Abstract
The tumor suppressor protein kinase LKB1 exerts its effects by phosphorylating and activating AMP-activated protein kinase (AMPK) and members of the AMPK-related kinase family, such as the brain-specific kinases BRSK1/BRSK2 (SAD-B/SAD-A). LKB1 contains a conserved serine residue near the C terminus (Ser-431 in mouse LKB1) that is phosphorylated by cyclic AMP-dependent protein kinase and p90-RSK. Although some studies suggest that LKB1 is constitutively active and is not rate-limiting for activation of AMPK, others have suggested that phosphorylation of Ser-431 is necessary to allow LKB1 to phosphorylate and activate AMPK and other downstream kinases. Prompted by our discovery of an LKB1 splice variant (LKB1S) that lacks Ser-431, we have reinvestigated this question. In HeLa cells (which lack endogenous LKB1), co-expression with STRADα and MO25α of wild type LKB1, the S431A or S431E mutants of LKB1, or LKB1S gave equal levels of activation of endogenous AMPK. Similarly, recombinant STRADα·MO25α complexes containing these LKB1 variants were equally effective at phosphorylating and activating AMPK, BRSK1, and BRSK2 in cell-free assays. Finally, all four LKB1 variants and a truncated LKB1 lacking the C-terminal region altogether were equally effective at causing cell cycle arrest when co-expressed with STRADα and MO25α in the G361 melanoma cell line. Our results do not support the idea that phosphorylation of Ser-431 increases the ability of LKB1 to phosphorylate downstream targets.
The AMP-activated protein kinase (AMPK)3 is an energy-sensing system involved in regulating energy balance at both the cellular and the whole body levels (1, 2). It exists as heterotrimeric complexes composed of a catalytic α subunit and regulatory β and γ subunits. Metabolic stresses that inhibit ATP synthesis (e.g. hypoxia, hypoglycemia) or that stimulate ATP consumption (e.g. muscle contraction) cause an increase in the cellular ADP:ATP ratio that is amplified by adenylate kinase into a much larger increase in the AMP:ATP ratio. AMP and ATP bind antagonistically to two sites formed by the four tandem cystathionine β-synthase motifs on the γ subunit (3, 4). AMPK is only active after phosphorylation of a critical threonine residue (Thr-172) within the kinase domain by upstream kinases (5, 6), the major upstream kinase being a complex between the tumor suppressor LKB1 and two accessory subunits, STRAD and MO25 (7, 8). The calmodulin-dependent protein kinase kinases, especially calmodulin-dependent protein kinase kinase β, can act as alternate upstream kinases phosphorylating Thr-172 (9–11). This latter effect is triggered by a rise in cytosolic Ca2+ without requiring any increase in AMP.
The sequence around Thr-172 is highly conserved in a family of kinases whose catalytic domains are closely related to those of AMPK. These AMPK-related kinases (ARKs) also require phosphorylation at the threonine residue equivalent to Thr-172 before they become active. They include the brain-specific kinases BRSK1/2 (also known as SAD-A/-B), SIK1/2/3 (SIK/QIK/QSK), NUAK1/2 (ARK5/SNARK), MARK1/2/3/4, and the testis-specific kinase SNRK (12, 13). In LKB1–/– mouse embryo fibroblasts those ARKs that are expressed are dephosphorylated and inactive, whereas they are phosphorylated and active in wild type mouse embryo fibroblasts (12).
Although LKB1 is required for the activity of AMPK and the ARKs in most cells, several lines of evidence suggest that it is constitutively active. Thus, in wild type mouse embryo fibroblasts that are stimulated with phenformin or AICAR, the activities of LKB1 and the ARKs are constant, despite increased phosphorylation and activation of AMPK (12). Similarly, the activities of LKB1 and the ARKs are unchanged in muscle during contraction, although the phosphorylation and activation of AMPK increases (14). These results suggest that LKB1 is constitutively active and continually phosphorylates AMPK. The effects of AICAR, phenformin, or contraction to promote Thr-172 phosphorylation appear to be due instead to the ability of 5-aminoimidazole-4-carboxamide ribonucleoside monophosphate (which increases in response to AICAR (15)) or AMP (which increases in response to phenformin (9), or contraction (16)) to inhibit its dephosphorylation (17–19).
Other experiments suggest that the activity of LKB1 is not rate-limiting for AMPK activation, at least in striated muscles. In hypomorphic mouse mutants with loxP sites in the Lkb1 gene, the expression of the protein in skeletal muscle is reduced by ≈50% in the heterozygote and >90% in the homozygote. However, activation of the α2 isoform of AMPK by electrically stimulated contraction was unaffected in the heterozygote and only moderately reduced, by ∼50%, in the homozygote (16). Similar results were obtained for the effect of ischemia in cardiac muscle (20). It was only when LKB1 expression was totally eliminated, by crossing with a strain expressing Cre recombinase from a muscle-specific promoter, that activation of AMPK-α2 by contraction or ischemia was abolished (16, 20). These results suggest that LKB1 is not normally rate-limiting for AMPK activation in muscle and that further activation of LKB1 would have little effect.
Contrasting with this view are reports suggesting that the C-terminal region of LKB1, distal to the kinase domain, might serve a regulatory function. This region is phosphorylated in intact cells at Ser-431 in mouse LKB1 by both cyclic AMP-dependent protein kinase and p90-RSK, and an S431A mutation was reported to prevent the ability of LKB1 to reduce proliferation of G361 cells (21). Glucagon caused phosphorylation of LKB1 at Ser-431 in perfused rat liver, and this correlated with increased phosphorylation of AMPK at the activating site, Thr-172 (22). Phosphorylation of Ser-431 by protein kinase Cζ was reported to be required for phosphorylation of Thr-172 on AMPK in response to metformin treatment of bovine aortic endothelial cells (23). Phosphorylation of Ser-431 on LKB1 appears to be required for axon specification in the developing nervous system in response to agents such as brain-derived neurotrophic factor, by promoting the ability of LKB1 to activate BRSK1/BRSK2 (SAD-B/SAD-A) (24, 25). Finally, in Drosophila melanogaster the residue equivalent to Ser-431 (Ser-535) can also be phosphorylated (26). Loss-of-function mutations in the lkb1 gene caused defects in polarity of the oocyte, and this could be rescued by low level expression in the germ line of wild type LKB1 or the potentially phosphomimetic S535E mutant, but not by the nonphosphorylatable S535A mutant, although the latter was effective if overexpressed (26).
In view of the conflicting evidence as to whether LKB1 exerts a regulatory role in vivo, we felt that it was important to re-examine the role of Ser-431 phosphorylation. This study was also prompted by our recent findings that variants of LKB1 exist because of alternate splicing at the 3′ end of the mRNA. The newly described short variant (LKB1S) contains an unique 38-residue sequence at the C terminus and lacks the Ser-431 site (27).
EXPERIMENTAL PROCEDURES
Reagents and Proteins—All of the reagents were from Sigma-Aldrich, unless stated otherwise. [γ-32P]ATP and glutathione-Sepharose were from GE Healthcare. The glutathione S-transferase (GST) fusions of the kinase domains of AMPK-α1 (1–312 (28)), BRSK1, and BRSK2 (residues 1–400 (12)) were expressed in bacteria as described previously.
General Methods and Buffers—Restriction enzyme digests, DNA ligations, and other recombinant DNA procedures were performed using standard protocols. DNA constructs used for transfection were purified from Escherichia coli using the Qiagen Plasmid Maxi kit according to the manufacturers' instructions. The lysis buffer was 50 mm Tris-HCl, pH 7.2, 50 mm NaF, 1 mm sodium pyrophosphate, 1 mm EDTA, 1 mm EGTA, 1 mm dithiothreitol (DTT), 0.1 mm benzamidine, 0.1 mm phenylmethanesulfonyl fluoride, 5 μg/ml soybean trypsin inhibitor, 1% (v/v) Triton X-100. Buffer A contained 50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 5 mm DTT. Buffer B contained 50 mm Tris-HCl, pH 7.4, 1 mm EGTA, 1 mm DTT, 0.5% (v/v) Triton X-100. Buffer C contained 50 mm Tris-HCl pH 7.4, 1 mm EGTA, 0.27 m sucrose, 1 mm DTT, 0.5% (v/v) Triton X-100. The assay buffer was 50 mm Na-Hepes, 1 mm DTT, 0.02% Brij-35 containing 200 μm ATP and 5 mm MgCl2.
Antibodies—Antibodies recognizing AMPK-α1 and AMPKα2 (29), LKB1 residues 24–36 (anti-LKB1(N) (30), GST (31), ACC (pS79) (32)), and LKB1(pS431) (21)) have been described previously. Monoclonal antibodies recognizing β-actin and FLAG epitope tags were obtained from Sigma-Aldrich, anti-myc and AMPK-α (pT172) were from Cell Signaling Technology, and monoclonal antibody recognizing green fluorescent protein (GFP) was from Roche Applied Science. Secondary antibodies coupled to IRDye680 were from Molecular Probes. Secondary antibodies coupled to IRDye800 and streptavidin conjugated to IRDye800 were from Rockland Inc.
Plasmids—Plasmids encoding GST-LKB1L in the pEBG-2T vector and FLAG-STRADα, and myc-MO25α in the pCMV5 vector have been described previously (21, 33). Point mutations encoding the substitutions in LKB1L, i.e. D194A, S431A, and S431E were generated using the QuikChange II site-directed mutagenesis kit (Stratagene) according to the manufacturer's instructions. The C-terminal truncation of LKB1 (LKB11–343) was generated using site-directed mutagenesis to insert a stop codon after residue 343. Wild type, mutant, and truncated LKB1 constructs were amplified by PCR from pcDNA3.1zeo plasmids (sense, 5′-cggactagtccgatggacgtggctgacccccag-3′; LKB1L antisense, 5′-cggggtaccccgtcactgctgcttgcaggccga-3′, LKB1S antisense, 5′-cggggtaccccgtcacagtggacaaagctttat-3′, 1–343 antisense, cggggtaccccgtcagtcctccaggtagggcac3′) and inserted into the SpeI/KpnI sites of the pEBG-2T vector. Positive clones were confirmed by DNA sequencing. Wild type, mutant and truncated LKB1 constructs were amplified by PCR from pcDNA3.1zeo plasmids (sense 5′-ccggaattccggatggacgtggctgacccccag-3′; antisense primers as above) and inserted into the EcoRI/KpnI sites of the pEGFP-C2 vector (a gift from Dr. Nick Leslie, University of Dundee). Positive clones were determined by DNA sequencing. All of the DNA sequencing was performed by the Sequencing Service, College of Life Sciences, University of Dundee, using Applied Biosystems Big-Dye version 3.1 chemistry on an Applied Biosystems model 3730 automated capillary DNA sequencer.
Cell Culture—HeLa and HEK293 cells were maintained in Dulbecco's modified Eagle's medium containing 10% (v/v) fetal bovine serum. Human melanoma G361 cells were maintained in McCoy's 5A medium containing 2 mml-glutamine and 10% (v/v) fetal bovine serum. The generation and maintenance of HeLa cells stably expressing the wild type and S431A mutant of LKB1 has been described previously (30). For all experiments, the cells were cultured on 60- or 100-mm-diameter dishes and lysed in situ in 0.25–0.5 ml of ice-cold lysis buffer (34). The lysates were clarified by centrifugation at 14,000 × g for 10 min at 4 °C.
Expression of GST Fusion Proteins in HEK293 Cells and Affinity Purification—Dishes (10 cm) of HEK293 cells were transiently transfected with 3 μg of the pEBG-2T constructs together with FLAG-STRADα and myc-MO25α using the PEI method. After 36–48 h the cells were lysed, and the clarified lysates were incubated for 2 h on a rotating platform with glutathione-Sepharose (25 μl/dish of lysate) previously equilibrated in Buffer A. The beads were washed three times in Buffer A, twice with Buffer B, and twice with Buffer C. The resin was incubated with 1–1.5 volumes Buffer C containing 20 mm glutathione to elute the GST fusion proteins. The eluate was snap frozen and stored at –80 °C.
Kinase Assays—AMPK was assayed as described previously (34). For the cell-free assays, 1.5 μg of GST-AMPKα1, GST-BRSK1, or GST-BRSK2 kinase domains were incubated with the indicated amount of purified GST·LKB1 complex in assay buffer in a final volume of 20 μl. After incubation at 30 °C for 15 min, the activities of GST-AMPKα1, GST-BRSK1, or GST-BRSK2 kinase domains were determined by adding 10 μl of this reaction to an assay containing 200 μm [γ-32P]ATP, 5 mm MgCl2, and 200 μm AMARA peptide (34, 35).
Immunoblotting—SDS-PAGE utilized precast Bis-Tris 4–12% gradient polyacrylamide gels, in the MOPS buffer system (Invitrogen), except for analysis of acetyl-CoA carboxylase, where precast 3–8% Tris acetate gels were used (Invitrogen). Analysis of Western blots using dual labeling of phospho-specific and phosphorylation-independent probes has been described previously (7).
Cell Cycle Analysis—G361 cells were co-transfected with plasmids encoding FLAG-STRADα and myc-MO25α, together with GFP alone or the indicated GFP-LKB1 construct using Effectene transfection reagent (Qiagen). Post-transfection (36 h), the cells were treated with nocodazole (70 ng/ml) and grown for an additional 18 h to induce a G2/M block. The cells were harvested, fixed in 70% ethanol, washed twice in phosphate-buffered saline containing 0.1 mm EDTA and 1% (v/v) fetal bovine serum, treated with RNase (50 μg/ml; Qiagen), and stained with propidium iodide (50 μg/ml). The cell cycle profiles of GFP-positive cells were determined by flow cytometry.
RESULTS
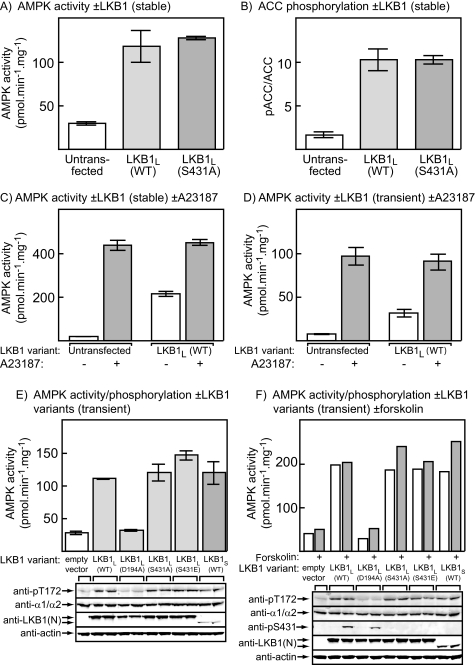
Mutation or Phosphorylation of Ser-431 Does Not Affect the Activation of AMPK in HeLa Cells—We initially utilized HeLa cells, which do not express LKB1 so that AMPK has a very low activity unless an ionophore or other agent that increases cellular Ca2+ is added to activate endogenous calmodulin-dependent protein kinase kinase β (7, 9) (Fig. 1, C and D). HeLa cells stably expressing wild type LKB1L or an S431A mutant of LKB1L have been previously generated (30). As expected, the cells expressing wild type LKB1L exhibited an increased AMPK activity compared with untransfected control cells, and this was associated with a large increase in phosphorylation of the downstream target of AMPK, ACC, at the AMPK site (Ser-79). However, there were identical increases in AMPK activity (Fig. 1A) and ACC phosphorylation (Fig. 1B) in the cells expressing the S431A mutant. The addition of the Ca2+ ionophore A23187 to activate calmodulin-dependent protein kinase kinase β, either to control cells or cells stably expressing wild type LKB1L (Fig. 1C) or to normal HeLa cells transiently transfected with DNAs encoding wild type LKB1L, STRADα, and MO25α (Fig. 1D), caused a larger increase in AMPK activity that was not additive with the effect of LKB1L expression. Next, we transiently transfected normal HeLa cells with DNAs encoding STRADα, MO25α, and wild type LKB1L, or several variants of it, including a kinase-inactive mutant (D194A), a nonphosphorylatable mutant (S431A), and a potentially phospho-mimetic mutant (S431E), as well as the short splice variant, LKB1S, that lacks the Ser-431 site. Fig. 1E shows that, by Western blotting using an antibody (anti-LKB1(N)) that recognizes an N-terminal epitope common to all variants, LKB1L and all mutants of it expressed at approximately equal levels, whereas LKB1S expressed at a significantly lower level. It also shows that all active variants, but not the D194A mutant, increased the activity of endogenous AMPK, and its phosphorylation at Thr-172, to an equal extent compared with untransfected control cells. LKB1S was just as effective as the three active LKB1L variants, even though its level of expression was much lower. These experiments were repeated with or without forskolin to activate endogenous cyclic AMP-dependent protein kinase, which is known to phosphorylate Ser-431 (21). As expected, Ser-431 was phosphorylated (assessed using a phosphospecific antibody) in response to forskolin only when LKB1L (or the kinase-inactive D194A mutant) and not when the S431A or S431E mutants of LKB1L, or LKB1S, were expressed. Even though Ser-431 phosphorylation was stimulated by forskolin using wild type LKB1L, the activation of endogenous AMPK, or its phosphorylation on Thr-172, was not significantly increased (Fig. 1F).
FIGURE 1.
Effect of expression of LKB1 variants in HeLa cells. A, the activities of AMPK measured in immunoprecipitates in lysates of control cells or cells stably expressing wild type LKB1L, or the S431A mutant of LKB1L. B, as A, but measuring the phosphorylation of Ser-79 on ACC, assessed as the ratio of signal obtained by Western blotting using a phosphospecific antibody and using streptavidin to detect total ACC. C, AMPK activity in untransfected cells or cells stably expressing inactive or wild type LKB1L treated with or without 10 μm A23187. D, AMPK activity in untransfected cells or cells transiently transfected with plasmids expressing wild type LKB1L, STRADα, and MO25α, treated with or without 10 μm A23187. E, activity of AMPK (top) and expression and phosphorylation of various proteins (bottom) in HeLa cells transiently transfected with empty vector or with plasmids expressing STRADα and MO25α with LKB1L, various mutants of LKB1L, or LKB1S. The phosphorylation of AMPK at Thr-172 was assessed by probing blots with a phosphospecific antibody, as was the total level of expression of AMPK and LKB1, the latter using an antibody recognizing an N-terminal epitope that recognizes both long and short splice variants. Actin expression was also assessed as a loading control. F as E, but cells were incubated with or without 20 μm forskolin. Phosphorylation of LKB1L at Ser-431 was assessed using a phosphospecific antibody. WT, wild type.
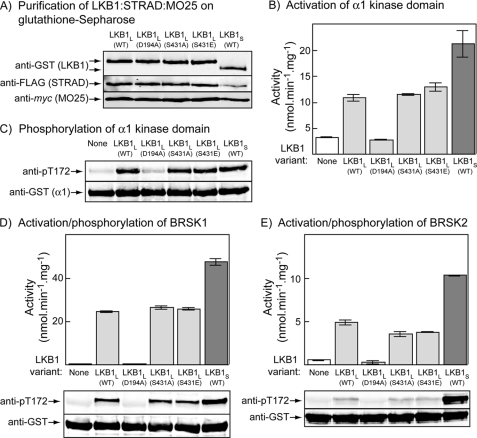
Phosphorylation of Ser-431 Does Not Stimulate Phosphorylation of AMPK, BRSK1, or BRSK2 in Cell-free Assays—Because it has been proposed that phosphorylation of Ser-431 on LKB1L promotes its ability to phosphorylate and activate BRSK1/2 and hence axon specification (24, 25), we also examined the role of this phosphorylation in the activation and phosphorylation of BRSK1 and BRSK2. Because these “brain-specific” kinases are not expressed in HeLa cells, this analysis was performed in cell-free assays after expression of GST fusions of LKB1 complexes in HEK-293 cells and of BRSK1 and BRSK2 in bacteria. Fig. 2A shows the results of purification of LKB1 complexes on glutathione-Sepharose, after co-expression of GST fusions of LKB1 variants with FLAG-tagged STRADα and myc-tagged MO25α in HEK-293 cells. All LKB1 variants, including wild type LKB1L, the D194A, S431A, and S431E mutants of LKB1L, and wild type LKB1S, expressed equally well and (unlike free GST, not shown) co-purified with FLAG-STRADα and myc-MO25α. We initially tested these complexes using a bacterially expressed GST fusion of the AMPK-α1 kinase domain, which is dephosphorylated and inactive because of the lack of upstream kinases in bacteria (28). The wild type, S431A, and S431E complexes of LKB1L all phosphorylated Thr-172 and activated the GST-α1 kinase domain fusion equally well, whereas the inactive D194A mutant did not increase phosphorylation and activity above that in a control without kinase, as expected (Fig. 2, B and C). Similar results were obtained for phosphorylation and activation of GST fusions of BRSK1 and BRSK2 (Fig. 2, D and E), although the level of phosphorylation and activation of BRSK2 was much less than that of BRSK1. The similar levels of activation obtained with the wild type, S431A, and S431E variants of LKB1S were not due to the assays being saturated with upstream kinase, because in every case LKB1S gave an even larger level of both phosphorylation and activation (Fig. 2, B–E).
FIGURE 2.
Phosphorylation and activation of AMPK, BRSK1, and BRSK2 by LKB1 variants in cell-free assays. A, purification of LKB1·STRADα·MO25α complexes from HEK-293 cells. Plasmids encoding FLAG-tagged STRADα and myc-tagged MO25α were co-expressed in HEK-293 cells with the indicated variants of GST-tagged LKB1. GST fusions were purified on glutathione-Sepharose, and the products were analyzed by Western blotting using anti-GST, anti-FLAG, or anti-myc antibodies. B–E, bacterially expressed GST fusions with the kinase domains of AMPK-α1 (B and C), BRSK1 (D), or BRSK2 (E) were incubated with MgATP and LKB1·STRADα·MO25α complexes (50 μg·ml–1) purified as in A. After 15 min the incubations were analyzed for activity of AMPK (B), BRSK1 (D), or BRSK2 (E) and for phosphorylation of the threonine residue equivalent to Thr-172 using anti-pT172 antibody (C–E). WT, wild type.
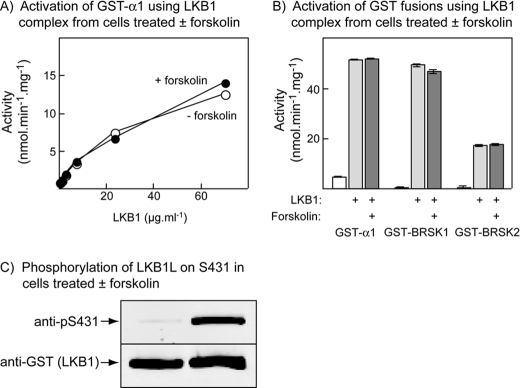
Phosphorylation of Ser-431 on LKB1 Does Not Affect the Activation of AMPK, BRSK1, or BRSK2—We also isolated the wild type GST·LKB1L complex from HEK-293 cells that had been treated with forskolin to cause phosphorylation of Ser-431. Using a phosphospecific antibody we could demonstrate that Ser-431 was phosphorylated if the cells had been treated with forskolin, but this had no effect on the activation of the bacterially expressed GST-α1 kinase domain, GST-BRSK1, or GST-BRSK2 (Fig. 3).
FIGURE 3.
Effect of Ser-431 phosphorylation on the activation of AMPK, BRSK1 and BRSK2 by LKB1 variants in cell-free assays. LKB1·STRADα· MO25α complexes were expressed in HEK-293 cells as for Fig. 2, except that some dishes of cells were treated with 20 μm forskolin for 20 min prior to lysis. A, a GST fusion of the AMPK-α1 kinase domain was incubated with MgATP and various concentrations of wild type LKB1L·STRADα·MO25α complex isolated from cells treated with or without forskolin for 15 min, and AMPK activity was determined. B, GST fusions of the AMPK-α1 kinase domain, BRSK1, and BRSK2 were incubated with MgATP and complexes containing STRADα, MO25α, and the indicated variant of LKB1 (80 μg·ml–1) for 15 min, and AMPK, BRSK1, and BRSK2 activities determined. C, samples of the purified LKB1L·STRADα·MO25α complexes from control and forskolin-treated cells were analyzed by Western blotting to assess the phosphorylation of Ser-431 and the total content of the GST-LKB1 fusion.
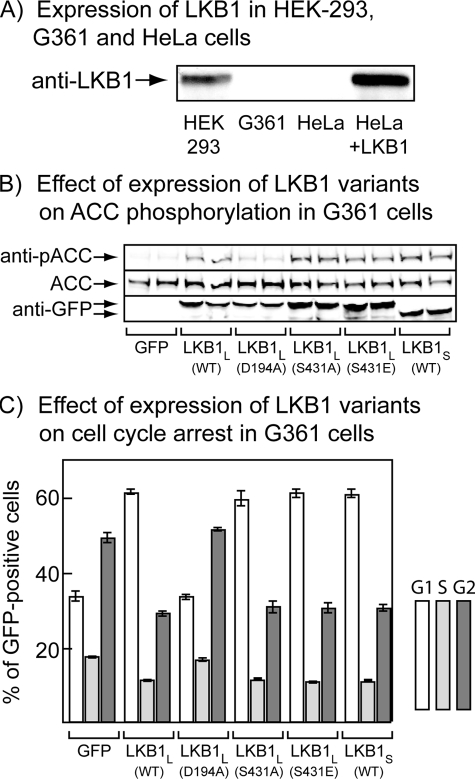
Phosphorylation of Ser-431 Does Not Affect Progress through the Cell Cycle—To assess the effect of Ser-431 phosphorylation on cell cycle arrest caused by LKB1, we co-transfected the G361 melanoma cell line with GFP-tagged LKB1 variants, FLAG-STRADα, and myc-MO25α. These cells, like HeLa cells, do not express endogenous LKB1 (Fig. 4A). The cells were transfected with plasmids encoding GFP or GFP fusions with wild type LKB1L, the D194A (kinase-inactive), S431A, or S431E mutants of LKB1L, or wild type LKB1S. After 36 h there was relatively uniform expression of all variants in the cells, assessed using anti-GFP antibody, and there was increased phosphorylation (compared with cells expressing GFP alone) of the AMPK target ACC at Ser-79 in cells expressing active LKB1 variants (wild type, S431A, S431E, or LKB1S) but not in cells expressing the kinase-inactive D194A mutant (Fig. 4B). The cells were then treated with nocodazole for a further 18 h to cause arrest of cells passing through G2-M phase, and we performed cell cycle analysis by flow cytometry of propidium iodide-labeled cells. The use of GFP-tagged LKB1 variants had the advantage that we could restrict analysis to cells expressing GFP, so we could be certain that all cells analyzed for their DNA content by propidium iodide staining were expressing the variants. The proportion of cells expressing GFP alone or those expressing kinase-inactive (D194A) LKB1L with G1, S and G2 phase DNA contents were very similar at ∼33, 17, and 50%, respectively. However, cells expressing the wild type, S431A, and S431E mutants of LKB1L showed clear evidence for a G1-S phase arrest, with a much higher proportion of cells in G1 (60%) and a lower proportion in S (10%) and G2 (30%). Cells expressing LKB1S gave very similar results (Fig. 4C).
FIGURE 4.
Effect of expression of LKB1 variants on progress through the cell cycle in G361 melanoma cells. A, expression of LKB1 in HEK-293, G361, and HeLa cells, and in HeLa transiently transfected with LKB1L (see Fig. 1) using anti-LKB1 antibody. LKB1 was immunoprecipitated from cell lysates using the anti-LKB1(N) antibody, and the immunoprecipitates were analyzed by Western blotting using the same antibody. B, phosphorylation of the AMPK target, ACC, in G361 cells 36 h after transfection with plasmids encoding STRADα, MO25α, and the indicated variant of LKB1. For this experiment the LKB1 variants had GFP fused at the N terminus and plasmids expressing GFP, STRADα, and MO25α were co-transfected in the control incubation (labeled GFP). The expression level of free GFP in this control was similar to that of the GFP-LKB1 fusions (not shown). C as B, but 36 h after transfection nocodazole was added to arrest cells passing at the G2-M boundary. After incubation for a further 18 h, the cells were fixed in 70% ethanol, stained with propidium iodide, and analyzed by flow cytometry. The cells expressing GFP were selected, and the proportions with G1, S, and G2 phase DNA contents are shown. WT, wild type.
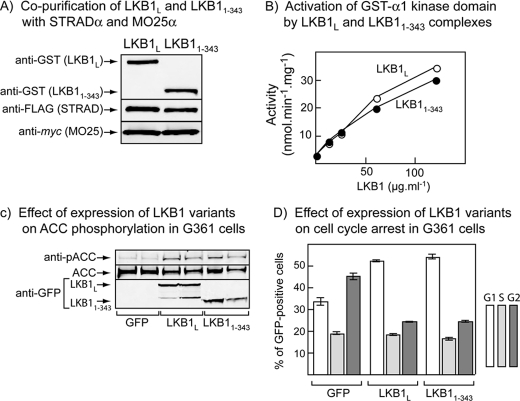
We also wished to test whether the C termini of LKB1L or LKB1S were necessary for AMPK activation and cell cycle arrest. For these experiments we utilized GFP fusions containing only the first 343 residues of LKB1, which is the smallest C-terminally truncated form that has been shown to still bind STRAD and MO25 (36). The 1–343 construct contains most, but not all, of the region common to LKB1L and LKB1S and completely lacks the Ser-431 site. We initially co-expressed GST fusions of wild type LKB1L and LKB11–343 with FLAG-STRADα and myc-MO25α in HEK-293 cells. After purification on glutathione-Sepharose, we obtained equal yields of full-length and truncated LKB1L, and both co-purified with FLAG-STRADα and myc-MO25α as expected (Fig. 5A). The LKB1L and LKB11–343 complexes activated the bacterially expressed GST fusion of the AMPK-α1 kinase domain equally well (Fig. 5B). We next co-expressed GFP alone, GFP-LKB1L, or GFP-LKB11–343 with FLAG-STRADα and myc-MO25α in G361 cells and carried out analysis of expression and ACC phosphorylation, as well as cell cycle analysis of GFP-expressing cells as before. The cells expressing GFP-LKB1L and GFP-LKB11–343 displayed a similar expression of the LKB1 constructs, a similar increased phosphorylation of the AMPK target ACC at Ser-79, and very similar cell cycle arrest relative to cells expressing GFP alone (Fig. 5, C and D).
FIGURE 5.
Effect of C-terminal truncation of LKB1 on AMPK activation in cell-free assays and ACC phosphorylation and cell cycle progress in G361 melanoma cells. A, plasmids encoding GST fusions of wild type LKB1L and a C-terminal truncation (1–343) were co-expressed with FLAG-STRADα and myc-MO25α in HEK-293 cells and purified on glutathione-Sepharose. The purified products were analyzed by Western blotting using anti-GST, anti-FLAG, and anti-myc antibodies. B, a bacterially expressed GST fusion of the AMPK-α1 kinase domain was incubated with MgATP and various concentrations of GST-LKB1·FLAG-STRADα·myc-MO25α complex purified as in A, and AMPK activity was determined after 15 min. C, phosphorylation of the AMPK target, ACC, total ACC, and expression of GFP-LKB1 assessed using an anti-GFP antibody, in G361 cells co-expressing STRADα and MO25α with free GFP (control) or GFP fusions of wild type LKB1L and a C-terminally truncated mutant (1–343). D, cell cycle analysis of GFP-expressing cells treated as in Fig. 5C, 18 h after nocodazole treatment.
DISCUSSION
Our results support the idea that LKB1 is constitutively active, and cast serious doubt on the idea that Ser-431 phosphorylation has a direct role in regulating LKB1 activity. The evidence in favor of this view may be summarized as follows: 1) Co-expression of wild type LKB1L with STRADα and MO25α in HeLa cells, which lack endogenous LKB1, caused an increase in phosphorylation at Thr-172 and activity of AMPK, and this was unaffected by mutation of Ser-431 to an nonphosphorylatable alanine residue (S431A) or a potentially phospho-mimetic glutamate residue (S431E). Phosphorylation and activation of AMPK was also the same when the short splice variant LKB1S was expressed, even though this lacks the Ser-431 site, and expression occurred at a lower level. The effect did, however, require the kinase activity of LKB1, because no increase in phosphorylation and activation of AMPK was observed when a kinase-inactive (D194A) mutant of LKB1L was expressed. 2) The results were not altered when the cells were treated with forskolin to activate cyclic AMP-dependent protein kinase. As expected, a strong signal was obtained using a phosphospecific antibody against Ser-431 in response to forskolin treatment when wild type LKB1L or the inactive D194A mutant of LKB1L had been expressed, but not when the S431A or S431E mutants of LKB1L, or LKB1S (which lacks the Ser-431 site) had been expressed. 3) When variants of GST-LKB1, including the wild type and S431A and S431E mutants of LKB1L and LKB1S were co-expressed with FLAG-STRADα and MO25α in HEK-293 cells and purified on glutathione-Sepharose, they all formed heterotrimeric complexes and in cell-free assays phosphorylated and activated the AMPK-α1 kinase domain, BRSK1, and BRSK2 equally well. Wild type GST-LKB1L·FLAG-STRADα·myc-MO25α complexes also phosphorylated and activated the AMPK-α1 kinase domain, BRSK1, and BRSK2 equally well, irrespective of whether they were phosphorylated at Ser-431 by prior treatment of the HEK-293 cells with forskolin.
Sapkota et al. (21) found that phosphorylation of LKB1 at Ser-431 did not directly affect its kinase activity, although at that time a physiological substrate had not been identified, so its activity was monitored either by autophosphorylation or by using an artificial substrate, p53. We have now confirmed that phosphorylation at Ser-431 does not alter LKB1 activity using three physiological targets, i.e. the kinase domain of AMPK-α1, BRSK1, and BRSK2. Sapkota et al. (21) did report that the S431A and S431D mutants of LKB1 were much less effective than the wild type at suppressing cell growth in G361 cells, a melanoma cell line that does not express endogenous LKB1. Their approach was to co-transfect plasmids encoding LKB1 variants with a vector that provided resistance to the antibiotic G418. Four days after transfection, the expression of wild type LKB1 (LKB1L) and the D194A, S431A, and S431D mutants was similar. The cells were then grown in G418 for 16 days, and the number of colonies formed was counted. By this approach, all three mutants seemed to be much less effective than the wild type in suppressing colony formation. We adopted a different approach in which GFP-tagged LKB1 variants were co-expressed with STRADα and MO25α (which had not been identified as accessory subunits for LKB1 at the time of the study by Sapkota et al. (21)). We added nocodazole to arrest cells passing through the G2-M boundary and analyzed the DNA content of GFP-expressing cells 18 h later by flow cytometry using propidium iodide staining. The advantage of this method is that we could be certain that the cells in which we conducted cell cycle analysis were expressing LKB1 and also that we were studying short term effects on the cell cycle rather than much longer term (16 day) effects on cell growth and proliferation, where secondary effects could have occurred. Our results do not support the idea that the S431A or S431E mutation or the phosphorylation of Ser-431 in the wild type had any effect on progress through the cell cycle.
How can we reconcile our results with those in previous studies where regulatory effects of phosphorylation of Ser-431 on LKB1 have been claimed? Kimball et al. (22) reported that perfusion of rat liver with glucagon caused increased phosphorylation of Ser-431 on LKB1, increased phosphorylation of AMPK on Thr-172, and repression of the mTOR pathway as judged by decreased phosphorylation of the mTOR substrates, S6 kinase I and 4E-binding protein 1. They appear to have assumed that the phosphorylation of LKB1 on Ser-431 and the associated activation of AMPK were causally related but in fact never showed that there was any change in LKB1 activity. Other studies have used genetic approaches to show that substitution of the residue equivalent to Ser-431 by alanine reduces the ability of LKB1 to rescue defects in cell polarity in the Drosophila oocyte (26) and to promote development of axons rather than dendrites in cultured embryonic hippocampal neurones, an effect that appears to require its phosphorylation of BRSK1/BRSK2 (SAD-B/-A) (24, 25). However, in none of these studies was the activity of LKB1 or of any of it downstream targets measured. Our results clearly demonstrate that phosphorylation of Ser-431 does not alter the intrinsic ability of LKB1·STRAD·MO25 complexes to phosphorylate and activate AMPK, BRSK1, or BRSK2, either in cell-free assays or, in the case of AMPK, in intact HeLa cells. They do not rule out more complex mechanisms; phosphorylation of the residue equivalent to Ser-431 could, for example, alter the localization of LKB1 and therefore affect its ability to phosphorylate downstream targets such as BRSK1 and BRSK2 in specific subcellular locations. Indeed, evidence that Ser-428 phosphorylation causes a change in the localization of human LKB1L has already been provided (23).
Author's Choice—Final version full access.
This work was also supported by a Wellcome Trust Programme Grant, by EXGENESIS Integrated Project Grant LSHM-CT-2004-005272 of the European Commission, and by the pharmaceutical companies supporting the Division of Signal Transduction Therapy at Dundee (AstraZeneca, Boehringer-Ingelheim, GlaxoSmithKline, Merck & Co. Inc, Merck KgaA and Pfizer). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: AMPK, AMP-activated protein kinase; AICAR, 5-aminoimidazole-4-carboxamide ribonucleoside; ACC, acetyl-CoA carboxylase; ARK, AMPK-related kinase; BRSK, brain-specific kinase; DTT, dithiothreitol; GFP, green fluorescent protein; GST, glutathione S-transferase; MOPS, 4-morpholinepropanesulfonic acid.
References
- 1.Kahn, B. B., Alquier, T., Carling, D., and Hardie, D. G. (2005) Cell Metab. 1 15–25 [DOI] [PubMed] [Google Scholar]
- 2.Hardie, D. G. (2007) Nat. Rev. Mol. Cell. Biol. 8 774–785 [DOI] [PubMed] [Google Scholar]
- 3.Scott, J. W., Hawley, S. A., Green, K. A., Anis, M., Stewart, G., Scullion, G. A., Norman, D. G., and Hardie, D. G. (2004) J. Clin. Investig. 113 274–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xiao, B., Heath, R., Saiu, P., Leiper, F. C., Leone, P., Jing, C., Walker, P. A., Haire, L., Eccleston, J. F., Davis, C. T., Martin, S. R., Carling, D., and Gamblin, S. J. (2007) Nature 449 496–500 [DOI] [PubMed] [Google Scholar]
- 5.Hawley, S. A., Davison, M., Woods, A., Davies, S. P., Beri, R. K., Carling, D., and Hardie, D. G. (1996) J. Biol. Chem. 271 27879–27887 [DOI] [PubMed] [Google Scholar]
- 6.Stein, S. C., Woods, A., Jones, N. A., Davison, M. D., and Carling, D. (2000) Biochem. J. 345 437–443 [PMC free article] [PubMed] [Google Scholar]
- 7.Hawley, S. A., Boudeau, J., Reid, J. L., Mustard, K. J., Udd, L., Makela, T. P., Alessi, D. R., and Hardie, D. G. (2003) J. Biol. 2 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Woods, A., Johnstone, S. R., Dickerson, K., Leiper, F. C., Fryer, L. G., Neumann, D., Schlattner, U., Wallimann, T., Carlson, M., and Carling, D. (2003) Curr. Biol. 13 2004–2008 [DOI] [PubMed] [Google Scholar]
- 9.Hawley, S. A., Pan, D. A., Mustard, K. J., Ross, L., Bain, J., Edelman, A. M., Frenguelli, B. G., and Hardie, D. G. (2005) Cell Metab. 2 9–19 [DOI] [PubMed] [Google Scholar]
- 10.Woods, A., Dickerson, K., Heath, R., Hong, S. P., Momcilovic, M., Johnstone, S. R., Carlson, M., and Carling, D. (2005) Cell Metab. 2 21–33 [DOI] [PubMed] [Google Scholar]
- 11.Hurley, R. L., Anderson, K. A., Franzone, J. M., Kemp, B. E., Means, A. R., and Witters, L. A. (2005) J. Biol. Chem. 280 29060–29066 [DOI] [PubMed] [Google Scholar]
- 12.Lizcano, J. M., Göransson, O., Toth, R., Deak, M., Morrice, N. A., Boudeau, J., Hawley, S. A., Udd, L., Mäkelä, T. P., Hardie, D. G., and Alessi, D. R. (2004) EMBO J. 23 833–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jaleel, M., McBride, A., Lizcano, J. M., Deak, M., Toth, R., Morrice, N. A., and Alessi, D. R. (2005) FEBS Lett. 579 1417–1423 [DOI] [PubMed] [Google Scholar]
- 14.Sakamoto, K., Goransson, O., Hardie, D. G., and Alessi, D. R. (2004) Am. J. Physiol. 287 E310–E317 [DOI] [PubMed] [Google Scholar]
- 15.Corton, J. M., Gillespie, J. G., Hawley, S. A., and Hardie, D. G. (1995) Eur. J. Biochem. 229 558–565 [DOI] [PubMed] [Google Scholar]
- 16.Sakamoto, K., McCarthy, A., Smith, D., Green, K. A., Hardie, D. G., Ashworth, A., and Alessi, D. R. (2005) EMBO J. 24 1810–1820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davies, S. P., Helps, N. R., Cohen, P. T. W., and Hardie, D. G. (1995) FEBS Lett. 377 421–425 [DOI] [PubMed] [Google Scholar]
- 18.Sanders, M. J., Grondin, P. O., Hegarty, B. D., Snowden, M. A., and Carling, D. (2007) Biochem. J. 403 139–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Suter, M., Riek, U., Tuerk, R., Schlattner, U., Wallimann, T., and Neumann, D. (2006) J. Biol. Chem. 281 32207–32216 [DOI] [PubMed] [Google Scholar]
- 20.Sakamoto, K., Zarrinpashneh, E., Budas, G. R., Pouleur, A. C., Dutta, A., Prescott, A. R., Vanoverschelde, J. L., Ashworth, A., Jovanovic, A., Alessi, D. R., and Bertrand, L. (2006) Am. J. Physiol. 290 E780–E788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sapkota, G. P., Kieloch, A., Lizcano, J. M., Lain, S., Arthur, J. S., Williams, M. R., Morrice, N., Deak, M., and Alessi, D. R. (2001) J. Biol. Chem. 276 19469–19482 [DOI] [PubMed] [Google Scholar]
- 22.Kimball, S. R., Siegfried, B. A., and Jefferson, L. S. (2004) J. Biol. Chem. 279 54103–54109 [DOI] [PubMed] [Google Scholar]
- 23.Xie, Z., Dong, Y., Scholz, R., Neumann, D., and Zou, M. H. (2008) Circulation 117 952–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barnes, A. P., Lilley, B. N., Pan, Y. A., Plummer, L. J., Powell, A. W., Raines, A. N., Sanes, J. R., and Polleux, F. (2007) Cell 129 549–563 [DOI] [PubMed] [Google Scholar]
- 25.Shelly, M., Cancedda, L., Heilshorn, S., Sumbre, G., and Poo, M. M. (2007) Cell 129 565–577 [DOI] [PubMed] [Google Scholar]
- 26.Martin, S. G., and St. Johnston, D. (2003) Nature 421 379–384 [DOI] [PubMed] [Google Scholar]
- 27.Towler, M. C., Fogarty, S., Hawley, S. A., Pan, D. A., Martin, D. M. A., Morrice, N., McCarthy, A., Galardo, M. N., Merino, S. B., Cigorraga, S. B., Ashworth, A., Sakamoto, K., and Hardie, D. G. (2008) Biochem. J. 416 1–14 [DOI] [PubMed] [Google Scholar]
- 28.Scott, J. W., Norman, D. G., Hawley, S. A., Kontogiannis, L., and Hardie, D. G. (2002) J. Mol. Biol. 317 309–323 [DOI] [PubMed] [Google Scholar]
- 29.Woods, A., Salt, I., Scott, J., Hardie, D. G., and Carling, D. (1996) FEBS Lett. 397 347–351 [DOI] [PubMed] [Google Scholar]
- 30.Sapkota, G. P., Deak, M., Kieloch, A., Morrice, N., Goodarzi, A. A., Smythe, C., Shiloh, Y., Lees-Miller, S. P., and Alessi, D. R. (2002) Biochem. J. 368 507–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goransson, O., McBride, A., Hawley, S. A., Ross, F. A., Shpiro, N., Foretz, M., Viollet, B., Hardie, D. G., and Sakamoto, K. (2007) J. Biol. Chem. 282 32549–32560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hawley, S. A., Gadalla, A. E., Olsen, G. S., and Hardie, D. G. (2002) Diabetes 51 2420–2425 [DOI] [PubMed] [Google Scholar]
- 33.Boudeau, J., Baas, A. F., Deak, M., Morrice, N. A., Kieloch, A., Schutkowski, M., Prescott, A. R., Clevers, H. C., and Alessi, D. R. (2003) EMBO J. 22 5102–5114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hardie, D. G., Salt, I. P., and Davies, S. P. (2000) Methods Mol. Biol. 99 63–75 [DOI] [PubMed] [Google Scholar]
- 35.Dale, S., Wilson, W. A., Edelman, A. M., and Hardie, D. G. (1995) FEBS Lett. 361 191–195 [DOI] [PubMed] [Google Scholar]
- 36.Baas, A. F., Boudeau, J., Sapkota, G. P., Smit, L., Medema, R., Morrice, N. A., Alessi, D. R., and Clevers, H. C. (2003) EMBO J. 22 3062–3072 [DOI] [PMC free article] [PubMed] [Google Scholar]