L., Elia, A., Itie, A., Mirtsos, C., Cheung, A., Hahn, S., Wakeham, A., Schwartz, L., Kern, S. E., Rossant, J., and Mak, T. W. (1998) Genes Dev. 12 107-119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang, X., Li, C., Xu, X., and Deng, C. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 3667-3672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Galvin, K. M., Donovan, M. J., Lynch, C. A., Meyer, R. I., Paul, R. J., Lorenz, J. N., Fairchild-Huntress, V., Dixon, K. L., Dunmore, J. H., Gimbrone, M. A., Jr., Falb, D., and Huszar, D. (2000) Nat. Genet. 24 171-174 [DOI] [PubMed] [Google Scholar]
- 15.Boulay, J. L., Mild, G., Lowy, A., Reuter, J., Lagrange, M., Terracciano, L., Laffer, U., Herrmann, R., and Rochlitz, C. (2003) Int. J. Cancer 104 446-449 [DOI] [PubMed] [Google Scholar]
- 16.Arnold, N. B., Ketterer, K., Kleeff, J., Friess, H., Buchler, M. W., and Korc, M. (2004) Cancer Res. 64 3599-3606 [DOI] [PubMed] [Google Scholar]
- 17.Dong, C., Zhu, S., Wang, T., Yoon, W., Li, Z., Alvarez, R. J., ten Dijke, P., White, B., Wigley, F. M., and Goldschmidt-Clermont, P. J. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 3908-3913 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18.Asano, Y., Ihn, H., Yamane, K., Kubo, M., and Tamaki, K. (2004) J. Clin. Invest. 113 253-264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Monteleone, G., Kumberova, A., Croft, N. M., McKenzie, C., Steer, H. W., and MacDonald, T. T. (2001) J. Clin. Invest. 108 601-609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li, R., Rosendahl, A., Brodin, G., Cheng, A. M., Ahgren, A., Sundquist, C., Kulkarni, S., Pawson, T., Heldin, C. H., and Heuchel, R. L. (2006) J. Immunol. 176 6777-6784 [DOI] [PubMed] [Google Scholar]
- 21.Mochizuki, T., Miyazaki, H., Hara, T., Furuya, T., Imamura, T., Watabe, T., and Miyazono, K. (2004) J. Biol. Chem. 279 31568-31574 [DOI] [PubMed] [Google Scholar]
- 22.Zwijsen, A., van Rooijen, M. A., Goumans, M. J., Dewulf, N., Bosman, E. A., ten Dijke, P., Mummery, C. L., and Huylebroeck, D. (2000) Dev. Dyn. 218 663-670 [DOI] [PubMed] [Google Scholar]
- 23.Liu, X., Chen, Q., Kuang, C., Zhang, M., Ruan, Y., Xu, Z. C., Wang, Z., and Chen, Y. (2007) Biochim. Biophys. Acta 1769 149-152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shen, W. H., Chen, Z., Shi, S., Chen, H., Zhu, W., Penner, A., Bu, G., Li, W., Boyle, D. W., Rubart, M., Field, L. J., Abraham, R., Liechty, E. A., and Shou, W. (2008) J. Biol. Chem. 283 13842-13849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koni, P. A., Joshi, S. K., Temann, U. A., Olson, D., Burkly, L., and Flavell, R. A. (2001) J. Exp. Med. 193 741-754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lakso, M., Pichel, J. G., Gorman, J. R., Sauer, B., Okamoto, Y., Lee, E., Alt, F. W., and Westphal, H. (1996) Proc. Natl. Acad. Sci. U. S. A. 93 5860-5865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stankunas, K., Hang, C. T., Tsun, Z. Y., Chen, H., Lee, N. V., Wu, J. I., Shang, C., Bayle, J. H., Shou, W., Iruela-Arispe, M. L., and Chang, C. P. (2008) Dev. Cell 14 298-311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hsieh, P. C., Davis, M. E., Lisowski, L. K., and Lee, R. T. (2006) Annu. Rev. Physiol. 68 51-66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gassmann, M., Casagranda, F., Orioli, D., Simon, H., Lai, C., Klein, R., and Lemke, G. (1995) Nature 378 390-394 [DOI] [PubMed] [Google Scholar]
- 30.Kramer, R., Bucay, N., Kane, D. J., Martin, L. E., Tarpley, J. E., and Theill, L. E. (1996) Proc. Natl. Acad. Sci. U. S. A. 93 4833-4838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee, K. F., Simon, H., Chen, H., Bates, B., Hung, M. C., and Hauser, C. (1995) Nature 378 394-398 [DOI] [PubMed] [Google Scholar]
- 32.Bartram, U., Molin, D. G., Wisse, L. J., Mohamad, A., Sanford, L. P., Doetschman, T., Speer, C. P., Poelmann, R. E., and Gittenberger-de Groot, A. C. (2001) Circulation 103 2745-2752 [DOI] [PubMed] [Google Scholar]
- 33.Keyes, W. M., and Sanders, E. J. (2002) Am. J. Physiol. 282 C1348-C1360 [DOI] [PubMed] [Google Scholar]
- 34.Ng, C. M., Cheng, A., Myers, L. A., Martinez-Murillo, F., Jie, C., Bedja, D., Gabrielson, K. L., Hausladen, J. M., Mecham, R. P., Judge, D. P., and Dietz, H. C. (2004) J. Clin. Invest. 114 1586-1592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liang, X., Sun, Y., Schneider, J., Ding, J. H., Cheng, H., Ye, M., Bhattacharya, S., Rearden, A., Evans, S., and Chen, J. (2007) Circ. Res. 100 527-535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gaussin, V., Van de Putte, T., Mishina, Y., Hanks, M. C., Zwijsen, A., Huylebroeck, D., Behringer, R. R., and Schneider, M. D. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 2878-2883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Song, L., Fassler, R., Mishina, Y., Jiao, K., and Baldwin, H. S. (2007) Dev. Biol. 301 276-286 [DOI] [PubMed] [Google Scholar]
- 38.Mo, F. E., and Lau, L. F. (2006) Circ. Res. 99 961-969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen, H., Shi, S., Acosta, L., Li, W., Lu, J., Bao, S., Chen, Z., Yang, Z., Schneider, M. D., Chien, K. R., Conway, S. J., Yoder, M. C., Haneline, L. S., Franco, D., and Shou, W. (2004) Development 131 2219-2231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shou, W., Aghdasi, B., Armstrong, D. L., Guo, Q., Bao, S., Charng, M. J., Mathews, L. M., Schneider, M. D., Hamilton, S. L., and Matzuk, M. M. (1998) Nature 391 489-492 [DOI] [PubMed] [Google Scholar]
- 41.Stockwell, B. R., and Schreiber, S. L. (1998) Chem. Biol. 5 385-395 [DOI] [PubMed] [Google Scholar]
- 42.Yamaguchi, T., Kurisaki, A., Yamakawa, N., Minakuchi, K., and Sugino, H. (2006) J. Mol. Endocrinol. 36 569-579 [DOI] [PubMed] [Google Scholar]
- 43.Chin, T. K., Perloff, J. K., Williams, R. G., Jue, K., and Mohrmann, R. (1990) Circulation 82 507-513 [DOI] [PubMed] [Google Scholar]
- 44.Luukko, K., Ylikorkala, A., and Makela, T. P. (2001) Mech. Dev. 101 209-212 [DOI] [PubMed] [Google Scholar]
- 45.de Sousa Lopes, S. M., Carvalho, R. L., van den Driesche, S., Goumans, M. J., ten Dijke, P., and Mummery, C. L. (2003) Gene Expr. Patterns 3 355-360 [DOI] [PubMed] [Google Scholar]
- 46.Schuster, N., and Krieglstein, K. (2002) Cell Tissue Res. 307 1-14 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.