Abstract
Copper metabolism Murr1 domain 1 (COMMD1) is a 21-kDa protein involved in copper export from the liver, NF-κB signaling, HIV infection, and sodium transport. The precise function of COMMD and the mechanism through which COMMD1 performs its multiple roles are not understood. Recombinant COMMD1 is a soluble protein, yet in cells COMMD1 is largely seen as targeted to cellular membranes. Using co-localization with organelle markers and cell fractionation, we determined that COMMD1 is located in the vesicles of the endocytic pathway, whereas little COMMD1 is detected in either the trans-Golgi network or lysosomes. The mechanism of COMMD1 recruitment to cell membranes was investigated using lipidspotted arrays and liposomes. COMMD1 specifically binds phosphatidylinositol 4,5-bisphosphate (PtdIns(4,5)P2) in the absence of other proteins and does not bind structural lipids; the phosphorylation of PtdIns at position 4 is essential for COMMD1 binding. Proteolytic sensitivity and molecular modeling experiments identified two distinct domains in the structure of COMMD1. The C-terminal domain appears sufficient for lipid binding, because both the full-length and C-terminal domain proteins bind to PtdIns(4,5)P2. In native conditions, endogenous COMMD1 forms large oligomeric complexes both in the cytosol and at the membrane; interaction with PtdIns(4,5)P2 increases the stability of oligomers. Altogether, our results suggest that COMMD1 is a scaffold protein in a distinct sub-compartment of endocytic pathway and offer first clues to its role as a regulator of structurally unrelated membrane transporters.
COMMD1 (Murr1) is the founding member of a recently discovered family of COMMD (Copper metabolism Murr1 domain)3 proteins (1). COMMD1 was identified as the product of a gene mutated in dogs with severe hepatic copper toxicosis (2). In several breeds of dogs, especially in Bedlington terriers, a mis-splicing event in the Murr1 gene results in the loss of protein expression (3) and massive copper accumulation in the liver that can reach remarkably high concentrations of 10,000-12,000 μg/g and lead to acute organ failure (4). The identification of Murr1 (COMMD1) by van de Sluis and co-workers (2) generated a great deal of interest and helped to create tools for genetic screening. However, the molecular mechanism through which COMMD1 regulates copper metabolism remains unknown.
COMMD1 was shown to bind Cu(II) in vitro (5), but whether this binding represents an in vivo property is uncertain. The observation, that in dogs lacking COMMD1 hepatic copper uptake is unperturbed but copper accumulates in dense lysosomal granules, suggested a defect in biliary copper export (6). The role of COMMD1 in copper export was further supported by the increased retention of copper in cultured cells in which COMMD1 was down-regulated with small interference RNA (7). However, COMMD1 is a small soluble protein (see below) and cannot directly mediate transmembrane copper transport. It was proposed that COMMD1 controls copper export by regulating the intracellular localization of the copper-transporting ATPase ATP7B, which is chiefly responsible for the removal of excess copper from the liver into the bile. In support of this hypothesis, recombinant COMMD1 was shown to interact with the N-terminal domain of ATP7B in vitro and in cell lysates (8). However, down-regulation of COMMD1 has no apparent effect on the ability of ATP7B to traffic from the trans-Golgi network (TGN) to exocytic vesicles (Ref. 9 and our data, not shown), suggesting that the effect of COMMD1 is downstream of the TGN. More recently, the observed increase in interactions between ATP7B mutants and COMMD1 led to the alternative hypothesis that COMMD1 may be a part of a quality-control mechanism for ATP7B folding (9).
A further layer of complexity (and uncertainly) about COMMD1 function was added by studies demonstrating that the genetic knockout of COMMD1 in mice leads to embryonic lethality between days 9.5 and 10.5 (10). Such a severe and early phenotype in rodents but not in dogs indicates that the functional significance of COMMD1 is species-dependent and may extend beyond hepatic copper export, a suggestion supported by the expression of Murr1 transcripts in all major tissues and cell types (3). Furthermore, various protein-protein interaction experiments uncovered the ability of COMMD1 to dimerize with several members of COMMD family (11, 12) and to form complexes with a growing number of other structurally and functionally unrelated proteins (3). Evaluations of these interactions revealed that COMMD1 levels influence HIV infection, NF-κB signaling, sodium transport, and proteasome-mediated protein degradation (11-14). Today, the importance of COMMD1 in mammalian physiology is well established, whereas the mechanism behind COMMD1 activity, particularly at a cell membrane, remains elusive.
Human COMMD1, characterized in this study, is a soluble 21-kDa protein, which is 88% identical to its canine orthologue. Members of the COMMD family show only modest homology to each other, sharing a single invariant residue (Trp-124 in COMMD1). Structural studies of the full-length COMMD1 have been complicated by protein instability and aggregation. However, the structure of the N-terminal fragment of COMMD1 (residues 1-108) was recently solved using NMR (15). These studies revealed a helical structure and two positive patches at the surface of the fragment, but structural motifs that might offer mechanistic clues to COMMD1 activity were not apparent.
We were in trigued by the observation that several COMMD1-dependent effects involve membrane proteins (copper transporter ATP7B and sodium channel ENaC) or membrane-dependent events (HIV infection and viral escape). Although it is thought that COMMD1 is a cytosolic protein, available data on the staining of endogenous COMMD1 in cells shows vesicular rather than cytosolic pattern (3). Therefore, to better understand the biochemical function of COMMD1 we characterized in detail recombinant COMMD1, determined the localization of endogenous COMMD1 in hepatocytes, and generated a molecular model of this protein. We discovered that COMMD1 binds with high specificity to an important signaling and regulatory lipid, phosphatidylinositol 4,5-bisphosphate (PtdIns(4,5)P2), localizes to the endosomal compartment, and forms higher order oligomers, which are stabilized by interactions with lipid. These properties suggest a mechanism by which COMMD1 can be recruited to the endocytic membranes and act as a scaffold/adaptor protein contributing to protein/vesicle sorting.
EXPERIMENTAL PROCEDURES
Antibodies for COMMD1 Detection—Mouse monoclonal anti-COMMD1 antibody (Ab) was obtained from Abnova (Taipei, Taiwan). Polyclonal anti-COMMD1 Abs were obtained after serial immunization of chickens with purified non-tagged COMMD1 and purification of immune IgY from egg yolk (Aves Laboratories, Tigard, OR). Monoclonal anti-COMMD1 and chicken polyclonal anti-COMMD1 detect equally the monomeric COMMD1, but recognize various oligomeric forms of COMMD1 with different affinities (not shown).
Immunofluorescence Detection of COMMD1 in HepG2 Cells—Cells were seeded onto lysine-coated coverslips and cultured in MEM+Glutamax, 10% fetal bovine serum, 100 units/ml penicillin, and 100 μg/ml streptomycin. Coverslips were washed with phosphate-buffered saline (PBS), pH 7.4, and then cells were fixed in 4% paraformaldehyde in PBS. Blocking and permeabilization were done simultaneously with 3% bovine serum albumin, 1% gelatin, and 0.1% Tween 20. Primary Abs were diluted in PBS with 3% bovine serum albumin, and secondary antibodies were diluted in PBS with 0.1% Tween 20. Primary antibody dilutions were as follows: mouse anti-COMMD1 IgG2, 1:100, chicken anti-COMMD1 polyclonal, 1:500, mouse anti-EEA1 IgG1 (Abcam, Cambridge, MA) 1:100, anti-CHMP2B (Abcam), 1:100, rat anti-LAMP1 (DHSB, University of Iowa), 1:100, anti-Golgin-97 (Molecular Probes, Eugene, OR), 1:100, rat anti-ATP7B (16) 1:1000. Anti-Chicken HiLyte 555 (Anaspec, San Jose, CA) was used at 1 μg/ml. All other secondary Abs were from Invitrogen and used at 2 μg/ml and conjugated with either AlexaFluor 488 (shown in green) or AlexaFluor 555 (shown in red). Confocal microscopy was done utilizing a Zeiss LSM 500 laser-scanning microscope. Treatment of cells with 200 μm BCS or 50 μm CuCl2 was for 8 h in the media described above. The extent of co-localization, i.e. the proportion of overlapping pixels, was measured with the NIH ImageJ “Co-localization Test” plug-in using the Costes method of randomization for 25 iterations (17).
Immunodetection of COMMD1 in Fractionated HepG2 Cell Lysates—HepG2 cells were seeded on 10-cm Petri dishes and allowed to grow for 3 days in the growth medium composed of MEM+Glutamax, 10% fetal bovine serum, penicillin, and streptomycin. Four hours prior to harvest, growth medium was replaced with fresh media containing 200 μm BCS or 50 μm CuSO4. Cells were washed with PBS before harvest and scraped from the plate in a lysis buffer (20 mm HEPES, pH 7.5, 1.0 mm EDTA, 1.0 mm EGTA, 1.0 mm 4-(2-aminoethyl)benzenesulfonylfluoride hydrochloride, and a Roche Complete protease inhibitor mixture). Cells were disrupted in a Dounce homogenizer with a loose-fitting pestle, incubated on ice for 5 min, made isotonic by addition of 0.1 volumes of 2.5 m sucrose, and homogenized by 25 strokes with a tight-fitting pestle. The cell lysates were then centrifuged at 6300 × g for 10 min to pellet nuclei and cell debris. The postnuclear supernatant was centrifuged for 30 min at 100,000 × g to separate the microsomal membranes from a soluble cytosolic fraction. The nuclei were purified from the initial 6300 × g pellet by twice re-suspending in TSE buffer (10 mm Tris-HCl, pH 7.6, 300 mm sucrose, 1.0 mm EDTA, 0.1% Igepal (Sigma-Aldrich)) and centrifuging for 5 min at 4000 × g.
Membrane and nuclear fractions were then solubilized in TSE buffer containing 0.2% Triton X-100, and protein concentration was determined by Bradford assay (18). Equal amounts of protein from nuclear, soluble, and membrane fractions were separated by SDS-PAGE on a 4-20% gradient gel (Pierce) under reducing conditions. Proteins were transferred to nitrocellulose by Western blot, and the membrane was blocked in Aquablock (EastCoastBio, North Berwick, ME) for 1 h at room temperature. Antibody incubations for immunodetection were used at following dilutions: mouse anti-COMMD1 (1:1000), chicken anti-COMMD1 (1:5000). Secondary antibodies, conjugated with either IRDye 700 or IRDye 800, were obtained from Rockland Immunochemicals (Gilbertsville, PA). Fluorescence detection of bound secondary Ab was performed with an Odyssey Scanner (Licor, Lincoln, NE). The same 21-kDa band corresponding to the endogenous COMMD1 was detected by different antibodies, immunodetection with chicken polyclonal anti-COMMD1 shown.
Density Gradient Fractionation of Postnuclear Fraction—HepG2 cells were grown as described above. Fractionation was done essentially as described in a previous study (19). Cells were washed once on the plate with PBS containing 2.0 mm CaCl2 and 0.9 mm MgCl2. Cells were scraped into PBS and centrifuged at 200 × g for 10 min. The cell pellet was resuspended in 3 volumes of 20 mm HEPES, pH 7.4, 150 mm NaCl, and protease inhibitors (Complete, Roche Applied Science). HepG2 cells were lysed by 15 passes through a 27-guage needle. Nuclei and unbroken cells were pelleted by two centrifugation steps at 1000 × g for 10 min. The postnuclear supernatant was loaded onto the top of a 20% Percoll cushion made isotonic with Tris-buffered saline and including protease inhibitors. Centrifugation was carried out in a Beckman Type 60 Ti ultracentrifuge rotor at 20,000 × g for 55 min. Fractions were collected from the bottom of the tube (∼950 μl each), to which CHAPS was added to 10 mm to solubilize membranes. After 1 h on ice with CHAPS, Percoll was sedimented by centrifugation at 100,000 × g for 60 min. Proteins were precipitated with 15% trichloroacetic acid in the presence of 0.12% deoxycholic acid. Denatured proteins were sedimented at 20,000 × g for 20 min, washed with 90% acetone, and air-dried. Denatured proteins were resuspended in 62.5 mm Tris, pH 6.8, 5% SDS, and 10% glycerol, then heated to 95 °C for 5 min. Urea was added to 2.5 m, and solubilized proteins were loaded onto a 12.5% gel for SDS-PAGE.
Cloning, Expression, and Purification of Recombinant COMMD1 and Its N-terminal and C-terminal Domains—Human COMMD1 cDNA (American Type Culture Collection, Manassas, VA) was cloned into pTYB12 (New England Biolabs, Ipswich, MA) to generate a fusion protein with the chitin-binding domain/intein moiety or into the pET28b vector (Novagen/EMD Biosciences, Darmstadt, Germany) to add an N-terminal His6 tag (construct pET28b-COMMD1). In either case the cloning sites, NdeI (5′) and SalI (3′), were introduced by PCR using proofreading Pfu DNA polymerase (Roche Applied Science) and the forward 5′-CATATGGCAGCAGGCGAGCTTGAGGGTGGGCAAA and reverse 5′-GTCGACCTATCAGTTAGGCTGGCTGATCAGTGT primers, respectively. The N-terminal domain of COMMD1 (residues Met-1 through Gly-121, VARD) was cloned into pET28b utilizing pET28b-COMMD1 as a template, the same forward primer as for cloning of the full-length COMMD1, and the reverse primer 5′-GCCATCAAGTCTCGCGCTCAGGCCCCG to generate an SalI restriction site. The C-terminal domain, CTD (COMMD) of COMMD1 was cloned into pET28b from pET28b-COMMD1 template. The forward primer 5′-CATATGAGAGTTGATGGCAAG containing the NdeI restriction site was used along with the reverse primer described for cloning the full-length COMMD1.
Expression and Purification—Initially, recombinant COMMD1 was expressed as a fusion with an intein and a chitin-binding domain in Escherichia coli ER2566 cells and purified in a nontagged form following dithiothreitol-induced excision from the fusion protein. In this purification system COMMD1 showed unusual properties. Specifically, despite good expression and solubility of the fusion protein, the yield of purified untagged COMMD1 was low because COMMD1 remained tightly bound to chitin (a polymer of N-acetylglucosamine) even after cleavage from the fusion portion. The protein could only be eluted from the resin using 4-6 m guanidine chloride. The small amount of non-tagged COMMD1 that was eluted from chitin beads showed rapid proteolysis to a stable ∼13-kDa fragment. Consequently, the non-tagged COMMD1 was only used to produce a polyclonal antibody (see above) and to confirm the identity of the 13-kDa product. For all functional studies, the His6-tagged variant was utilized.
To produce recombinant His-tagged proteins (COMMD1 or individual domains), overnight cultures of E. coli BL21(DE3) cells transformed with the appropriate plasmid were diluted 1:200 to into 1-liter volumes of LB in baffled Erlenmeyer flasks and grown at 37 °C to an A600 of 0.8-1.0. Cultures were cooled to 16 °C before induction with 1 mm isopropyl 1-thio-β-d-galactopyranoside. Cells were harvested after 4 h of growth at 16 °C. Cell pellets were resuspended in Buffer A (50 mm sodium phosphate, pH 7.8, 300 mm NaCl, 10 mm imidazole, 1 mm 4-(2-aminoethyl)benzenesulfonyl fluoride hydrochloride, and a mixture of protease inhibitors (Complete Protease Inhibitors, Roche Applied Science). Cells were lysed in a French press, centrifuged at 20,000 × g for 30 min, and the soluble fraction was passed through a 27-guage needle to reduce viscosity. The soluble lysate was applied to a column of either nickel-nitrilotriacetic acid (Qiagen) or Talon Cobalt resin (Clontech, Mountain View, CA) equilibrated with buffer A. The column was washed by gravity flow with 20 column bed volumes of buffer A, then 20 volumes buffer A containing 0.1% Triton X-100, and another 20 volumes of buffer A. COMMD1 was eluted with steps of buffer B (50 mm sodium phosphate, pH 7.8, 300 mm NaCl) containing 50 mm, 100 mm, 150 mm, and 500 mm imidazole. Most of the full-length COMMD1 elutes with 150 mm imidazole, whereas degradation products elute at 50-100 mm imidazole (supplemental Fig. S1A). The VARD and CTD were purified using the same methods and eluted with 150 mm imidazole. For the liposome floatation assay, purified proteins were dialyzed to liposome buffer (20 mm HEPES, 50 mm KAc, 1 mm EDTA, pH 7.4).
Mass spectrometry of purified COMMD1 and of the stable degradation fragment was performed at the Oregon Health & Science University Metal Ion Core by Dr. M. Ralle. N-terminal sequencing was performed at the Microchemical Facility at Emory University by Dr. J. Pohl.
PIPstrip Assays—PIPstrips (Echelon, Salt Lake City, UT) were blocked in SuperBlock T-20 PBS (Pierce) for 2 h at room temperature and incubated with 1.0 μg/ml COMMD1, CTD, VARD, or the His6-tagged ATP-binding domain of ATP7B ATP-BD (16) (used as a negative control) for 2 h in SuperBlock (Pierce). After washing with PBS-T, the strips were incubated with anti-6His monoclonal antibody (Abcam, Cambridge, UK) or chicken anti-COMMD1 Ab and visualized with appropriate secondary IRDye 800 Ab (Rockland) using an Odyssey scanner. Both primary Abs gave similar results.
Liposome Binding/Enzyme-linked Immunosorbent Assay Plate Assay—COMMD1, ATP-BD, alcohol dehydrogenase (Sigma-Aldrich), and PH domain of phospholipase Cδ (PIP2grip, Echelon Inc., Salt Lake City, UT) were bound to a Corning Costar 3591 polystyrene (medium binding) plate by incubating 1.0, 0.5, and 0.1 μg of each protein in 100 μl of PBS overnight at 4 °C. Unbound protein was removed, and wells were then blocked for 2 h at room temperature with 200 μl of blocking buffer (3% bovine serum albumin in PBS). Liposomes (PIPosome, Echelon), containing 1% of phosphorylated or non-phosphorylated phosphatidylinositol, were added at a total lipid concentration of 1 mm in 50 μl of blocking buffer and incubated for 1 h at room temperature with gentle rocking. The liposome solution was removed, and the wells were washed twice with 200 μl of PBS, then 4 times 10 min with blocking buffer. Bound liposomes were detected with Alexa-fluor 680-streptavidin (Molecular Probes, Eugene, OR) using the Odyssey scanner. Fluorescence was quantified with Odyssey 2.0 software. Data are shown for 0.5 μg of protein bound to the plate.
Testing Specificity of Lipid Binding in Solution Using a Liposome Floatation Assay—Lipids were obtained as chloroform solutions from Avanti Polar Lipids (Alabaster, AL). Phosphatidylcholine was mixed with target lipids in glass tubes at indicated percentages (w/w) for 2 mg of total lipid. Chloroform was removed under a stream of nitrogen gas. The dried lipids were dissolved in 200 μl of hexane, dried under nitrogen, and then dried for 45 min under vacuum. The dried lipid film was resuspended in liposome buffer (20 mm HEPES, 50 mm KAc, 1 mm EDTA, pH 7.4) as described by Knödler and Mayinger (20) and sonicated in a cup-horn sonicator for 15 min at 130-140 watts.
For liposome binding, 100 μl of 2.0 mg/ml liposomes was mixed with purified COMMD1, VARD, or CTD at a final volume of 150 μl and allowed to bind at room temperature for 15 min. 450 μl of 40% sucrose in liposome buffer was added to a final sucrose concentration of 30%, and the mixture was layered onto a 700-μl 45% sucrose cushion. The liposome-protein-30% sucrose step was overlaid with 600 μl of 15% sucrose in liposome buffer, then with 150 μl of liposome buffer. The resultant sucrose step gradients were centrifuged at 40,000 rpm in a Beckman TLS-55 rotor for 20 min. Liposomes and bound protein were removed from the buffer phase (100 μl), and the protein was precipitated by addition of 400 μl of methanol. 100 μl of chloroform was added, then 300 μl of water. The mixture was centrifuged at 13,000 rpm for 3 min in a tabletop microcentrifuge, and the aqueous upper phase was removed, leaving the precipitated protein at the interphase. 400 μl of methanol was added to reduce density, and proteins were pelleted by centrifugation at 13,000 rpm for 5 min. Protein pellets were air-dried and dissolved in SDS-PAGE sample buffer. Proteins were separated by reducing SDS-PAGE and visualized by Coomassie Brilliant Blue staining or by Western blot as described above.
BN-PAGE—HepG2 cells were cultured in 10-cm plates as described above and harvested by scraping into BN buffer (50 mm Bis-Tris, 500 mm ε-amino caproic acid, 2.5% glycerol, Roche Complete protease inhibitors, pH 7.0). Cells were lysed by 25 passages through a 27-gauge needle. Nuclei and cell debris were pelleted by centrifugation at 5,000 × g. The postnuclear supernatant was centrifuged at 100,000 × g for 30 min to separate soluble and membrane fractions. Membrane pellets were dissolved in BN buffer with 2% dodecyl maltoside DDM and incubated on ice for 1 h. Solubilized membrane fractions were centrifuged at 20,000 × g for 20 min, and the supernatants were used for BN-PAGE. Soluble and membrane fractions were loaded onto a 4-20% NativePage gel (Invitrogen) in BN buffer containing 2% dodecyl maltoside and 0.5% Coomassie G-250. For Western blot analysis, the gel was equilibrated in 10 mm CAPS, pH 11.0, and transferred to a polyvinylidene difluoride membrane in the same buffer for 3 h at 180 mA. Immunodetection was performed as described above utilizing polyclonal chicken anti-COMMD1 and secondary detection with horse-radish peroxidase-conjugated donkey anti-Chicken (Aves Labs, Tigard, OR).
Molecular Modeling of COMMD1 Structure—Ab initio structure prediction was carried out on a locally installed Rosetta ab initio software version 2.0 licensed through the University of Washington (the web-based version of this program is known as “Robetta”); the fragment libraries were generated using the web version of the Rosetta fragment server. In the fragment selection, the “homs” option was included while all remaining parameters were set as defaults. Using “ab initio” mode after decoy population filtering 1,000 structures had been obtained. In the case of the “constrained” mode, the distances between the Cα atoms for the N-terminal part of COMMD1 (2H2M) were calculated using a Shell script. Cα distances >8 but <12 Å were used to create a constraint file, which was used for decoy population filtering. The 1000 structures thus obtained were clustered using the Rosetta clustering program. The center of the most populated cluster was selected and minimized using CharmM and then validated as previously described (21).
RESULTS
COMMD1 Is Localized to Endosomal Vesicles in Hepatic Cells—The original genetics experiments revealed the important physiological role of COMMD1 in copper export from the liver, however the intracellular localization of COMMD1 in polarized hepatocytes has not been characterized. Consequently, we first examined the distribution of endogenous COMMD1 in cell compartments using subcellular fractionation. HepG2 cells were allowed to polarize (22), and then cell homogenates were fractionated by differential centrifugation into nuclear, cytosolic, and microsomal membrane fractions. COMMD1 was detected by Western blot analysis in the soluble fraction and in microsomal membranes (Fig. 1A). Because COMMD1 was previously implicated in copper metabolism, we examined whether the amount of COMMD1 in cells or the relative abundance in these sub-cellular fractions can be altered by addition of excess copper to growth media. The distribution of COMMD1 between fractions did not change appreciably in response to copper treatment, suggesting that the proportion of COMMD1 associated with the intracellular membrane in hepatic cells is not dependent on cellular copper concentration.
FIGURE 1.
Subcellular fractionation of COMMD1 in hepatic cells. A, immunodetection of COMMD1 in sub-cellular fractions of HepG2 grown under copper-limiting (BCS) and 50 μm copper (Cu) conditions. Fractions are: N, nuclei; S, soluble cytosol; M, pelleted membranes. B, immunodetection of Golgin-97, CHMP2B, and COMMD1 in fractions from Percoll density gradient.
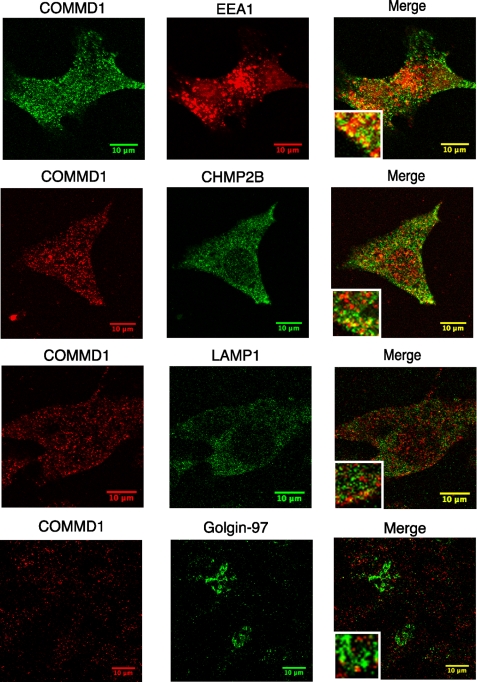
A significant portion of COMMD1 was associated with the microsomal membranes; therefore, we used indirect immunofluorescence with an anti-COMMD1 monoclonal antibody to visualize cell compartments that contain COMMD1. These studies revealed the presence of COMMD1 in punctate vesicles throughout the cytosol (Fig. 2). A similar pattern was obtained using chicken polyclonal antibodies; either antibody was used for subsequent experiments. To characterize the identity of COMMD1-containing vesicles, we carried out co-localization experiments using antibodies against resident proteins of Golgi (Golgin-97), early endosomes (EEA1), late endosomes/multive-sicular bodies (ESCRT-III subunit CHMP2B), and lysosomes (LAMP1). Little co-localization was observed with the Golgi marker Golgin-97 (Fig. 2), suggesting that the primary location of COMMD1 is not at the Golgi network. Some co-staining was detected with the lysosomal marker (LAMP1), however the percent of co-localization was low (Fig. 2 and supplemental Fig. S2). The highest co-localization was observed with the markers for early endosomes (EEA1) and late endosomes/multivesicular bodies, MVB (ESCRT-III subunit CHMP2B (23) (Fig. 2 and supplemental Fig. S2); this co-localization was also partial.
FIGURE 2.
Endogenous COMMD1 is present in endocytic/MVB pathway. Confocal immunofluorescence microscopy of COMMD1 with markers for EEA1 (early endosomes), RAB9 (late endosomes), CHMP2B (late endosome/multivesicular bodies), LAMP1 (lysosomes), Golgin-97 (Golgi), and ATP7B (trans-Golgi network).
The enrichment of COMMD1 in the endosomal/MVB membrane was confirmed by separating the HepG2 membranes by centrifugation on an in situ generated Percoll gradient (19). Starting with the heaviest (high density) fraction and finishing with the lightest fraction, the presence of COMMD1 and the TGN and endosomal/MVB markers in fractions was detected on a Western blot using appropriate antibodies. Consistent with the result of immunofluorescence, we find both COMMD1 and CHMP2B peak in the same fraction in the upper portion of the gradient (Fig. 1B). The Golgi-associated protein Golgin-97 showed wider distribution with its peak intensity in a fraction distinctly separated from COMMD1. Altogether, the results of immunofluorescence and subcellular fractionation place COMMD1 in a post-Golgi vesicular compartment that overlaps with the endosomal trafficking/recycling/degradation pathway.
Previously, copper-induced trafficking from the Golgi to the vesicular compartment (in the vicinity of the apical membrane) was shown for the copper transporter ATP7B (24). ATP7B and COMMD1 can be co-immunoprecipitated (8), however in which cellular compartment these proteins interact remains unclear. COMMD1 localization in the endocytic pathway excludes interactions with ATP7B at the TGN (and explains why COMMD1 down-regulation does not affect the ATP7B exit from the TGN). However, it is possible that COMMD1 binds to ATP7B when copper levels increase and ATP7B passes through the endocytic pathway toward the plasma membrane. Alternatively, COMMD1 targeting may change in response to copper elevation and interaction with ATP7B may take place at the apical membrane.
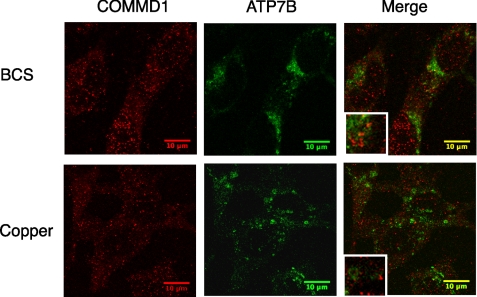
We tested these possibilities by co-immunostaining COMMD1 and ATP7B in non-polarized as well as polarized HepG2 in low copper (BCS-treated cells) and elevated copper conditions (Fig. 3). In response to copper, the trafficking of ATP7B was apparent, whereas the COMMD pattern remained largely unchanged in either polarized or non-polarized cells. The co-localization for COMMD1 and ATP7B was low under both high and low copper (Fig. 3). Thus, the intracellular localization of COMMD1 is not altered by copper in a noticeable way, and only a small percentage of total COMMD1 is involved in interactions with ATP7B. In other words, the function of COMMD1 at the membrane is unlikely to be restricted to regulation of copper transport.
FIGURE 3.
Intracellular localization of COMMD1 and ATP7B in HepG2 cells following BCS or CuCl2 treatment. Confocal immunofluorescence microscopy of COMMD1 and ATP7B was performed after HepG2 cells are treated for 8 h with either 200 μm BCS or 50 μm CuCl2. Non-polarized HepG2 cells are shown; the pattern of COMMD1 in polarized cells was very similar.
Recombinant COMMD1 Interacts Directly and Specifically with Lipids—Although membrane association of COMMD1 is clear and published phenotypic data indicate function in membrane-localized processes, it is not known how COMMD1 is recruited to membranes. Recombinant COMMD1 is a soluble protein; therefore, in cells the membrane association of COMMD1 could be either due to recruitment by membrane proteins and/or due to specific interactions with membrane lipids. Binding of COMMD1 to two membrane proteins, ATP7B and δENaC, has been previously reported (8, 13). However, at steady state neither ATP7B nor δENaC are endosomal proteins and they only traverse the endocytic pathway on their way toward the apical plasma membrane. Consequently, we hypothesized that the observed steady-state localization of COMMD1 in the endocytic vesicles is due to direct interaction of COMMD1 with lipids. To test this hypothesis we first employed a biochemical approach, which allows analysis of such interactions under well defined conditions. Recombinant COMMD1 was produced in a soluble form in Escherichia coli, and its biochemical properties were investigated. The mass of purified COMMD1 (23211.7 Da, obtained by mass spectroscopy) corresponded closely to the theoretical mass of 23211.56 (supplemental Fig. S1B), indicating a lack of modifications.
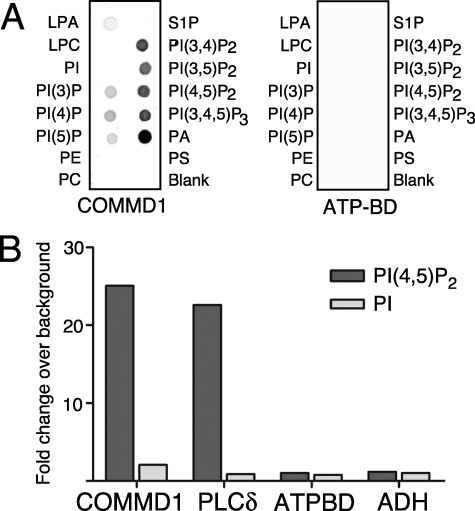
To determine whether COMMD1 can interact with membrane lipids in the absence of other proteins, we first utilized lipid spotted arrays (PIPstrips), whereby COMMD1 reproducibly showed binding to several lipids (Fig. 4A, left). The binding was strongest for phosphatidylinositols phosphorylated at two (phosphatidylinositol 1,4,5-bisphosphate) or three (phosphatidylinositol 1,4,5-trisphosphate) positions, as well as phosphatidic acid (PA), whereas monophosphorylated phosphatidylinositols showed much weaker interaction. COMMD1 did not bind to non-phosphorylated PtdIns, phosphatidylserine, phosphatidylcholine, and other lipids, indicating strong preference for phosphoinositol moiety. The His-tagged ATP-binding domain of ATP7B (ATP-BD), a protein of similar size, was used as a negative control and showed no binding to lipids (Fig. 4A, right).
FIGURE 4.
COMMD1 binds phospholipids. A, PIP strip assay for the full-length HisTag-COMMD1 and a negative control HisTag-ATP-BD. Immunodetection with anti-His6 antibody is shown. B, liposome binding to 0.5 μg of each immobilized protein: COMMD1; PH-phospholipase Cδ is positive control, ATP-BD and ADH are negative controls. COMMD1 or control proteins were bound to a 96-well plate and incubated with biotinylated liposomes containing PtdIns(4,5)P2 or non-phosphorylated PtdIns. Liposome retention is determined by fluorescence of IRDye-conjugated streptavidin.
High concentrations of lipids on PIPstrips may allow for promiscuous protein binding, while performing measurements in a membrane-like environment is expected to better reveal specific protein-lipid interactions, particularly at lower concentrations of lipid. Consequently, the ability of COMMD1 to bind phosphorylated phosphatidylinositols was verified using PtdIns(4,5)P2-containing liposomes (Fig. 4B). COMMD1 strongly retained liposomes containing PtdIns(4,5)P2, showing binding that was comparable to the binding of the well characterized PtdIns(4,5)P2-binding protein, PH-phospholipase Cδ, used as a positive control. No interaction with PtdIns(4,5)P2-containing liposomes was observed for His-tagged ATP-BD or alcohol dehydrogenase (negative controls). Similarly, no interaction was detected between COMMD1 and liposomes containing non-phosphorylated phosphatidylinositols, further confirming the specificity of COMMD1-lipid interactions.
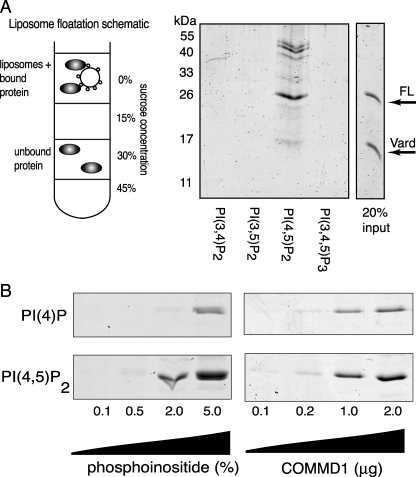
PtdIns(4,5)P2-containing Liposomes Recruit COMMD1 and Stabilize COMMD1 Dimers—The above data suggested that the presence of COMMD1 in cellular membranes was a result of recruitment by specific signaling lipids, such as PtdIns(4,5)P2 or PtdIns(3,4,5)P3, rather than simple charge-complementation interaction with the membrane surface. To test this hypothesis, the binding of COMMD1 to liposomes containing different bis- and tris-phosphatidylinositols was evaluated under equilibrium conditions. COMMD1 was incubated with phosphatidylcholine liposomes containing 2% of one of the following phosphatidylinositols: PtdIns(3,4)P2, PtdIns(3,5)P2, PtdIns(4,5)P2, and PtdIns(3,4,5)P3. Protein recruited to liposomes was recovered following liposome floatation on a sucrose step gradient (Fig. 5A, schematic). COMMD1 was found associated only with PtdIns(4,5)P2-containing liposomes (Fig. 5A).
FIGURE 5.
Equilibrium binding of COMMD1 to liposomes demonstrates preference to the bis-phosphorylated phosphatidylinositol. In A: Left, schematic of liposome floatation assay, liposomes and proteins are loaded in the 30% sucrose layer and liposomes with bound protein are recovered from the 15% sucrose/buffer interface. Unbound protein remains in the 30% sucrose layer. Right, SDS-PAGE of COMMD1 recovered with liposomes containing 2% of indicated bis- and tri-phosphorylated liposomes. Vard is the stable degradation product of COMMD1 (residues 1-121). In B: Left, COMMD1 recovered with liposomes containing increasing concentrations of PtdIns(4)P and PtdIns(4,5)P2. Right, increasing amounts of COMMD1 recovered with 2% PtdIns(4)P or PtdIns(4,5)P2.
It was also apparent that the binding to these liposomes was accompanied by a significant stabilization of the COMMD1 dimer. Prior to incubation with PtdIns(4,5)P2-containing liposomes, the protein ran on reducing SDS-PAGE as a monomer (FL in Fig. 5A); after binding to liposomes, a significant proportion of COMMD1 did not fully dissociate and migrated in bands corresponding to the size of the dimer. It should be noted that following interaction with liposomes more than one form of a dimer appeared (see multiple bands). These high molecular weight bands were not impurities, because they were absent in the sample prior to incubation with liposomes (see input lane).
COMMD1-PtdIns(4,5)P2 Interactions—The observed marked difference in the binding of COMMD1 to PtdIns(4,5)P2 and PtdIns(3,5)P2 liposomes (Fig. 5A, above) suggested that the phosphate at the position 4 of PtdIns(4,5)P2 plays an important role in the COMMD1-lipid interaction. To test this prediction, we measured binding of COMMD1 to PtdIns(4)P-containing liposomes and compared the relative affinity of COMMD1 for PtdIns(4)P and PtdIns(4,5)P2 (Fig. 5B). In liposomes containing 2% PtdIns(4)P or PtdIns(4,5)P2, COMMD1 was bound to either lipid, indicating that phosphorylation at position 4 is sufficient for COMMD1-lipid binding. At the same time, binding to PtdIns(4,5)P2 was stronger compared with PtdIns(4)P at all tested protein concentrations (Fig. 5B, right panel). Preferential binding of COMMD1 to PtdIns(4,5)P2 was further corroborated in experiments where protein concentration was kept the same, while target lipid content in liposomes was varied (Fig. 6B, left panel). Thus, phosphorylation at position 4 is sufficient for COMMD1 recruitment in these conditions, whereas addition of a phosphate group at position 5 appears to increase affinity of COMMD1 for PtdIns(4,5)P2.
FIGURE 6.
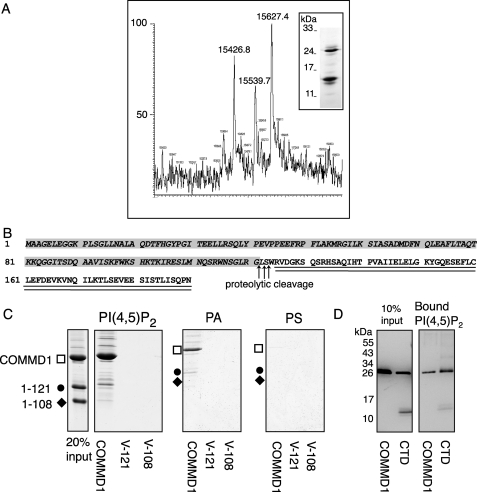
COMMD1 proteolytic sensitivity and domain organization. A, COMMD1 degrades to a 15-kDa cleavage product (VARD, lower band) yielding three possible masses depending on cleavage site. B, amino acid sequence of COMMD1 indicating domain organization; the experimentally determined VARD is italicized. C, full-length COMMD1, but not the N-terminal domain is recruited to liposomes. COMMD1, V-121, and V-108 binding to liposomes containing 10% phosphatidic acid (PA), phosphatidylserine (PS), or PtdIns(4,5)P2. D, comparison of the COMMD1 and CTD binding to liposomes containing 10% PtdIns(4,5)P2 demonstrates that CTD alone is sufficient for interactions with the lipid. Immunodetection with anti-COMMD1 Ab is shown to confirm the identity of oligomeric bands in the CTD sample.
The C-terminal Domain of COMMD1 Is Required for PtdIns(4,5)P2 Binding—Purified soluble COMMD1 showed significant proteolytic sensitivity seen as degradation to a stable 15-kDa fragment when left overnight at 4 °C (Fig. 6A, inset). We carried out the N-terminal sequencing and mass spectrometry analysis of this fragment and determined that the stable 15-kDa fragment includes the entire N terminus of COMMD1 and has Gly-121 or Ser-123 at the C terminus (Fig. 6A). These results suggested a two-domain structure for COMMD1, where the independently folded N- and C-terminal domains were connected by a proteolytically sensitive linker (Fig. 6B). Such architecture is consistent with the previously published results of multiple alignments that revealed a more conserved C-terminal portion for the COMMD proteins and a more variable, subfamilies-specific, N terminus (11).
Recently, the structure of the N-terminal fragment of COMMD1 (residues 1-108) was determined by NMR and revealed a large positive patch on its surface (15). Therefore, it was interesting to examine whether the N-terminal domain can function in lipid binding and alone be recruited to PtdIns(4,5)P2 liposomes. To address this question we generated the recombinant N-terminal domain (VARiable Domain, VARD, residues 1-121) and compared the binding of VARD and the full-size COMMD1 to liposomes. We also used the shorter N-terminal fragment (residues 1-108), which was previously utilized for the structure determination. COMMD1, VARD (residues 1-121), or V1-108 were incubated with liposomes containing 90% phosphatidylcholine and 10% of PA, phosphatidylserine, or PtdIns (4,5)P2 to maximize possibility of binding (Fig. 6C). Consistent with our other results, COMMD1 was bound strongly to liposomes containing PtdIns(4,5)P2, however neither VARD nor V1-108 were bound to the liposomes. Thus, the presence the N-terminal (VARD) domain alone is not sufficient for stable interactions of COMMD1 with PtdIns(4,5)P2. A significant amount of the full-length COMMD1 was recovered with PA-containing liposomes, but not with phosphatidylserine-containing liposomes (Fig. 6C), suggesting that PA may also be a relevant interacting partner of COMMD1.
To test the contribution of CTD (residues 125-190, Fig. 6B) to lipid binding, we generated a recombinant CTD and tested its binding to liposomes containing 10% PtdIns(4,5)P2. CTD was bound to liposomes; furthermore, it was recovered predominantly in an oligomeric form that was stable in reducing SDS-PAGE (Fig. 6D). Higher molecular weight forms were also apparent; the identity of all bands as CTD was confirmed by immunostaining.
COMMD1 Forms Stable, Multimeric Complexes in Solution and at the Membrane—Altogether, the cellular and biochemical characterization revealed specific lipid-mediated anchoring of COMMD1 to the endocytic compartment (a compartment involved in the delivery of membrane proteins to the plasma membrane and their sorting between the recycling and degradation pathways). It was also apparent that interaction with PtdIns(4,5)P2 led to the formation of stable oligomeric complexes of COMMD1 at the membrane surface. We also noticed the propensity of COMMD1 to aggregate upon storage and bind tightly to many surfaces (even to glass), in agreement with the previously reported difficulties with obtaining uniform COMMD1 for structural studies (15). Although experimentally very inconvenient, these properties were nevertheless interesting because our BLAST searches revealed a homology of COMMD1 to the segment of the endoplasmic reticulum-microtubule targeting protein CLIMP-63 (CKAP-4) (supplemental Fig. S3). This region of CKAP-4 is known to oligomerize and form elongated rods in solution; in cells, it determines the immobility of CKAP-4 within the membrane (25). Consequently, we hypothesized that COMMD1 at the membrane may form oligomeric structures, i.e. act as a scaffold protein.
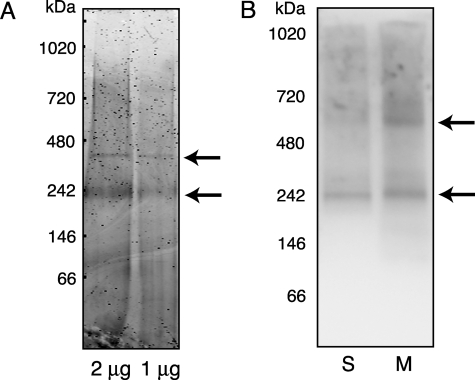
To test this hypothesis, we utilized BN-PAGE and investigated the oligomerization state of both the recombinant and endogenous COMMD1 under non-denaturing conditions. Purified soluble COMMD1 runs as a complex of ∼240 kDa and to a lesser extent as a 400-kDa complex (Fig. 7A), demonstrating that COMMD1 can form stable higher order structures in the absence of other proteins or lipids. Immunodetection of endogenous COMMD1 in HepG2 fractions separated by BN-PAGE revealed a band at ∼240 kDa in both soluble and microsomal fractions, which was similar to the recombinant COMMD1 oligomer (Fig. 7B). The membrane fraction contained an additional COMMD1 band at ∼600 kDa, which possibly reflects the recruitment of other proteins to the COMMD1 oligomer at the membrane.
FIGURE 7.
Endogenous and recombinant COMMD1 form large oligomeric complexes. A, BN-PAGE of 2.0 μg and 1.0 μg of purified recombinant COMMD1. B, BN-PAGE of soluble (S) and membrane (M) fractions from HepG2 cells. Immunodetection with the anti-COMMD1 Ab is shown. The COMMD1-containing complexes are indicated by the arrows.
We then studied the mechanism of COMMD1 oligomerization in more detail. Previously, the fragment 60-190 of COMMD1 was shown to be sufficient for dimerization (5, 12). Our data on COMMD1 proteolysis indicate that the N-terminal VARD encompasses residues 1-121, whereas the CTD includes residues 125-190 (Fig. 6A), i.e. the previously identified dimerization region includes a half of the N-terminal domain and the entire C-terminal domain. Our studies on interactions with PtdIns(4,5)P2 revealed the ability of CTD alone to form oligomers particularly in the presence of liposomes. To better define the role of the N-terminal domain, we expressed VARD and CTD individually and investigated their ability to form oligomers as compared with COMMD1.
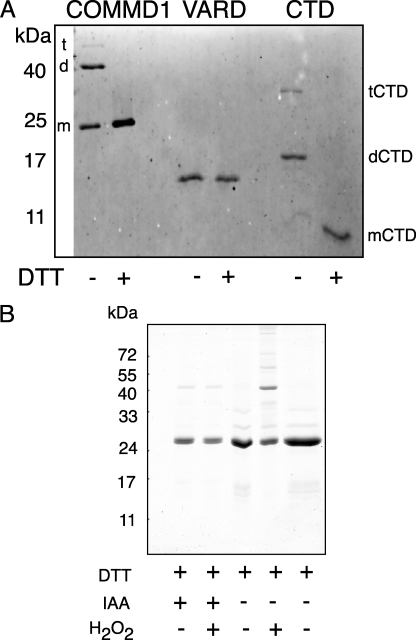
In non-reducing SDS-PAGE, the full-length COMMD1 behaved as a mixture of monomers and dimers with a small portion running as a tetramer (Fig. 8A). Addition of dithiothreitol produced mostly monomeric COMMD1, in agreement with earlier results (5). VARD is a monomer under either nonreducing or reducing conditions, indicating that this domain alone is not sufficient for SDS-PAGE-resistant dimerization. The CTD runs mostly as a homodimer with smaller fraction migrating as a tetramer. CTD contains a single Cys residue and is converted to a monomer by treatment with dithiothreitol (Fig. 8A). Formation of higher order oligomers in COMMD1 can be greatly facilitated by oxidation using brief treatment with 10 mm H2O2, but this can be significantly reduced by modifying the cysteine residue with iodoacetamide (Fig. 8B).
FIGURE 8.
CTD is sufficient for oligomerization. VARD and CTD were separated on a 15% acrylamide Laemmli gel and transferred to nitrocellulose for immunodetection. The samples were loaded with or without 20 mm dithiothreitol, as indicated. COMMD1 monomer, dimer, and tetramer sizes are labeled with the letters “m,” “d,” and “t”(left), respectively. CTD species are indicated with as “mCTD,” “dCTD,” and “tCTD”(right). B, redox regulation of COMMD1 oligomerization with indicated treatments.
Ab Initio Modeling of COMMD1 Predicts the Two-domain Structure with Homology to Myosin and Sec7 Domains Containing Proteins—To better understand how COMMD1 can form an oligomeric scaffold, structural information is necessary. Current structural data on COMMD1 are limited to the NMR structure of the N-terminal 1-108 fragment (15). Consequently, we utilized Rosetta ab initio modeling (26) to gain information about possible structural organization of the full-length COMMD1. We initially generated a completely ab initio model with no constraints. The highest scoring model of the full-length COMMD1 showed two distinct domains connected by a flexible region (Fig. 9A). The identified proteolytic cleavage site (indicated by the arrow in Fig. 9D) was located within the predicted flexible linker and provided the first experimental evidence for the COMMD1 model. The relative accuracy of this initial modeling was further illustrated by the comparison of the Rosetta-generated model (Fig. 9A) and the NMR structure for the 1-108 fragment of COMMD1 (Fig. 9B) (15), which appeared after the model was generated. The NMR structure and the corresponding region of the model have the same secondary structure, the same number of α-helices, and similar overall fold. (An overlay of the solution structure and residues 1-108 of the initial Rosetta model is shown in Fig. 9C).
FIGURE 9.
Structural model of COMMD1. A, ab initio model with experimental VARD in green and the empirically determined CTD in orange. B, NMR solution structure of residues 1-108. C, overlay of Ab initio modeled COMMD1 residues 1-108 (green) and the NMR solution structure (cyan). D, COMMD1 model with solution structure of residues 1-108 as initial constraint. Residues 1-108 are shown in cyan, 109-129 in green, and the remainder of the empirically determined CTD is orange. The proteolytic site is indicated by the arrow.
To further refine the model, we used the NMR-derived coordinates for the 1-108 region as experimental constraints and carried out another round of modeling. The refined model for the full-length COMMD1 retains a two-domain organization and a flexible linker, residues 119-128 (Fig. 9D; the PDB coordinates for the full-length COMMD1 model are available to interested investigators upon request).
The N-terminal domain in the full-length COMMD1 remains entirely α-helical. The C-terminal domain contains three β-strands and has two short α-helices. (The helical nature of VARD and the presence of β-strands in the full-length COMMD1 were confirmed experimentally by CD spectroscopy; our data, not shown). Interestingly, the only Cys in CTD is located in one of the β-strands facing outwards and thus can easily participate in the disulfide-bond formation/dimerization that we detect under oxidizing conditions. The most striking structural feature of the COMMD1 model is helix 5 (the last helix of the N-terminal domain), which is very long and highly amphipathic, having one positively charged surface.
The generated model was then utilized for structural homology searches using the Dali server. Strikingly, the top scoring structures involve various myosins (z-score of 3.1) and Arno (z-score of 2.6): the proteins known to form oligomeric complexes and participate in protein and vesicle trafficking. We conclude that COMMD1 is a novel scaffold protein that is targeted to a specific subset of PtdIns(4)P- or PtdIns(4,5)P2-containing endosomes, where it forms large oligomeric complexes and may regulate the intracellular fate of membrane proteins.
DISCUSSION
COMMD1 is a small soluble protein that participates in a number of important physiological processes. Interacting partners for COMMD1 are both cytosolic and intrinsic membrane proteins. The involvement of COMMD1 in diverse protein-protein interactions has received significant experimental support (7, 8, 12, 13), and downstream consequences of some of these interactions are becoming more apparent (9, 14). However, so far there has been no information on the mechanism through which COMMD1 acts at the membrane and interacts (often simultaneously) with various proteins. Our studies uncovered new structural and functional properties of COMMD1, which explain the presence of COMMD1 in cell membranes and suggest the mechanism of COMMD1 action.
Specific interaction of COMMD1 with phosphatidylinositols is a very exciting new finding. COMMD1 shows strong preference for PtdIns(4,5)P2, and lower, yet significant interaction with PtdIns(4)P and PA. PtdIns(4,5)P2 is an important regulatory and membrane-anchoring molecule with a well described role in vesicular trafficking (27), modulation of transporter activity, and establishment and maintenance of apical-basolateral polarity in epithelial cells (28). Identification of specific interactions with this important lipid revealed the mechanism through which COMMD1 binds to specific cellular compartments. Given recently described role of PtdIns(4,5)P2 in generating and maintaining the apical membrane domain in polarized cells (28), it is particularly interesting that both known COMMD1-interacting partners ATP7B and ENaC traffic toward apical membranes in polarized cells. In this study we have not considered soluble targets of COMMD1, but it is conceivable that the interaction of COMMD1 with PtdIns(4,5)P2 may affect COMMD1-dependent signaling events beyond cell membrane.
Another novel and important property of COMMD1 is its ability to form stable high order oligomers. The presence of 240- and 600-kDa bands in BN-PAGE suggests that the oligomeric COMMD1 forms a scaffold that recruits other proteins into complexes while present at membrane surface. Functioning as a higher order complex, COMMD1 may resemble the hexameric Hrs protein, another phosphatidylinositol-binding molecule, which orchestrates vesicle sorting by providing multiple sites for diverse protein-protein interactions (29). Oligomerization may also increase binding specificity of COMMD1 to PtdIns(4,5)P2 as was described for the PH domain of dynamin (30) and as suggested by our observation of increased oligomer stability in gels.
Our modeling studies, proteolysis data, and the characterization of recombinant VARD and CTD provide direct experimental evidence for the organization of COMMD1 as a two-domain protein. The two-domain structure of COMMD1 family has been predicted by sequence alignments (11), and it was proposed that most COMMD proteins, with the exception of COMMD6, have such organization. COMMD6 was thought not to have the N-terminal domain. However, more accurate definition of the domain borders obtained in our experiments indicates that COMMD6 is likely to be a typical COMMD protein with a very small N-terminal domain (11).
The C-terminal residues 125-190 of COMMD1 are sufficient for the oligomerization of COMMD1. Because each COMMD1 monomer contains only one Cys residue (Cys-160 in the CTD), the presence of tetramers suggests self-interaction that occurs in two ways: with and without participation of Cys. (It should be noted that, while Cys residue is involved in dimer stabilization, it is not essential and can be mutated to Ala without negative effect on dimer formation (5).) Although the N-terminal domain alone does not form stable oligomers, the banding pattern of our gels suggest that the N-terminal fragment can stably bind to a dimer, i.e. both domains are likely to contribute to the formation of high order oligomers.
The intracellular location of COMMD1 established in our study helps to better understand the phenotypic consequence of COMMD1 inactivation, i.e. defective copper export into the bile (2, 3, 31). It is yet unclear whether copper alters levels of PtdIns. We did not observe the effect of copper on either the intracellular localization of COMMD1 or its recruitment to liposomes. However, the COMMD1 transcript levels are decreased by prolonged copper treatment (32). Further, the copper-binding ubiquitin ligase X-linked apoptosis protein has recently been implicated in modulating the COMMD1 levels in cells (33). Thus, it appears that the presence of COMMD1 in the secretory pathway is controlled by at least two mechanisms. The copper-dependent mechanisms regulate the total abundance of COMMD1, whereas copper-independent interaction with PtdIns ensures the targeting of COMMD1 to the appropriate compartment.
The presence of COMMD1 in compartments enriched in PtdIns(4)P and PtdIns(4,5)P2 may serve as an important signal to direct the routing of copper transporter ATP7B when it passes through the secretory pathway toward sub-apical vesicles of hepatocytes, allowing eventual fusion of copper-containing vesicles with the canalicular membrane. Alternatively, the presence of COMMD1 in endosomes/MVB may influence the fate of ATP7B by shifting equilibrium from recycling to targeting to multivesicular bodies and lysosomal degradation. The potential role of COMMD1 in membrane protein recycling is supported by the recent observation of K63-linked ubiquitination of COMMD1 (34), which is a modification implicated in targeting the apical membrane proteins into the endosomal recycling pathway (35). The observation that immunofluorescence shows little COMMD1/ATP7B co-localization is consistent with other studies (9) and is likely to reflect a more general role of COMMD1 as a scaffold for various proteins (Fig. 10). Weak co-localization may also be due to a transient nature of ATP7B-COMMD1 interaction. As shown by immunoprecipitation, the interaction between COMMD1 and ATP7B is weaker than the interaction between the copper chaperone Atox1 and ATP7B (8). This latter interaction was shown to be transient by NMR, as it serves to transfer ligand from one protein to another.
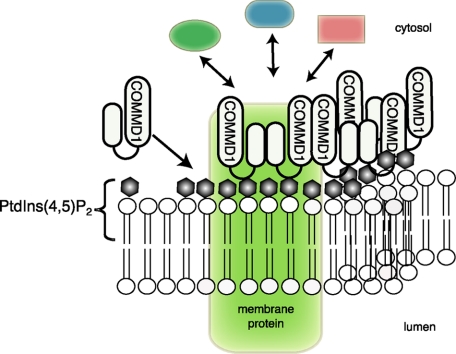
FIGURE 10.
Schematic of COMMD1 complex formation at the membrane. COMMD1 is recruited to the membrane by PtdIns(4,5)P2. Complex formation with the membrane and intrinsic membrane proteins signals recruitment of additional effectors.
A similar regulatory mechanism (Fig. 10) may be responsible for the COMMD1-mediated regulation of sodium channel δENaC. Dual interaction with δENaC (13) and PtdIns(4,5)P2 (this study), a known modulator of ENaC (36), may provide a binary code for COMMD1 assembly and facilitate recruitment of other ENaC regulators. For example, COMMD1 was proposed to interact with ubiquitin ligases (14). The plasma membrane location of ENaC is regulated by the ubiquitin ligase Nedd4-2 (37). Interestingly Nedd4-2 does not interact directly with the C-terminal tail of δENaC, while COMMD1 does, suggesting that COMMD1 may provide a platform for recruitment of δENaC regulators. The experimental protocols and a molecular model of COMMD1 generated in our study offer necessary experimental framework to test these new hypotheses.
Supplementary Material
Acknowledgments
We thank Dr. Amy Rosenzweig for providing structural data and materials ahead of publication, Dr. Ezra Burstein for sharing an antibody used to confirm specificity of ours reagents, Dr. Peter Mayinger and Andreas Knoedler for their assistance with the initial characterization of phosphatidylinositol binding, and Dr. Vinzenz Unger for helpful comments and advice with the liposome binding assay. The Oregon Health & Science University Metal Ion Core under the direction of Dr. Martina Ralle performed mass spectrometry studies.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 DK071865 (to S. L.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1-S3.
Footnotes
The abbreviations used are: COMMD, copper metabolism Murr1 domain; TGN, trans-Golgi network; HIV, human immunodeficiency virus; Ab, antibody; PBS, phosphate-buffered saline; CHAPS, 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid; VARD, residues Met-1 through Gly-121; Bis-Tris, 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol; CAPS, 3-(cyclohexylamino)propanesulfonic acid; BCS, bathocuproine disulfonate; PA, phosphatidic acid; PtdIns(4,5)P2, phosphatidylinositol 4,5-bisphosphate; CTD, C-terminal domain; BN-PAGE, Blue-native PAGE.
References
- 1.Maine, G. N., and Burstein, E. (2007) Cell Mol. Life Sci. 64 1997-2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van De Sluis, B., Rothuizen, J., Pearson, P. L., van Oost, B. A., and Wijmenga, C. (2002) Hum. Mol. Genet. 11 165-173 [DOI] [PubMed] [Google Scholar]
- 3.Klomp, A. E., van de Sluis, B., Klomp, L. W., and Wijmenga, C. (2003) J. Hepatol. 39 703-709 [DOI] [PubMed] [Google Scholar]
- 4.Kawamura, M., Takahashi, I., and Kaneko, J. J. (2002) Vet. Pathol. 39 747-750 [DOI] [PubMed] [Google Scholar]
- 5.Narindrasorasak, S., Kulkarni, P., Deschamps, P., She, Y. M., and Sarkar, B. (2007) Biochemistry 46 3116-3128 [DOI] [PubMed] [Google Scholar]
- 6.Johnson, G. F., Morell, A. G., Stockert, R. J., and Sternlieb, I. (1981) Hepatology 1 243-248 [DOI] [PubMed] [Google Scholar]
- 7.Burstein, E., Ganesh, L., Dick, R. D., van De Sluis, B., Wilkinson, J. C., Klomp, L. W., Wijmenga, C., Brewer, G. J., Nabel, G. J., and Duckett, C. S. (2004) EMBO J. 23 244-254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tao, T. Y., Liu, F., Klomp, L., Wijmenga, C., and Gitlin, J. D. (2003) J. Biol. Chem. 278 41593-41596 [DOI] [PubMed] [Google Scholar]
- 9.de Bie, P., van de Sluis, B., Burstein, E., van de Berghe, P. V., Muller, P., Berger, R., Gitlin, J. D., Wijmenga, C., and Klomp, L. W. (2007) Gastroenterology 133 1316-1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van de Sluis, B., Muller, P., Duran, K., Chen, A., Groot, A. J., Klomp, L. W., Liu, P. P., and Wijmenga, C. (2007) Mol. Cell Biol. 27 4142-4156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burstein, E., Hoberg, J. E., Wilkinson, A. S., Rumble, J. M., Csomos, R. A., Komarck, C. M., Maine, G. N., Wilkinson, J. C., Mayo, M. W., and Duckett, C. S. (2005) J. Biol. Chem. 280 22222-22232 [DOI] [PubMed] [Google Scholar]
- 12.de Bie, P., van de Sluis, B., Burstein, E., Duran, K. J., Berger, R., Duckett, C. S., Wijmenga, C., and Klomp, L. W. (2006) Biochem. J. 398 63-71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Biasio, W., Chang, T., McIntosh, C. J., and McDonald, F. J. (2004) J. Biol. Chem. 279 5429-5434 [DOI] [PubMed] [Google Scholar]
- 14.Maine, G. N., Mao, X., Komarck, C. M., and Burstein, E. (2007) EMBO J. 26 436-447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sommerhalter, M., Zhang, Y., and Rosenzweig, A. C. (2007) J. Mol. Biol. 365 715-721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tsivkovskii, R., MacArthur, B. C., and Lutsenko, S. (2001) J. Biol. Chem. 276 2234-2242 [DOI] [PubMed] [Google Scholar]
- 17.Costes, S. V., Daelemans, D., Cho, E. H., Dobbin, Z., Pavlakis, G., and Lockett, S. (2004) Biophys. J. 86 3993-4003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bradford, M. M. (1976) Anal. Biochem. 72 248-254 [DOI] [PubMed] [Google Scholar]
- 19.Press, B., Feng, Y., Hoflack, B., and Wandinger-Ness, A. (1998) J. Cell Biol. 140 1075-1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Knodler, A., and Mayinger, P. (2005) BioTechniques 38 858, 860, 862 [DOI] [PubMed] [Google Scholar]
- 21.Subbian, E., Yabuta, Y., and Shinde, U. (2004) Biochemistry 43 14348-14360 [DOI] [PubMed] [Google Scholar]
- 22.Sormunen, R., Eskelinen, S., and Lehto, V. P. (1993) Lab. Invest. 68 652-662 [PubMed] [Google Scholar]
- 23.Williams, R. L., and Urbe, S. (2007) Nat. Rev. Mol. Cell Biol. 8 355-368 [DOI] [PubMed] [Google Scholar]
- 24.Schaefer, M., Hopkins, R. G., Failla, M. L., and Gitlin, J. D. (1999) Am. J. Physiol. 276 G639-G646 [DOI] [PubMed] [Google Scholar]
- 25.Klopfenstein, D. R., Klumperman, J., Lustig, A., Kammerer, R. A., Oorschot, V., and Hauri, H. P. (2001) J. Cell Biol. 153 1287-1300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim, D. E., Chivian, D., and Baker, D. (2004) Nucleic Acids Res. 32 W526-W231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yin, H. L., and Janmey, P. A. (2003) Annu. Rev. Physiol. 65 761-789 [DOI] [PubMed] [Google Scholar]
- 28.Martin-Belmonte, F., Gassama, A., Datta, A., Yu, W., Rescher, U., Gerke, V., and Mostov, K. (2007) Cell 128 383-397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pullan, L., Mullapudi, S., Huang, Z., Baldwin, P. R., Chin, C., Sun, W., Tsujimoto, S., Kolodziej, S. J., Stoops, J. K., Lee, J. C., Waxham, M. N., Bean, A. J., and Penczek, P. A. (2006) Structure 14 661-671 [DOI] [PubMed] [Google Scholar]
- 30.Klein, D. E., Lee, A., Frank, D. W., Marks, M. S., and Lemmon, M. A. (1998) J. Biol. Chem. 273 27725-27733 [DOI] [PubMed] [Google Scholar]
- 31.Twedt, D. C., Sternlieb, I., and Gilbertson, S. R. (1979) J. Am. Vet. Med. Assoc. 175 269-275 [PubMed] [Google Scholar]
- 32.Muller, P., van Bakel, H., van de Sluis, B., Holstege, F., Wijmenga, C., and Klomp, L. W. (2007) J. Biol. Inorg. Chem. 12 495-507 [DOI] [PubMed] [Google Scholar]
- 33.Maine, G. N., Mao, X., Muller, P. A., Komarck, C. M., Klomp, L. W., and Burstein, E. (2008) Biochem. J., in press [DOI] [PMC free article] [PubMed]
- 34.Huang, Y., Wu, M., and Li, H. Y. (2008) J. Biol. Chem. 283 11453-11460 [DOI] [PubMed] [Google Scholar]
- 35.Kamsteeg, E. J., Hendriks, G., Boone, M., Konings, I. B., Oorschot, V., van der Sluijs, P., Klumperman, J., and Deen, P. M. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 18344-18349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pochynyuk, O., Tong, Q., Staruschenko, A., Ma, H. P., and Stockand, J. D. (2006) Am. J. Physiol. 290 F949-F957 [DOI] [PubMed] [Google Scholar]
- 37.Lu, C., Pribanic, S., Debonneville, A., Jiang, C., and Rotin, D. (2007) Traffic 8 1246-1264 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.