Abstract
RNase R and RNase II are the two representatives from the RNR family of processive, 3′ to 5′ exoribonucleases in Escherichia coli. Although RNase II is specific for single-stranded RNA, RNase R readily degrades through structured RNA. Furthermore, RNase R appears to be the only known 3′ to 5′ exoribonuclease that is able to degrade through double-stranded RNA without the aid of a helicase activity. Consequently, its functional domains and mechanism of action are of great interest. Using a series of truncated RNase R proteins we show that the cold-shock and S1 domains contribute to substrate binding. The cold-shock domains appear to play a role in substrate recruitment, whereas the S1 domain is most likely required to position substrates for efficient catalysis. Most importantly, the nuclease domain alone, devoid of the cold-shock and S1 domains, is sufficient for RNase R to bind and degrade structured RNAs. Moreover, this is a unique property of the nuclease domain of RNase R because this domain in RNase II stalls as it approaches a duplex. We also show that the nuclease domain of RNase R binds RNA more tightly than the nuclease domain of RNase II. This tighter binding may help to explain the difference in catalytic properties between RNase R and RNase II.
Ribonucleases (RNases) play important roles in RNA metabolism. They are responsible for the maturation of stable RNA and the degradation of RNA molecules that are defective or no longer required by the cell. Both maturation and degradation are initiated by endoribonucleolytic cleavage(s) and completed by the action of exoribonucleases (1). In Escherichia coli, three, relatively nonspecific, 3′ to 5′ processive exoribonucleases are responsible for degradation of RNA: RNase II, RNase R, and polynucleotide phosphorylase (PNPase).3 RNase II and PNPase appear to be primarily responsible for mRNA decay (2), although their precise functions may differ (3). However, mRNAs containing extensive secondary structure, such as repetitive extragenic palindromic sequences, are degraded by PNPase (4, 5) or RNase R (5). Likewise, degradation of highly structured regions of rRNA (6) and tRNA (7),4 is carried out by PNPase and/or RNase R. These findings suggest that PNPase and RNase R are the universal degraders of structured RNAs in vivo, leaving RNase II to act on relatively unstructured RNAs.
Whether or not an RNase acts upon a particular RNA appears to depend upon the specificity of the RNase and the accessibility of the RNA to that RNase (1). Purified RNase R readily degrades both single- and double-stranded RNA molecules (5, 8), and it is the only known 3′ to 5′ exoribonuclease able to degrade through double-stranded RNA without the aid of helicase activity. To degrade RNA molecules containing double-stranded regions, RNase R requires a 3′ single-stranded overhang at least 5 nucleotides long to serve as a binding site from which degradation can be initiated (5, 8, 9).5 How RNase R then proceeds through the RNA duplex is of great interest. An important step toward elucidating the mechanism of action of RNase R is to determine the contribution that each of its domains makes to substrate binding and exoribonuclease activity.
Despite differences in their physiological roles and intrinsic substrate specificities, RNase R and RNase II both belong to the widely distributed RNR family of exoribonucleases (10–12). RNR family members are all large multidomain proteins with processive 3′ to 5′ hydrolytic exoribonuclease activity that share a common linear domain organization. RNase R contains two cold-shock domains (CSD1 and CSD2) near its N terminus, a central nuclease, or RNB domain, an S1 domain near the C terminus, and a low complexity, highly basic region at the C terminus (Fig. 1A). The nuclease domain contains four highly conserved sequence motifs (10, 11). Motif I contains four conserved aspartate residues that are thought to coordinate two divalent metal ions that facilitate a two-metal ion mechanism similar to that of DEDD family exoribonucleases and the proofreading domains of many polymerases (13, 14). CSDs (15–17) and S1 domains (18, 19) are well known examples of RNA-binding domains. Interestingly, there are reports that both of these domains can act as nucleic acid chaperones and unwind RNA (20–29), providing a possible explanation for the ability of RNase R to degrade structured RNAs. The role of the basic region at the C terminus of RNase R is unknown, but it may act as an RNA-binding domain and/or a mediator of protein-protein interactions.
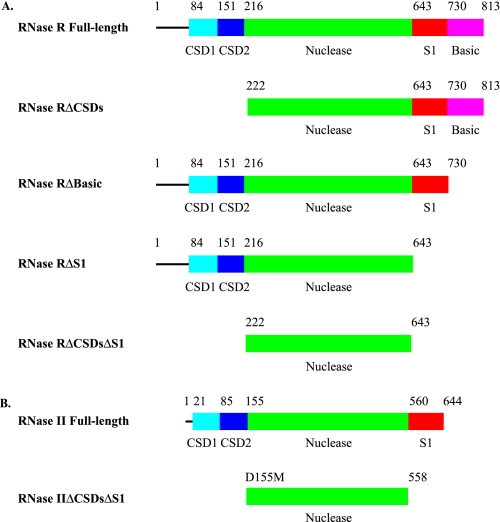
FIGURE 1.
Linear domain organization of RNase R and RNase II proteins. The CSDs are colored in cyan and blue for CSD1 and CSD2, respectively, the nuclease domains are in green, the S1 domains are red, and the low complexity, highly basic region, found in RNase R only, is in magenta. A, RNase R. RNase R full-length is the full-length wild-type RNase R protein. RNase RΔCSDs lacks both CSD1 and CSD2. RNase RΔBasic is missing the low complexity, highly basic region. RNase RΔS1 is missing both the S1 domain and the low complexity, highly basic region. RNase RΔCSDsΔS1 consists of the nuclease domain alone. B, RNase II. RNase II full-length is the full-length wild-type RNase II protein. RNase IIΔCSDsΔS1 contains the nuclease domain alone.
Crystal structures of E. coli wild-type RNase II and a D209N catalytic site mutant in complex with single-stranded RNA have recently been solved (14, 30). In these structures the two CSDs and the S1 domain come together to form an RNA-binding clamp that directs RNA to the catalytic center at the base of a narrow, basic channel within the nuclease domain (14, 30). Only single-stranded RNA can be accommodated by the RNA-binding clamp and the nuclease domain channel, which explains the single strand specificity of RNase II. It is expected that RNase R will adopt a similar structure.
In this study, we determine the contribution that each of the domains of RNase R makes to RNA-binding and exoribonuclease activity. We show that the CSDs and the S1 domain are important for substrate binding, although their roles differ. Of most interest, we show that the nuclease domain alone of RNase R is sufficient to degrade through double-stranded RNA, whereas the nuclease domain of RNase II is unable to carry out this reaction. The nuclease domain of RNase R also binds RNA more tightly, which may explain the difference in catalytic properties between RNase R and RNase II.
EXPERIMENTAL PROCEDURES
Materials—DNA primers were synthesized and purified by Sigma Genosys. pET15b and the KOD Hot Start DNA Polymerase Kit were from Novagen. DpnI, NcoI, BamHI, EcoRI, T4 DNA ligase, and T4 polynucleotide kinase were from New England Biolabs, Inc. pETRNB (14, 30) and purified RNase RΔBasic (31) and RNase II (30) were from Dr. A. Malhotra (University of Miami, Miami, FL). RNA oligoribonucleotides were synthesized and purified by Dharmacon Inc. [γ-32P]ATP was from PerkinElmer Life Sciences and poly[8-3H]adenylic acid was purchased from Amersham Biosciences. SequaGel solutions for denaturing urea-polyacrylamide gels were obtained from National Diagnostics. Nitrocellulose and Biodyne Plus nylon membranes were from Pall Corp. All other chemicals were reagent grade.
Cloning of RNase R Truncations—pETRΔS1 was created by inserting two stop codons and a frameshift after the codon corresponding to amino acid 643 of RNase R in pETR (8) by standard site-directed mutagenesis protocols. An EcoRI site was also inserted at this position for screening purposes. The forward primer used was 5′-g aag tgt gac ttc atg ctc gac cag gta tag tga att cgg taa cgt ctt taa agg cgt aat ttc cag c-3′, and the reverse primer, 5′-g ctg gaa att acg cct tta aag acg tta ccg aat tca cta tac ctg gtc gag cat gaa gtc aca ctt c-3′ (the stop codons are shown in bold and the EcoRI site is underlined). pETRΔCSDs and pETRΔCSDsΔS1 were generated by amplifying the DNA sequence coding for amino acids 222 to 813 of RNase R from pETR (8) or pETRΔS1 (thereby retaining the stop codons and frameshift following the codon for amino acid 643), respectively, and inserting it into the NcoI-BamHI site of pET15b. The forward primer was 5′-atc aat cat cca tgg cgg ttg ata tcg ctc tgc gta ccc-3′ (the NcoI restriction site is underlined and the start codon is in bold). The reverse primer was 5′-tgt atg cat tgg atc ctc act ctg cca ctt ttt tct tcg ccg cac g-3′ (the BamHI restriction site is underlined).
Cloning of RNase R D278N—pETRD278N was generated by converting the GAC codon corresponding to amino acid 278 in pETR (8) to AAC by standard site-directed mutagenesis protocols. The forward primer was 5′-ggg gaa gac gcc cgt aac ttt gac gat gca gtt tac tgc g-3′ and the reverse primer, 5′-c gca gta aac tgc atc gtc aaa gtt acg ggc gtc ttc gcc-3′ (the mutated codon is shown in bold).
Cloning of RNase IIΔCSDsΔS1—pETIIΔCSDsΔS1(D155M) was generated by amplifying the DNA sequence coding for amino acids 156 to 558 of RNase II from pETRNB (30) and inserting it into the NcoI-BamHI site of pET15b. An initiating methionine codon was inserted before amino acid 156 and a stop codon was inserted after amino acid 558. The forward primer was 5′-aat cat cca tgg atc act ttg tac cgt ggt ggg tt-3′ (the NcoI restriction site is underlined and the start codon is in bold) and reverse primer, 5′-tgc att gga ttc tta ggc ttt gtc ttt cag gaa gcg tgc g-3′ (the BamHI site is underlined and the stop codon is in bold).
Overexpression of RNase R and RNase II Constructs—BL21II-R-(DE3)pLysS harboring pET44R (9), pETRΔCSDs, pETRΔS1, pETRΔCSDsΔS1, pETRD278N, or pETIIΔCSD1ΔS1 (D155M) was grown at 37 °C with shaking to an A600 ≈ 0.6 in 500 ml of yeast-Tryptone medium supplemented with 100 μg/ml ampicillin, 34 μg/ml chloramphenicol, 25 μg/ml kanamycin, and 10 μg/ml tetracycline. Expression was induced by the addition of isopropyl β-d-thiogalactopyranoside to a final concentration of 1 mm and cells were allowed to grow for a further 2 h at 37 °C. Cells were harvested by centrifugation at 5,000 × g at 4 °C. The resulting cell pellet was stored frozen at -80 °C.
Purification of RNase R and RNase II Constructs—All steps were carried out at 4 °C and all purification columns were loaded by ÄKTAbasic 100 FPLC (Amersham Biosciences) unless stated otherwise.
Purification of Full-length Wild-type RNase R and RNase R D278N—Full-length wild-type RNase R and RNase R D278N were purified as described previously for full-length wild-type RNase R (9).
Purification of RNase RΔCSDs—The frozen cell pellet for BL21II-R-(DE3)pLysS, pETRΔCSDs, was thawed on ice and resuspended in 25 ml of 50 mm HEPES (pH 7.5), 500 mm KCl, 0.5 mm EDTA, 5 mm DTT, and 0.1 mm PMSF. Cells were disrupted by three passes through an EmulsiFlex-C3 high-pressure homogenizer (Avestin) and the lysate was centrifuged at 25,000 × g for 10 min. The resulting supernatant fraction was again centrifuged at 35,000 × g for 20 min and the S35 fraction was loaded onto a 5-ml HiTrap Blue HP affinity column (Amersham Biosciences) equilibrated with 50 mm HEPES (pH 7.5), 500 mm KCl, 0.5 mm EDTA, 5 mm DTT, and 0.1 mm PMSF. RNase RΔCSDs was eluted in a single step to 2 m KCl. The KCl concentration was reduced to 300 mm with a HiPrep 26/10 Desalting column (Amersham Biosciences) equilibrated in 50 mm HEPES (pH 7.5), 300 mm KCl, 0.5 mm EDTA, 5 mm DTT, and 0.1 mm PMSF. Buffer-exchanged RNase RΔCSDs was then applied to a Mono S HR 10/10 cation-exchange column (Amersham Biosciences) equilibrated with 50 mm HEPES (pH 7.5), 300 mm KCl, 0.5 mm EDTA, 5 mm DTT, and 0.1 mm PMSF. RNase RΔCSDs was eluted upon application of a linear gradient from 300 to 600 mm KCl. Fractions containing RNase RΔCSDs were pooled, concentrated to a volume of 1 ml in a Vivaspin 6-ml ultrafiltration concentrator with a 30-kDa molecular mass cut off (Sartorious North America, Inc.), and applied to a HiLoad 16/60 Superdex 200 pg size-exclusion column (Amersham Biosciences) equilibrated with 50 mm HEPES (pH 7.5), 500 mm KCl, 0.5 mm EDTA, 5 mm DTT, 0.1 mm PMSF, and 10% glycerol. Fractions containing RNase RΔCSDs were pooled, concentrated, aliquoted, and stored frozen at -80 °C.
Purification of RNase RΔS1—The frozen cell pellet for BL21II-R-(DE3)pLysS, pETRΔS1, was thawed on ice and resuspended in 10 ml of 50 mm Tris-HCl (pH 8.0), 500 mm KCl, 0.5 mm EDTA, 5 mm DTT, and 0.1 mm PMSF. Cells were disrupted by three passes through an Aminco French press at 16,000 p.s.i. The lysate was centrifuged at 150,000 × g for 2 h. The resulting S150 fraction was loaded onto a 5-ml HiTrap Blue HP affinity column equilibrated with 50 mm Tris-HCl (pH 8.0), 500 mm KCl, 0.5 mm EDTA, 5 mm DTT, and 0.1 mm PMSF. RNase RΔS1 was eluted in a single step to 2 m KCl. The KCl concentration was reduced to 100 mm with a HiPrep 26/10 Desalting column equilibrated with 50 mm Tris-HCl (pH 8.0), 100 mm KCl, 0.5 mm EDTA, 5 mm DTT, and 0.1 mm PMSF. Buffer-exchanged RNase RΔS1 was applied to a Mono Q HR 10/10 anion-exchange column (Amersham Biosciences) equilibrated with 50 mm Tris-HCl (pH 8.0), 100 mm KCl, 0.5 mm EDTA, 5 mm DTT, and 0.1 mm PMSF. RNase RΔS1 was eluted upon application of a linear gradient from 100 to 500 mm KCl. Glycerol was added to fractions containing RNase RΔS1 to a final concentration of 10%. The fractions were then pooled, aliquoted, and stored frozen at -80 °C.
Purification of RNase RΔCSDsΔS1 and RNase IIΔCSDsΔS1—The frozen cell pellets for BL21II-R-(DE3)pLysS harboring pETRΔCSDsΔS1 or pETIIΔCSDsΔS1(D155M) were thawed on ice and resuspended in 10 ml of 50 mm Tris-HCl (pH 8.0), 500 mm KCl, 0.5 mm EDTA, 5 mm DTT, and 0.1 mm PMSF. Cells were disrupted by three passes through an Aminco French press at 16,000 p.s.i. The lysate was centrifuged at 150,000 × g for 2 h. The S150 was diluted 10-fold with 50 mm Tris-HCl (pH 8.0), 0.5 mm EDTA, 5 mm DTT, and 0.1 mm PMSF to reduce the KCl concentration to 50 mm and loaded onto a HiPrep 16/10 Heparin FF column (Amersham Biosciences) equilibrated with 50 mm Tris-HCl (pH 8.0), 50 mm KCl, 0.5 mm EDTA, 5 mm DTT, and 0.1 mm PMSF. RNase RΔCSDsΔS1 or RNase IIΔCSDsΔS1 were eluted upon application of a linear gradient from 50 to 500 mm KCl. Fractions containing RNase RΔCSDsΔS1 or RNase IIΔCSDsΔS1 were pooled and applied to a HiPrep 26/10 Desalting column equilibrated with 50 mm Tris-HCl (pH 8.0), 100 mm KCl, 0.5 mm EDTA, 5 mm DTT, and 0.1 mm PMSF to bring the KCl concentration to 100 mm. Buffer-exchanged RNase RΔCSDsΔS1 or RNase IIΔCSDsΔS1 was applied to a Mono Q HR 10/10 anion-exchange column equilibrated with 50 mm Tris-HCl (pH 8.0), 100 mm KCl, 0.5 mm EDTA, 5 mm DTT, and 0.1 mm PMSF. RNase RΔCSDsΔS1 or RNase IIΔCSDsΔS1 were eluted upon application of a linear gradient from 100 to 500 mm KCl. Glycerol was added to fractions containing RNase IIΔCSDsΔS1 to a final concentration of 10%. The fractions were then pooled, concentrated, aliquoted, and stored frozen at -80 °C. Fractions containing RNase RΔCSDsΔS1 were concentrated down to a volume of 1 ml and applied to a HiLoad 16/60 Superdex 75 pg size-exclusion column (Amersham Biosciences) equilibrated with 50 mm Tris-HCl (pH 8.0), 500 mm KCl, 0.5 mm EDTA, 5 mm DTT, 0.1 mm PMSF, and 10% glycerol. Fractions containing RNase R ΔCSDsΔS1 were pooled, concentrated, aliquoted, and stored frozen at -80 °C.
Preparation of Oligoribonucleotide Substrates—Oligoribonucleotides were deprotected according to the manufacturer's instructions. The single-stranded oligoribonucleotide substrates used were A4, A17, and ss17-A17 (5′-CCCCACCACCAUCACUUA17-3′). These were 5′-labeled with 32P using T4 polynucleotide kinase and [γ-32P]ATP. A substrate consisting of a 17-base pair duplex with a 17-nucleotide 3′ overhang (ds17-A17) was prepared by mixing 5′-32P-labeled ss17-A17 with the non-radioactive complementary oligoribonucleotide (5′-AAGUGAUGGUGGUGGGG-3′) in a 1:1.2 molar ratio in the presence of 10 mm Tris-HCl (pH 8.0) and 20 mm KCl, heating the mixture in a boiling water bath for 5 min, and then allowing the solution to cool slowly to room temperature.
Acid Soluble Activity Assays—Assays were carried out in 50-μl reaction mixtures containing 50 mm Tris-HCl (pH 8.0), 300 mm KCl, 0.25 mm MgCl2, 5 mm DTT, and 30 μg[3H]polyadenylic acid substrate (100 cpm nmol-1). The concentration of purified enzyme was adjusted to ensure that less than 25% of the substrate was degraded. Reaction mixtures were incubated at 37 °C for 15 min. The reaction was stopped by the addition of 150 μl of 0.5% (w/v) yeast RNA and 200 μl of 20% (w/v) trichloroacetic acid. This was incubated on ice for 15 min and then centrifuged at 16,000 × g for 10 min at 4 °C. The acid-soluble counts in 200 μl of the supernatant fraction were determined by liquid scintillation counting using a LS 6500 multipurpose scintillation counter (Beckman Coulter, Inc.). Activities were normalized according to the moles of protein in each sample.
Electrophoretic Activity Assays—RNase R assays were typically carried out in 30-μl reaction mixtures containing 50 mm Tris-HCl (pH 8.0), 300 mm KCl, 0.25 mm MgCl2, 5 mm DTT, and 10 μm oligoribonucleotide substrate. RNase II assays were carried out in 30-μl reaction mixtures containing 50 mm Tris-HCl (pH 8.0), 100 mm KCl, 1 mm MgCl2, 5 mm DTT, and 10 μm oligoribonucleotide substrate. The amount of purified enzyme was as indicated in the figures or, for calculation of the initial rates, was selected to ensure that less than 25% of the substrate was degraded. Reaction mixtures were incubated at 37 °C, portions were taken at the indicated times or at regular intervals for determination of the initial rates, and the reaction was terminated by the addition of 2 volumes of gel loading buffer (95% formamide, 20 mm EDTA, 0.025% bromphenol blue, and 0.025% xylene cyanol). Reaction products were resolved on denaturing 7.5 m urea, 20% polyacrylamide gels and visualized using a STORM 840 PhosphorImager (GE Healthcare). Quantification was carried out using ImageJ (NIH) (32), initial rates were calculated from at least four points taken from the linear region of the degradation curve.
Binding Assays—The double-filter nucleic acid-binding assay developed by Wong and Lohman (33) and adapted by Tanaka and Schwer (34) was used. Nitrocellulose and nylon membranes were washed as described by Vincent and Deutscher (9) and equilibrated in binding buffer (50 mm Tris-HCl, pH 8.0, 100 mm KCl, 5 mm DTT, 10 mm EDTA, and 10% glycerol) for at least 1 h prior to use. Twenty-μl reaction mixtures containing 50 mm Tris-HCl (pH 8.0), 100 mm KCl, 5 mm DTT, 10 mm EDTA, 10% glycerol, 200 pm 32P-labeled oligoribonucleotide substrate, and varying amounts of purified enzyme were incubated on ice for 30 min. No degradation of the RNA occurs in the absence of Mg2+. A 96-well dot-blot apparatus (Bio-Rad) was used to apply the sample to a nitrocellulose membrane placed above a nylon membrane. Each well was washed with 100 μl of ice-cold binding buffer just prior to loading the sample and immediately following sample application. The apparatus was disassembled, the membranes were allowed to air dry and were visualized using a PhosphorImager. Quantification was carried out in ImageJ (NIH) (32) and the fraction of RNA bound was determined from signalnitrocellulose/(signalnitrocellulose + signalnylon). The fraction of RNA bound was plotted against the total enzyme concentration and the Kd was determined by non-linear regression analysis using a one-site binding hyperbola in Prism (GraphPad Software, Inc.).
RESULTS
Construction of Truncated RNase R Proteins—Based upon sequence analysis, RNase R contains a central nuclease domain and four putative RNA-binding domains: two cold-shock domains (CSD1 and CSD2) near the N terminus of the protein, an S1 domain near the C terminus of the protein, and a low complexity, highly basic region at the C terminus (Fig. 1A). To investigate the relative contribution that each of these domains makes to substrate binding and exoribonuclease activity, a series of truncated RNase R proteins was constructed. The domain organization of the truncated proteins is shown in Fig. 1A. RNase RΔCSDs is missing the first 221 amino acids of RNase R, which include CSD1 and CSD2. RNase RΔBasic lacks the 83 amino acids from the C terminus, which comprise the low complexity, highly basic region. RNase RΔS1 is truncated 170 amino acids from the C terminus to remove both the S1 domain and the low complexity, highly basic region. Finally, RNase RΔCSDsΔS1 consists of the nuclease domain alone, and, therefore, lacks all of the putative RNA-binding domains.
With the exception of RNase RΔBasic (31), which was obtained from Dr. A. Malhotra (University of Miami, Miami, FL), the RNase R truncations were expressed and purified from an R-/II- strain as described under “Experimental Procedures.” Homogeneous preparations were obtained for each of the RNase R truncations as determined by SDS-PAGE (data not shown).
RNA Binding by Truncated RNase R Proteins—To assess the contribution that each domain of RNase R makes to substrate binding, a double-filter binding assay was used to determine the Kd for binding of single-stranded A17 to each of the truncated RNase R proteins. In this assay, protein-bound RNA is retained on the nitrocellulose membrane, whereas unbound RNA is trapped on the lower nylon membrane, thus allowing determination of the fraction of RNA bound. Binding assays were carried out in the absence of Mg2+ and in the presence of 10 mm EDTA to prevent degradation of the RNA upon binding to RNase R.
As shown in Table 1, A17 bound tightly to full-length RNase R with a Kd of 0.8 nm. This is consistent with previous data (9). A17 also bound tightly to RNase RΔBasic with a Kd below the detection limit of the filter binding assay. This increase in affinity of RNA for RNase RΔBasic relative to the full-length protein indicates that the low complexity, highly basic region can affect RNA binding, although how it does so, and whether it binds RNA itself, is unclear.
TABLE 1.
RNA binding to RNase R-truncated proteins The Kd for each substrate-truncated protein pair was determined by a filter binding assay as described under “Experimental Procedures.” Each value represents the mean of at least two experiments.
|
Substrate
|
Kd
|
||||
|---|---|---|---|---|---|
| Full-length | ΔBasic | ΔCSDs | ΔS1 | ΔCSDsΔS1 | |
| nm | |||||
| A17 | 0.8 ± 0.1 | <0.2 | 19 ± 2.5 | 14 ± 1.7 | >10,000a |
| A4 | 1,200 ± 80 | 590 ± 110 | 820 ± 180 | 1,700 ± 310 | >10,000a |
Estimate based upon the highest concentration of truncated RNase R protein tested as saturation was not achieved
In contrast, RNase R-truncated proteins missing either the CSDs or the S1 domain bound A17 with Kd values ∼20-fold higher than full-length RNase R (Table 1). This decrease in affinity upon deletion of either the CSDs or the S1 domain suggests that these regions actually function as RNA-binding domains. This conclusion is supported by the finding that RNase RΔCSDsΔS1, which lacks all of the putative RNA-binding domains, did not detectably bind A17 even when the enzyme was present at a concentration of 10 μm.
To extend these findings, we also measured the Kd for binding of an A4 tetranucleotide to each of the truncated RNase R proteins. If A4 binds in the correct position for exoribonucleolytic cleavage then, based on the RNase II crystal structures (14, 30), binding should be entirely within the channel of the nuclease domain of RNase R and may not be affected by the RNA binding domains. Thus, because each of the truncated RNase R proteins contains an intact nuclease domain, we might expect that they would each bind A4 with a similar Kd.
As shown in Table 1, the Kd for A4 binding to full-length RNase R is 1,200 nm, a value 1,500-fold greater than the Kd for A17. This observation supports the conclusion that the additional contacts made to longer RNA substrates by the CSDs and/or S1 domain contribute significantly to overall substrate binding. Nevertheless, a Kd of 1,200 nm still represents relatively tight binding. In fact, based on these data, as much as 66% of the total binding energy comes from binding within the channel. However, it should also be noted that substrate binding in the channel may be greatly increased by Mg2+ ions that would coordinate between the substrate and the catalytic site residues. Consequently, because these binding assays were carried out in the absence of Mg2+, the actual contribution of the channel to overall binding may be even greater.
Consistent with our hypothesis that each of the truncated RNase R proteins should bind A4 with a similar Kd, full-length RNase R, RNase RΔCSDs, RNase RΔBasic, and RNase RΔS1 all have similar Kd values within the range of ∼600 to 1,700 nm (Table 1). However, as observed with A17, A4 binding to RNase RΔCSDsΔS1 could not be detected (Table 1). This suggests that even though most of the binding energy may be derived from contacts within the channel, the RNA binding domains can have an effect on binding solely within the channel. This point will be considered under “Discussion.”
Exoribonuclease Activity of Truncated RNase R Proteins on Single-stranded RNA—To investigate the role that each of the domains of RNase R plays in the exoribonuclease activity, we measured the activity of each of the truncated RNase R proteins on three single-stranded substrates: poly(A) (estimated to be ≥200 nucleotides long), A17, and A4. RNase R acts processively on all three substrates releasing AMP molecules and limit products of di- and trinucleotides. Activity on poly(A) was determined using an acid soluble assay as described under “Experimental Procedures.” In this assay the release of acid-soluble AMP molecules from the acid-precipitable polymer chain is monitored. In contrast, activity on A17 and A4 was determined using the electrophoretic assay described under “Experimental Procedures.” In this assay the reaction products are analyzed by denaturing PAGE. Because the substrate label is at the 5′ end of the RNA, and RNase R degrades in the 3′ to 5′ direction, only the full-length starting substrate and the di- and trinucleotide limit products can be detected, although AMP would also be released. Therefore, the disappearance of the starting oligoribonucleotide is monitored. It is possible to estimate the AMP released by multiplying this value by the length of the substrate.
Full-length RNase R has similar activity on both poly(A) and A17 substrates, despite the apparent difference shown in Table 2, when the differences between the methods used for measurement of activity are taken into consideration. Thus, if the activity determined for A17 (Table 2) is multiplied by the 17 nmol of AMP present per nmol of A17 to account for total nucleotide released, as in the acid soluble assay, a similar value is obtained for both substrates. A4, on the other hand, is a much poorer substrate, and is degraded ∼400-fold more slowly than A17 based on the calculated nucleotide released (∼100-fold difference based on the electrophoretic assay shown in Table 2).
TABLE 2.
Activity of RNase R-truncated proteins on single-stranded RNA Activity assays were performed as described under “Experimental Procedures.” Each value represents the mean of at least three experiments.
|
Substrate
|
Activitya
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Full-length | ΔBasic | ΔCSDs | ΔS1 | ΔCSDsΔS1 | |||||
| nmol min–1 nmol–1 | |||||||||
| Poly(A) | 460 ± 27 | 980 ± 84 | 330 ± 39 | 12 ± 3.0 | 2.0 ± 1.0 | ||||
| A17 | 33 ± 9.9 | 95 ± 36 | 2.8 ± 1.8 | 0.7 ± 0.1 | 0.007 ± 0.002 | ||||
| A4 | 0.3 ± 0.2 | 0.6 ± 0.1 | 0.2 ± 0.1 | 0.1 ± 0.01 | 0.2 ± 0.1 | ||||
For poly(A) this value represents nmol of AMP released per min per nmol of RNase R. For A17 and A4 this value represents nmol of substrate oligoribonucleotide degraded per min per nmol RNase R
As also shown in Table 2, RNase RΔBasic displays ∼2-fold higher activity than full-length wild-type RNase R on all three substrates tested. This may be related to the observation above, in which this form of RNase R also displayed a higher affinity for the substrates. However, the explanation for the negative effect of the C-terminal, basic region on both binding and activity is not yet understood.
RNase RΔCSDs lost ∼30% of the activity of full-length RNase R on poly(A) (Table 2); however, its activity was reduced ∼90% on the shorter A17 substrate (Table 2). A possible explanation for these data is that the role of the CSDs may be to help recruit the RNA substrate into the catalytic channel. With the longer poly(A) substrate, during the course of an assay, each RNase R, which is a highly processive enzyme, may need to bind and initiate degradation of a new RNA molecule only a few times. Thus, RNase RΔCSDs, even with a deficiency in substrate recruitment, would be expected to have only slightly lower activity than full-length RNase R, as was found. In contrast, degradation of the shorter A17 substrate would require many more binding/initiation events throughout the assay. In this case, the deficiency in substrate recruitment would lead to a significant decrease in activity for RNase RΔCSDs.
Deletion of the S1 domain resulted in a dramatic 40- to 50-fold decrease in activity on both poly(A) and A17 (Table 2). Because the loss in activity was not dependent on substrate length, the S1 domain does not appear to participate in substrate recruitment. Rather, it may function to stabilize and orient the substrate to enable positioning of the 3′ end for efficient catalysis.
The RNase RΔCSDsΔS1 truncated protein retained only ∼0.5% activity of the full-length protein on poly(A), and only 0.02% activity on A17 (Table 2). The combined deletion of the CSDs and the S1 domain most likely renders this protein deficient in both the substrate recruitment and substrate orientation components of RNA binding, resulting in low activity. Inasmuch as RNA binding is a prerequisite for exoribonuclease activity, the fact that some activity can be detected with RNase RΔCSDsΔS1 indicates that the nuclease domain of RNase R must be able to bind to RNA. Nevertheless, poor substrate binding may at least partly account for the extremely low levels of activity observed. Binding of A17 could not be detected in the filter binding assay at an RΔCSDsΔS1 concentration of 10 mm (Table 1) and assays were performed at a substrate concentration of 10 mm. Also, increasing the substrate concentration from 10 to 25 mm for a substrate consisting of a 17-base pair duplex with a 17-nucleotide 3′ single-stranded overhang (ds17-A17) enhanced the rate (data not shown).
The apparent discrepancy between the filter binding assay and the activity assays for RNase RΔCSDsΔS1 is best explained by the fact that Mg2+ is absent in the filter binding assays, but present in the activity assays. Two Mg2+ are expected to bind at the catalytic center of RNase R and participate in a two-metal ion exonuclease mechanism (13, 14). The Mg2+ ions provide additional contacts between the protein and the RNA substrate in the activity assays relative to the filter binding assays resulting in tighter binding in the former assays. This contribution to substrate binding appears to be critical for the RNase RΔCSDsΔS1 truncated protein in which the usual stabilization by the RNA-binding domains would be lacking.
To confirm this hypothesis, we compared the activity of each of the RNase R-truncated proteins on A4. As discussed earlier, A4 should be bound entirely within the channel of the nuclease domain of RNase R, and only the nuclease domain should contribute to binding and activity. Consistent with this idea, all of the RNase R-truncated proteins, including RNase RΔCSDsΔS1, have comparable activity on A4 (Table 2). This suggests that the presence of Mg2+ is important for substrate binding in the channel as well as for catalysis.
Furthermore, a D278N mutation at the catalytic center of RNase R is inactive on A4, but retains ∼4% activity of wild-type RNase R on poly(A) and A17 (data not shown). Asp-278 is predicted to coordinate a Mg2+ at the catalytic center, and mutation of this residue would disrupt Mg2+ binding. The fact that this effect is greater for the shorter substrate, which is bound entirely within the nuclease domain, supports the importance of Mg2+ for binding within the channel.
Exoribonuclease Activity of Truncated RNase R Proteins on Substrates Containing a Double-stranded Region—Full-length wild-type RNase R can degrade through a double-stranded region of RNA provided there is a 3′ single-stranded overhang at least 5 nucleotides long to serve as a binding site from which exoribonuclease activity can be initiated (5, 9).5 RNase R is the only known 3′ to 5′ exoribonuclease able to degrade through extensive secondary structure without the aid of a helicase activity. Consequently, the mechanism that it uses to proceed through structured regions of RNA is of great interest. As both CSDs and S1 domains have been reported to have nucleic acid chaperone activity (20–29), we reasoned that the CSDs and/or S1 domain of RNase R might be required for the degradation of highly structured substrates.
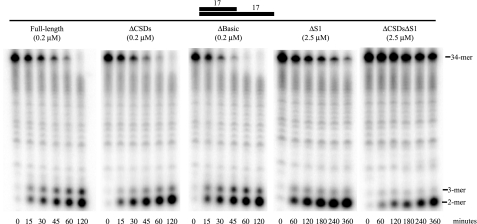
To test this, we compared the action of each of the truncated RNase R proteins on a substrate consisting of a 17-base pair, GC-rich duplex with a 17-nucleotide poly(A) 3′ overhang (ds17-A17). Fig. 2 shows that all of the truncated RNase R proteins, including RNase RΔCSDsΔS1, digest the duplex-containing substrate to the limit products of di- and trinucleotides, indicating that each of the truncated proteins is capable of degrading double-stranded RNA. Consequently, if there is any RNA-chaperone activity associated with either the CSDs and/or the S1 domain, it cannot be responsible for the ability of RNase R to degrade structured RNAs. Rather, degradation of double-stranded RNA is a property of the nuclease domain alone.
FIGURE 2.
Exoribonuclease activity of truncated RNase R proteins on a substrate containing a duplex. Assays were carried out as described under “Experimental Procedures” with 10 μm ds17-A17 substrate and the indicated enzyme concentrations. Aliquots were taken at the indicated times and analyzed by denaturing PAGE.
The activity of each of the truncated RNase R proteins on the duplex-containing substrate (ds17-A17) and also on the corresponding single-stranded 34-mer (ss17-A17) was quantified. As shown in Table 3, the activity on ds17-A17 was 10-fold lower than on the corresponding single-stranded substrate with full-length RNase R. This has been observed previously (5), and apparently reflects the additional time required for RNase R to degrade through the double-stranded region. A similar reduction in activity on the duplex-containing substrate was observed for RNase RΔBasic. The activity on ds17-A17 relative to ss17-A17 was a further 3-fold lower for RNase RΔS1 compared with full-length RNase R, suggesting that the S1 domain may assist in the degradation of structured substrates. In contrast, RNase RΔCSDs and RNase RΔCSDsΔS1 degraded ds17-A17 and ss17-A17 at similar rates. These data imply that the CSDs impede the degradation through a double-stranded region.
TABLE 3.
Activity of truncated RNase R proteins on a substrate containing a duplex Activity assays were performed as described under “Experimental Procedures.” Each value represents the mean of at least three experiments.
|
Substrate
|
Activitya
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Full-length | ΔBasic | ΔCSDs | ΔS1 | ΔCSDsΔS1 | |||||
| nmol min–1 nmol–1 | |||||||||
| ss17-A17 | 5.5 ± 2.6 | 11 ± 0.7 | 0.5 ± 0.1 | 0.6 ± 0.1 | 0.005 ± 0.0002 | ||||
| ds17-A17 | 0.5 ± 0.2 | 0.7 ± 0.4 | 0.7 ± 0.2 | 0.02 ± 0.01 | 0.003 ± 0.0009 | ||||
This value represents nmol of substrate oligoribonucleotide degraded per min per nmol of RNase R
Comparison of the Nuclease Domains of RNase R and RNase II—RNase R and RNase II both belong to the RNR family of exoribonucleases (10, 11), and consequently, they share a common linear domain organization and probably adopt similar three-dimensional structures. Despite this, RNase II is specific for single-stranded RNA, stalling 6 to 11 nucleotides before a duplex (5, 8, 35–37), whereas RNase R readily degrades both single- and double-stranded RNA molecules (5, 8).
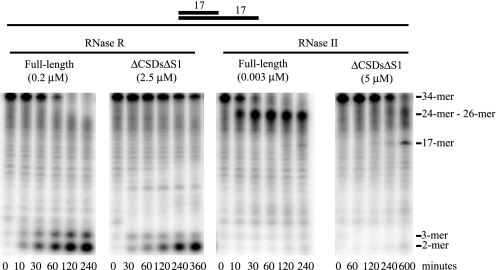
It has been reported that for RNase II the initial barrier to degradation of duplex RNA is the RNA-binding clamp formed by the CSDs and S1 domain (30, 37). A truncated RNase II protein containing only the nuclease domain and part of CSD2 still stalls 4 to 6 nucleotides before a double-stranded region suggesting that the nuclease domain itself also is unable to degrade through structured RNA (37). To confirm this, we constructed a truncated RNase II protein consisting of the nuclease domain alone, but lacking any part of CSD2 (Fig. 1B), and examined its activity on a 17-base pair duplex with a 17-nucleotide overhang (ds17-A17). This protein was compared with the nuclease domain of RNase R.
As shown in Fig. 3, full-length RNase R and RNase RΔCSDsΔS1 both digest ds17-A17 to the limit products of di- and trinucleotides. Full-length RNase II, on the other hand, stalls 7 to 9 nucleotides before the double-stranded region, as reported earlier (5). In contrast, RNase IIΔCSDsΔS1 appears to degrade further, stalling at the single strand/double strand junction. These data indicate that removal of the RNA-binding domains does allow RNase II to proceed further. Yet, it is still unable to digest through a double strand.
FIGURE 3.
Comparison of RNase II and RNase R full-length proteins and nuclease domain-truncated proteins on a substrate containing a duplex. Assays were carried out as described under “Experimental Procedures” with 10 μm ds17-A17 substrate and the indicated enzyme concentrations. Aliquots were taken at the indicated times and analyzed by denaturing PAGE. The origin of the band at the approximate position for an 8-mer that first appears at the 30-min time point with RNase RΔCSDsΔS1 is unknown. However, it is not observed reproducibly.
It is difficult to make a direct quantitative comparison between the exoribonuclease activity of RNase R and that of RNase II on substrates such as ds17-A17 because RNase R completely degrades the substrate, whereas RNase II only degrades within the single-stranded region. Thus, to directly compare the activity of full-length RNase R and RNase II and their respective nuclease domains, we measured the exoribonuclease activity of each of the proteins on a single-stranded A17 substrate. As shown in Table 4, full-length RNase II is ∼20-fold more active on A17 than full-length RNase R. Similar results have been reported when poly(A) is used as a substrate (8). In contrast, the nuclease domain of RNase R and the nuclease domain of RNase II have similar activity on A17. This represents an ∼5,000-fold reduction in activity relative to the full-length protein for RNase R, but an ∼85,000-fold reduction for RNase II, and suggests that the RNA-binding domains of RNase II play a more important role in its exoribonuclease activity than they do in the activity of RNase R.
TABLE 4.
Comparison of full-length RNase R and RNase II and their respective nuclease domains Activity assays were performed as described under “Experimental Procedures.” Each value represents the mean of at least three experiments. The Kd values were determined by a filter binding assay as described under “Experimental Procedures.” Each value represents the mean of at least two experiments.
These data prompted us to determine the Kd values for single-stranded A17 and for A4 for the RNase R and RNase II full-length proteins and for the corresponding nuclease domains. As observed above with RNase R, in the absence of Mg2+, binding also could not be detected for A17 to the nuclease domain of RNase II. However, full-length RNase II did bind to A17 with a Kd of 7.1 nm (Table 4). This value is consistent with previous data (30, 37, 38), but interestingly, it is ∼10-fold higher than the Kd for the same substrate with full-length RNase R, implying that RNase R binds RNA more tightly than does RNase II.
To determine whether the weaker binding of RNase II is due to differences between the RNA-binding domains or between the nuclease domains, we measured the Kd for the A4 tetranucleotide. As A4 should be bound entirely within the nuclease domain of RNase R or RNase II, differences in the Kd with this substrate would reflect differences in RNA binding between the two nuclease domains. As shown in Table 4, whereas full-length RNase R bound to A4 with a Kd of 1,200 nm, binding to RNase II was beyond the limits of detection. Because RNase II does have activity on A4 (data not shown), it appears that Mg2+ also is critical for RNase II binding to a short oligoribonucleotide substrate. Nevertheless, taken together, these data indicate that the nuclease domain of RNase R binds RNA at least 10-fold more tightly than the nuclease domain of RNase II, and this may be related to the ability of RNase R, but not RNase II, to digest through double-stranded substrates.
DISCUSSION
Using a series of truncated RNase R proteins and a wide range of specifically chosen RNA substrates, we have determined the distinctive role that each of the domains of RNase R plays in RNA binding and catalysis. We found that the nuclease domain alone is sufficient for RNase R to bind and degrade RNA, including structured RNA. Although its activity on most substrates is reduced by more than 99% compared with full-length RNase R, the short A4 substrate, which should be bound entirely within the nuclease domain, is degraded at a similar rate by both proteins. These data indicate that the CSDs and/or the S1 domain are required only for the efficient degradation of longer RNA substrates, and not for catalysis per se.
Nevertheless, despite its low level of activity, RNase RΔCSDsΔS1 is able to completely degrade through the double-stranded region of the ds17-A17 substrate. Thus, degradation of extensively structured RNA is a property of the nuclease domain alone, and it is a unique property of the nuclease domain of RNase R. Neither full-length RNase II nor its nuclease domain are able to degrade through double-stranded RNA. It has been suggested that one barrier to degradation of duplex RNA by RNase II is the RNA-binding clamp formed by the CSDs and the S1 domain (14, 30, 37). Yet, our data indicate that RNase IIΔCSDsΔS1, in which the CSDs and S1 domain have been deleted, is still unable to degrade through double-stranded RNA. However, this truncated protein can now proceed as far as the single strand/double strand junction, indicating that the clamp does serve as an initial barrier to duplex RNA.
Based on the crystal structures of RNase II, its nuclease domain can only accommodate single-stranded RNA (14, 30). Therefore, to reach the catalytic center at the base of the nuclease channel, 5 single-stranded nucleotides would have to be bound within the nuclease domain. Consequently, to degrade right up to the RNA duplex, RNase IIΔCSDsΔS1 would need to partially access or open the double-stranded region. The fact that it cannot proceed through the duplex suggests that 5 nucleotides may be the limit in which the RNA duplex can transiently open by a thermal breathing mechanism or by the action of the protein.
Several pieces of evidence indicate that the nuclease domain of RNase R binds more tightly to RNA than the nuclease domain of RNase II. A17 and A4 bind to RNase II with higher Kd values than to RNase R. Also, the limit products of digestion are di- and trinucleotides for RNase R, but tetra- and pentanucleotides for RNase II, suggesting that very short oligoribonucleotides within the nuclease domain of RNase R bind more tightly than to RNase II, and, therefore, can be acted upon prior to dissociation. In addition, RNase II becomes distributive on substrates shorter than ∼10 nucleotides (8, 39). This suggests that RNase II requires the CSDs and/or the S1 domain to stabilize substrate binding; the nuclease domain itself is not sufficient. In contrast, RNase R remains processive on substrates as short as 6 nucleotides (8). Consistent with this, there was a much greater reduction in activity relative to the full-length proteins upon removal of the CSDs and S1 domain from RNase II compared with RNase R.
CSDs and S1 domains are examples of canonical RNA-binding domains, and the lowered activity of the RNase RΔCSDsΔS1 on longer substrates is presumably due to deficiencies in stabilization of substrate binding in the absence of these regions. In support of this idea, binding of A17 to RNase RΔCSDsΔS1 could not be detected. Unexpectedly, we were also unable to detect binding of A4 even though it should be bound exclusively within the nuclease domain; A4 did bind to full-length RNase R and each of the other truncated proteins. This indicates that the CSDs and/or S1 domain also play some role in the binding of short substrates. One possibility is that these domains undergo a conformational change that blocks the entrance to the nuclease domain channel and prevents dissociation of a substrate once it has bound. In fact, the crystal structures for RNase II suggest that the RNA-binding clamp formed by the CSDs and the S1 domain is flexible and narrows upon RNA binding (14).
Inasmuch as RNase RΔCSDsΔS1 displays activity on both A17 and A4, the nuclease domain alone must bind these RNAs. Most likely, Mg2+, which is present in the activity assay, but not in the binding assay, enhances substrate binding by coordinating between the catalytic site residues of the protein and the RNA molecule. We attempted, but were unable, to determine the Kd for binding of A4 to RNase RΔCSDsΔS1 in the presence of Mg2+ due to substrate degradation. Nevertheless, in support of the role of Mg2+, mutating Asp-278, one of the residues required for coordination of Mg2+, to asparagine abolishes activity on A4, and significantly reduces activity on A17 and poly(A).
Truncated RNase R proteins missing either the CSDs or the S1 domain each bound A17 with ∼20-fold higher Kd values, consistent with the conclusion that both domains play a role in RNA binding. However, the CSDs and S1 deletions differed in their effects on activity suggesting that their specific functions in the binding process may not be the same. Thus, deletion of the CSDs resulted in ∼90% lower activity on A17 and ss17-A17, but only a 30% reduction with poly(A), and no effect with ds17-A17. Removal of the S1 domain, on the other hand, led to ≥90% lower activity on all of these substrates. Activity on A4 was little affected by removal of either domain.
These observations can be reconciled if deletion of the CSDs impairs substrate recruitment, i.e. the initial binding of the substrate to the enzyme. In such a scenario, RNase RΔCSDs is able to degrade a longer substrate, such as poly(A), at a relatively faster rate because fewer initial binding events are required compared with a shorter substrate such as A17 or ss17-A17. The lack of an effect of CSD removal on the same length partially double-stranded substrate, ds17-A17, suggests that the CSDs may normally act as a barrier to double-stranded RNA, as is observed for the CSDs of RNase II (30, 37). In contrast, removal of the S1 domain affects all of the substrates, except A4, equally. A likely explanation is that the S1 domain anchors all of the longer substrates to orient them properly for efficient exoribonucleolytic activity. In fact, an “anchor” binding site was suggested to be present in RNase II (39, 40). We propose that the S1 domain may serve that role in both RNase R and RNase II, especially as it is known that the S1 domain of RNase R can substitute for the S1 domain of RNase II (41).
RNase R is the only known 3′ to 5′ exoribonuclease with the ability to degrade through structured RNA without the aid of a helicase activity. The data presented here provide important, new information regarding the mechanism of substrate recruitment, positioning, and degradation by RNase R. In particular, the characterization of the individual domains of RNase R has revealed critical determinants for substrate binding and catalysis. We anticipate that future studies will lead to an even more detailed understanding of RNase R catalysis.
Acknowledgments
We thank Dr. A. Malhotra, Dr. Y. Zuo, and Y. Wang for the pETRNB plasmid and for purified RNase RΔBasic and RNase II. We thank Dr. T. K. Harris for the use of the fast protein liquid chromatography system. We thank Dr. A. Malhotra and Dr. Y. Zuo for helpful discussions and comments.
This work was supported, in whole or in part, by National Institutes of Health Grant GM16317. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: PNPase, polynucleotide phosphorylase; CSD, cold-shock domain; DTT, dithiothreitol; PMSF, phenylmethylsulfonyl fluoride; ss, single-stranded; ds, double-stranded.
S. Chebolu, C. Kim, E. Quesada, and M. P. Deutscher, personal communication.
H. A. Vincent and M. P. Deutscher, unpublished observation.
References
- 1.Deutscher, M. P. (2006) Nucleic Acids Res. 34 659-666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Donovan, W. P., and Kushner, S. R. (1986) Proc. Natl. Acad. Sci. U. S. A. 83 120-124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mohanty, B. K., and Kushner, S. R. (2003) Mol. Microbiol. 50 645-658 [DOI] [PubMed] [Google Scholar]
- 4.Khemici, V., and Carpousis, A. J. (2004) Mol. Microbiol. 51 777-790 [DOI] [PubMed] [Google Scholar]
- 5.Cheng, Z. F., and Deutscher, M. P. (2005) Mol. Cell 17 313-318 [DOI] [PubMed] [Google Scholar]
- 6.Cheng, Z. F., and Deutscher, M. P. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 6388-6393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li, Z., Reimers, S., Pandit, S., and Deutscher, M. P. (2002) EMBO J. 21 1132-1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheng, Z. F., and Deutscher, M. P. (2002) J. Biol. Chem. 277 21624-21629 [DOI] [PubMed] [Google Scholar]
- 9.Vincent, H. A., and Deutscher, M. P. (2006) J. Biol. Chem. 281 29769-29775 [DOI] [PubMed] [Google Scholar]
- 10.Mian, I. S. (1997) Nucleic Acids Res. 25 3187-3195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zuo, Y., and Deutscher, M. P. (2001) Nucleic Acids Res. 29 1017-1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Condon, C., and Putzer, H. (2002) Nucleic Acids Res. 30 5339-5346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Steitz, T. A., and Steitz, J. A. (1993) Proc. Natl. Acad. Sci. U. S. A. 90 6498-6502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frazão, C., McVey, C. E., Amblar, M., Barbas, A., Vonrhein, C., Arraiano, C. M., and Carrondo, M. A. (2006) Nature 443 110-114 [DOI] [PubMed] [Google Scholar]
- 15.Phadtare, S., Alsina, J., and Inouye, M. (1999) Curr. Opin. Microbiol. 2 175-180 [DOI] [PubMed] [Google Scholar]
- 16.Schindelin, H., Jiang, W., Inouye, M., and Heinemann, U. (1994) Proc. Natl. Acad. Sci. U. S. A. 91 5119-5123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Newkirk, K., Feng, W., Jiang, W., Tejero, R., Emerson, S. D., Inouye, M., and Montelione, G. T. (1994) Proc. Natl. Acad. Sci. U. S. A. 91 5114-5118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Subramanian, A. R. (1983) Prog. Nucleic Acids Res. Mol. Biol. 28 101-142 [DOI] [PubMed] [Google Scholar]
- 19.Bycroft, M., Hubbard, T. J., Proctor, M., Freund, S. M., and Murzin, A. G. (1997) Cell 88 235-242 [DOI] [PubMed] [Google Scholar]
- 20.Szer, W., Hermoso, J. M., and Boublik, M. (1976) Biochem. Biophys. Res. Commun. 70 957-964 [DOI] [PubMed] [Google Scholar]
- 21.Bear, D. G., Ng, R., Van Derveer, D., Johnson, N. P., Thomas, G., Schleich, T., and Noller, H. F. (1976) Proc. Natl. Acad. Sci. U. S. A. 73 1824-1828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kolb, A., Hermoso, J. M., Thomas, J. O., and Szer, W. (1977) Proc. Natl. Acad. Sci. U. S. A. 74 2379-2383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thomas, J. O., Kolb, A., and Szer, W. (1978) J. Mol. Biol. 123 163-176 [DOI] [PubMed] [Google Scholar]
- 24.Jiang, W., Hou, Y., and Inouye, M. (1997) J. Biol. Chem. 272 196-202 [DOI] [PubMed] [Google Scholar]
- 25.Bae, W., Xia, B., Inouye, M., and Severinov, K. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 7784-7789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Phadtare, S., Inouye, M., and Severinov, K. (2002) J. Biol. Chem. 277 7239-7245 [DOI] [PubMed] [Google Scholar]
- 27.Phadtare, S., Tyagi, S., Inouye, M., and Severinov, K. (2002) J. Biol. Chem. 277 46706-46711 [DOI] [PubMed] [Google Scholar]
- 28.Phadtare, S., Inouye, M., and Severinov, K. (2004) J. Mol. Biol. 337 147-155 [DOI] [PubMed] [Google Scholar]
- 29.Phadtare, S., and Severinov, K. (2005) Nucleic Acids Res. 33 5583-5590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zuo, Y., Vincent, H. A., Zhang, J., Wang, Y., Deutscher, M. P., and Malhotra, A. (2006) Mol. Cell 24 149-156 [DOI] [PubMed] [Google Scholar]
- 31.Suzuki, H., Zuo, Y., Wang, J., Zhang, M. Q., Malhotra, A., and Mayeda, A. (2006) Nucleic Acids Res. 34 e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Abramoff, M. D., Magelhaes, P. J., and Ram, S. J. (2004) Biophotonics Int. 11 36-42 [Google Scholar]
- 33.Wong, I., and Lohman, T. M. (1993) Proc. Natl. Acad. Sci. U. S. A. 90 5428-5432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tanaka, N., and Schwer, B. (2005) Biochemistry 44 9795-9803 [DOI] [PubMed] [Google Scholar]
- 35.Coburn, G. A., and Mackie, G. A. (1996) J. Biol. Chem. 271 1048-1053 [DOI] [PubMed] [Google Scholar]
- 36.Spickler, C., and Mackie, G. A. (2000) J. Bacteriol. 182 2422-2427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Amblar, M., Barbas, A., Fialho, A. M., and Arraiano, C. M. (2006) J. Mol. Biol. 360 921-933 [DOI] [PubMed] [Google Scholar]
- 38.Barbas, A., Matos, R. G., Amblar, M., López-Viñas, E., Gomez-Puertas, P., and Arraiano, C. M. (2008) J. Biol. Chem. 283 13070-13076 [DOI] [PubMed] [Google Scholar]
- 39.Cannistraro, V. J., and Kennell, D. (1994) J. Mol. Biol. 243 930-943 [DOI] [PubMed] [Google Scholar]
- 40.Cannistraro, V. J., and Kennell, D. (1999) Biochim. Biophys. Acta 1433 170-187 [DOI] [PubMed] [Google Scholar]
- 41.Amblar, M., Barbas, A., Gomez-Puertas, P., and Arraiano, C. M. (2007) RNA (Cold Spring Harbor) 13 317-327 [DOI] [PMC free article] [PubMed] [Google Scholar]