Abstract
Microsomal prostaglandin E synthase type 1 (mPGES-1) converts prostaglandin endoperoxides, generated from arachidonic acid by cyclooxygenases, into prostaglandin E2. This enzyme belongs to the membrane-associated proteins in eicosanoid and glutathione metabolism (MAPEG) family of integral membrane proteins, and because of its link to inflammatory conditions and preferential coupling to cyclooxygenase 2, it has received considerable attention as a drug target. Based on the high resolution crystal structure of human leukotriene C4 synthase, a model of mPGES-1 has been constructed in which the tripeptide co-substrate glutathione is bound in a horseshoe-shaped conformation with its thiol group positioned in close proximity to Arg-126. Mutation of Arg-126 into an Ala or Gln strongly reduces the enzyme's prostaglandin E synthase activity (85–95%), whereas mutation of a neighboring Arg-122 does not have any significant effect. Interestingly, R126A and R126Q mPGES-1 exhibit a novel, glutathione-dependent, reductase activity, which allows conversion of prostaglandin H2 into prostaglandin F2α. Our data show that Arg-126 is a catalytic residue in mPGES-1 and suggest that MAPEG enzymes share significant structural components of their active sites.
Prostaglandin E2 (PGE2)3 is an abundant lipid mediator that signals via four receptors (EP1 to -4), resulting in an array of important biological actions in physiology as well as pathophysiology (1, 2). Biosynthesis of PGE2 proceeds from arachidonic acid, which is converted to the unstable endoperoxide, prostaglandin H2, by cyclooxygenase type 1 and 2. PGH2 is further isomerized into PGE2 by three distinct enzymes, cytosolic PGE synthase, microsomal PGE synthase type 1 (mPGES-1), and microsomal PGE synthase type 2 (3–5). Although cytosolic PGE synthase and microsomal PGE synthase type 2 seem to provide a basal synthesis of PGE2, mPGES-1 appears to account for PGE2 synthesis under proinflammatory conditions. Thus, mPGES-1 is up-regulated by mitogens and cytokines and is functionally coupled to cyclooxygenase type 2 (4, 6). Due to its key role in PGE2 synthesis, mPGES-1 has attracted attention as a potential drug target in the areas of inflammation, pain, fever, and cancer (7).
mPGES-1 is a member of the MAPEG superfamily of enzymes (8), which also includes two key proteins in the leukotriene cascade, viz. 5-lipoxygenase-activating protein and leukotriene C4 synthase (LTC4S). Since all MAPEG members are integral membrane proteins, structural information on this family has been scarce. Recently, however, significant progress has been made in this area, and several high and low resolution structures have been solved by three-dimensional as well as two-dimensional crystallography (9–12). The crystal structures of human LTC4S clearly provided the most detailed structural information, inter alia a unique, horse-shoe shaped binding conformation of GSH, a hydrophobic crevice presumably binding the lipid substrate leukotriene A4, and an Arg residue, possibly involved in the activation of the GSH thiol. Here we used a homology model, based on the LTC4S structure, and site-directed mutagenesis to identify Arg-126 as a key catalytic residue in mPGES-1. Our data are consistent with the notion that MAPEG members share a structurally similar binding site to accommodate a horseshoe-shaped GSH involved in different catalytic reactions.
EXPERIMENTAL PROCEDURES
Materials—The I.M.A.G.E. clones were obtained from the Medical Research Council Geneservice (Cambridge, UK). Enzymes, Escherichia coli TOP10 cells, and pPICZA were from Invitrogen. His6-pSP19T7LT was a kind gift from Dr. Sipra Saha. Anti-mPGES-1 antiserum was purchased from Cayman Chemical (Ann Arbor, MI). Platinum Pfx polymerase was from Invitrogen, and Pfu polymerase was from Promega (Madison, WI). PGH2 (purity in excess of 95%) was purchased from Larodane Fine Chemicals (Malmö, Sweden).
Cloning and Plasmid Construction—The human mPGES-1 gene was subcloned into pPICZA from the His6-pSP19T7LT vector. The coding part of the gene, supplemented with an N-terminal in frame sequence encoding a His6 tag, was PCR-amplified using the primer pair 5′-CGACAACTTGAGAAGATCAAAATGTCTCACCATCATCACCACCATCCTGCCCACAGCCTGG and 5′-GCAAGACCGGTCTTCTCTCACAGGTGGCGGGCCGCTTCCCAGAGGATCTGCAGAGCCAT and Pfx polymerase. Also, the linearized vector was PCR-amplified. For this Pfu polymerase, the primers 5′-GAGAAGACCGGTCTTGC and 5′-TTTGATCTTCTCAAGTTGTCG were used. The PCR products were co-transformed into CaCl2-competent E. coli TOP10 cells, utilizing the endogenous recombinase activity of E. coli to recombine the fragments (13). The protein coding part of the resulting expression vector, pPICZ-hisMPGES, was sequenced for verification.
Mutagenesis—Selected amino acids in pPICZ-hisMPGES were mutated using the QuikChange site-directed mutagenesis kit from Stratagene (La Jolla, CA).
Protein Expression and Microsome Preparation—Human recombinant mPGES-1 was overexpressed in Pichia pastoris. For most experiments, the strain KM71H was used, but strain X-33 also worked. The expression vector was transformed into competent P. pastoris cells using the Pichia EasyComp Transformation kit (Invitrogen). Recombinant cells were cultivated in baffled flasks in 2.5 liters of minimal yeast medium with glycerol (Invitrogen) at 27 °C. When A600 reached 10–12, the cells were resuspended in 0.5 liters of minimal yeast medium with 0.5% methanol. After another 72 h, cells were harvested by centrifugation (2,500 × g, 7 min) and resuspended in 15 mm Tris-HCl, pH 7.8, 0.25 m sucrose, and 1 mm GSH. Cells were homogenized with glass beads (0.5 mm), and the slurry was filtered through nylon filters (180 μm; Millipore) and centrifuged (5,000 × g, 10 min). The supernatant was ultracentrifuged (100,000 × g, 65 min), and microsomes were prepared from the pellet by homogenization in 20 mm Tris-HCl, pH 7.8, in a glass homogenizer. The microsome suspension was kept in aliquots at -20 °C. Prior to incubations with GSH analogues, the microsomes were washed from residual GSH: 100 μl of microsomes were mixed with 1 ml of 0.1 m Tris-HCl, pH 7.8, kept on ice for 15 min, and ultracentrifuged (100,000 × g, 30 min); this procedure was repeated 3-fold, and the final pellet was homogenized in 100 μl of buffer.
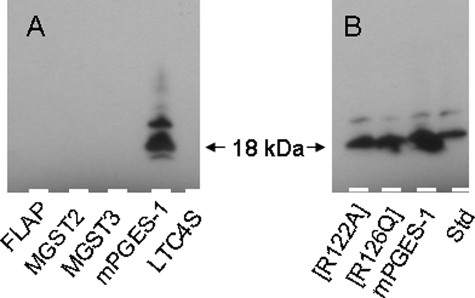
Expression of both nonmutated and mutated mPGES-1 was confirmed by Western blot analysis (Fig. 1) using a commercial antiserum (Cayman 160140) specific for mPGES-1. We could not observe any cross-reactivity with other recombinant MAPEG proteins produced in P. pastoris.
FIGURE 1.
Western blot of MAPEG proteins and mPGES-1. Shown is Western blot analysis of microsomes prepared from P. pastoris membranes in which the indicated proteins were overexpressed. A, various MAPEG proteins. Microsomes were diluted 20-fold, and 4 μl was loaded per lane. B, nonmutated and mutated mPGES-1 (∼3, 3, and 5 μg of total protein/lane). Std, 20 ng of purified mPGES-1.
Western Blot—The proteins were separated by SDS-PAGE and transferred to polyvinylidene difluoride membranes using a Pharmacia Phast system. Antiserum against mPGES-1 (catalog number 160140; Cayman) and horseradish peroxidase-linked anti-rabbit IgG (catalog number NA934; Amersham Biosciences) were used together with an ECL Plus detection kit (Amersham Biosciences) to visualize the proteins.
Enzyme Activity Assay—Microsomal suspensions (1.5 μl) were incubated on ice for 5 min with 10 μm PGH2 and 2.5 mm GSH in 300 μl of 0.1 m potassium phosphate buffer, pH 7.4. The reaction was terminated by the addition of 1.2 ml of 20 mm FeCl2 and 50 mm citric acid, pH 2–3. A solution of [3,3,4,4-2H4]PGE2 (1 μg) and [3,3,4,4-2H4]PGF2α (1 μg) in 200 μl of ethanol was added, and the mixture was applied to a 1-ml Chromabond C18 column for solid phase extraction. Material eluted with 80% methanol was esterified by treatment with diazomethane and trimethylsilylated by treatment with trimethylchlorosilane/hexamethyldisilazane/pyridine (2:1:2, v/v/v). Aliquots of the derivatized material were subjected to gas chromatography-mass spectrometry using a Hewlett-Packard model 5970B mass selective detector connected to a Hewlett-Packard model 5890 gas chromatograph equipped with a 5% phenylmethylsilicone capillary column (12 m, 0.33-μm film thickness). Helium was used as the carrier gas, and the column temperature was increased from 120 to 300 °C at a rate of 10 °C/min. The mass spectrometer was operated in the selected ion monitoring mode using the ions m/z 439 and 443 (unlabeled and deuterated PGE2, respectively) and m/z 423 and 427 (unlabeled and deuterated PGF2α, respectively). The amounts of PGE2 and PGF2α were calculated in the usual manner using ion intensity ratios and the appropriate standard curves. Unreacted PGH2 was decomposed into 12-hydroxy-heptadecatrienoic acid and malondialdehyde by FeCl2 in the stop solution.
Homology Modeling—To obtain the mPGES-1 model structure, kindly provided by Professor Pär Nordlund, structure-based sequence alignment of MAPEG members and homology modeling were carried out as described (12, 14). PGH2 was manually fitted in the active site of this model using the computer program O.
RESULTS AND DISCUSSION
Microsomal PGES-1 is critical for production of PGE2 during inflammatory reactions and has attracted considerable attention as a drug target. Since this enzyme is an integral membrane protein, information on structure-activity relationships has been scarce and essentially limited to a projection map obtained by two-dimensional crystallography, demonstrating a trimeric quaternary structure (15). However, a rapid and significant progress in this area was recently achieved when the crystal structures of MGST-1, 5-lipoxygenase-activating protein, and LTC4S were reported (9–12). In particular, the LTC4S structures allowed a detailed view of the enzyme's active center and revealed a hydrophobic cleft that accommodates the lipid substrate and a deeper pocket for GSH bound in a horseshoe-shaped conformation. In addition, an Arg residue seemed to be positioned for activation of the GSH thiol and formation of a thiolate anion.
A Model of mPGES-1 Identifies Arg-126 as a Potential Catalytic Residue—Based on these new three-dimensional structures of MAPEG members, a structure-based sequence alignment was performed (12), from which a model of mPGES-1 with bound GSH could be generated (14). When compared with the structure of LTC4S, the lipid binding crevice appears wider (Fig. 2), as one would expect from the chemistry of PGH2 as compared with leukotriene A4. The co-substrate GSH is bound in a horseshoe conformation, and Arg-126 has a position compatible with a role in catalysis (Fig. 2). In the primary structure, mPGES-1 contains two additional amino aids in the stretch Tyr-117 to Arg-126, as compared with LTC4S. This difference between mPGES-1 and LTC4S has been disregarded in the model, since the two residues do not appear to have any impact on the α-helical structure carrying Arg-126.
FIGURE 2.
Model structure of the active site of mPGES-1 and comparison with LTC4S. A, co-substrate GSH in its binding pocket in the x-ray structure of LTC4S (right) and in the homology model of mPGES-1 (left). B, estimated distance between the free thiol group of bound GSH and residue Arg-126 in mPGES-1. This figure is adapted from Ref. 14.
Mutation of Arg-126 Strongly Reduces the Isomerase Activity of mPGES-1—Arg-126 in human mPGES-1 was exchanged for an Ala or Gln residue by site-directed mutagenesis. As a control, a neighboring Arg-122 was also mutated into an Ala or Gln residue, respectively. Microsome preparations from yeast cells expressing nonmutated and mutated enzyme were assayed for PGE2 synthase activity. Nonmutated mPGES-1 converted almost all added PGH2 (10 μm) to PGE2 when incubated on ice for 5 min. However, when R126A or R126Q mPGES-1 were incubated with PGH2, a pronounced loss (85–95%; n = 6) of conversion into PGE2 was observed (Fig. 3A). In contrast, R122A and R122Q mPGES-1 retained the activity measured for wild type enzyme.
FIGURE 3.
PGE2 synthase activity of nonmutated and mutated mPGES-1. PGE2 production when microsomes (∼70 μg of total protein) were incubated in 0.3 ml with 10 μm PGH2 and 2.5 mm GSH for 5 min on ice (A) or microsomes (∼70 μg), after extensive washing to remove residual GSH, were incubated with 10 μm PGH2 and 2.5 mm GSH or the indicated GSH analogue (B). Each value is a mean of duplicates from one typical experiment (n = 3(A) and n = 2(B)).
Exchange of GSH for Its Ethylester Is Compatible with Catalysis, whereas Substitution for a Thiolester Does Not Allow any PGE2 Synthase Activity—To gain further mechanistic information, we exchanged GSH for various analogues during the incubation with PGH2 (Fig. 3B). S-methyl-GSH did not result in any PGE2 formation, indicating that the thiol group of GSH is crucial for the reaction. Incubations with GSH-ethylester (esterified at the Cα of Glu), on the other hand, did still result in some PGE2 production (about 28% of nonmutated), which suggests that minor changes of the GSH binding conformation, at least near its Glu carboxylate, still allows a productive position of the GSH free thiol. These data support the proposed role of the thiolate anion as the attacking species of GSH during conversion of PGH2 to PGE2.
Mutation of Arg-126 Allows a Reductase Activity That Converts PGH2 into PGF2α—Interestingly, when microsome preparations containing mutants of Arg-126 were incubated with PGH2, we detected a reductase activity rather than an isomerase activity, such that PGF2α appeared as the major product in the gas chromatography-mass spectrometry analysis (Figs. 4 and 5A). Both R126A and R126Q mPGES-1 converted PGH2 to PGF2α, a prostanoid with a distinct spectrum of bioactions, different from those of PGE2. This reductase activity also required the presence of GSH and its free thiol, as assessed from incubations with GSH and S-methyl-GSH (Fig. 5B).
FIGURE 4.
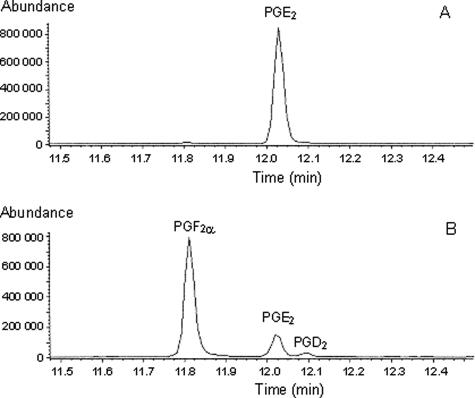
Gas chromatography-mass spectrometry profiling of prostaglandins formed from PGH2 by wild type and mutated mPGES-1. Incubations of wild type (A) and R126Q mPGES-1 (B) with 10 μm PGH2 were carried out at 0 °C for 30 min, and reaction products were isolated by extraction with ethyl acetate. The material obtained was methyl-esterified (diazomethane), trimethylsilylated, and subjected to gas chromatography-mass spectrometry analysis, as described under “Experimental Procedures.” Selected ions typical for PGE2, PGD2, and PGF2α were monitored. The absence of other prostaglandin derivatives was verified by analyses run in the full scan mode (m/z 50–600).
FIGURE 5.
PGF2α synthase activity of nonmutated and mutated mPGES-1. Shown is PGF2α production when microsomes were incubated with 10 μm PGH2 and 2.5 mm GSH (A) or microsomes, after extensive washing to remove residual GSH, were incubated with 10 μm PGH2 and 2.5 mm GSH or the indicated GSH analogue (B). Each value is a mean of duplicates from one typical experiment (n = 3(A) and n = 2(B)).
Putative Mechanism of mPGES-1—Conversion of PGH2 into PGE2 by mPGES-1 is GSH-dependent and involves cleavage of the endoperoxide O–O bond and elimination of the C-9 hydrogen as a proton (16). The mechanism of this conversion may involve glutathione thiolate as a base in the deprotonation step. Alternatively, it has been suggested that glutathione thiolate attacks the C-9 carbon to produce a thiohemiketal intermediate, which spontaneously rearranges to PGE2 (17). A third possibility would involve attack by glutathione thiolate on the C-9 endoperoxide oxygen, forming a mixed sulfide. Subsequent deprotonation at C-9 and cleavage of the O–S bond would lead to the formation of PGE2 and regeneration of the glutathione thiolate (Fig. 6). It is noteworthy that the mixed sulfide intermediate of this mechanism would be prone to reduction (e.g. it could be attacked by a second glutathione thiolate to produce PGF2α and oxidized glutathione) (Fig. 6). In light of our present data, it seems possible that an important function of the active site amino acid structure of mPGES-1 is to prevent such reduction from taking place and that the Arg-126 mutants are defective in this respect and therefore produce PGF2α rather than PGE2. This notion is also supported by the structural implications of replacing Arg-126, as illustrated in Fig. 7. Both R126A and R126Q mPGES-1 leave more space around the GSH thiol group, thereby allowing for a second GSH molecule to approach in agreement with the observed PGF2α formation. In this context, it may be mentioned that enzymatic conversion of PGH2 into PGF2α by a glutathione-dependent microsomal endoperoxide reductase has been reported (18), as well as the formation of PGE2 and PGF2α by glutathione S-transferase-promoted conversions of PGH2 (19).
FIGURE 6.
Suggested mechanisms for PGE2 synthase (top) and PGF2α synthase (bottom) activities of mPGES-1 and Arg-126 mutants, respectively. GS-, glutathione thiolate. The identity of the amino acid(s) represented by Enz-B is presently unclear. Arg-126 is a candidate, but since R126A and R126Q mPGES-1 retained PGE synthase activity and also exhibited a GSH-dependent PGF synthase activity, other residues must also be involved in the activation of the GSH thiol.
FIGURE 7.
Model of structural consequences caused by mutations of Arg-126 in mPGES-1. In the homology model, Arg-126 (green, left) has been replaced by Ala (orange, middle) or Gln (violet, right). In addition to GSH (white), the lipid substrate PGH2 (blue) has been manually fitted into the putative active site.
From the crystal structure of LTC4S in complex with GSH, it was suggested that Arg-104 is critical for the activation of the GSH thiol and stabilization of the thiolate anion (12). Since our data show that mutation of the corresponding Arg-126 is detrimental for the PGE2 synthase activity, it is tempting to speculate a similar role for this residue. However, the mutants R126A and R126Q mPGES-1 actually retained a very small, albeit detectable, isomerase activity and also exhibited a significant, GSH-dependent reductase activity, which is likely also to involve formation of a thiolate anion (Figs. 3 and 4). Hence, our data suggest that the primary role of Arg-126 is not activation of the GSH thiol. Further structure-function studies with crystallography and mutagenesis will hopefully clarify the precise role of Arg-126 and other residues involved in the molecular mechanism of mPGES-1 catalysis.
During the course of these investigations, Jegerschöld et al. (20) reported a structure of mPGES-1 at 3.5 Å resolution, obtained by electron crystallography. GSH was found to bind in a U-shaped conformation, and, interestingly, the authors could not find an access path for PGH2 to the bound GSH molecule, indicating that the protein was in a closed conformation and that dynamic changes, involving helices 1 and 4, occur at the active site during binding and turnover of the lipid substrate. A putative structure of an open conformation of the active site, with a critical Arg residue (Arg-126), was obtained through modeling against the crystal structure of human LTC4S. Hence, the mPGES-1 structure presented by Jegerschöld et al. (20) agrees very well with the results of our own investigation and suggests that further mechanistic studies must take into account the consequences of protein dynamics.
Acknowledgments
We thank Gunvor Hamberg for technical assistance.
The work was supported by Swedish Research Council Grants 10350, 20854, Linnéus Grant CERIC, and 2001-2553, the CIDaT consortium (Vinnova), the Söderberg Foundation, and European Commission FP6 Grant LSHM-CT-2004-005033. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.

Author's Choice—Final version full access.
Footnotes
The abbreviations used are: PGE2, prostaglandin E2 (11α,15S-dihydroxy-9-keto-prosta-5,13-dienoic acid); PGE, prostaglandin E; PGH2, prostaglandin H2 ((15S)-hydroxy-9α,11α-peroxidoprosta-5,13-dienoic acid); PGF2α, prostaglandin F2α (9α,11α,15S-trihydroxyprosta-5,13-dienoic acid); mPGES-1, microsomal PGE synthase-1; LTC4S, leukotriene C4 synthase; MAPEG, membrane-associated proteins in eicosanoid and glutathione metabolism.
References
- 1.Samuelsson, B., Goldyne, M., Granström, E., Hamberg, M., Hammarström, S., and Malmsten, C. (1978) Annu. Rev. Biochem. 47 997-1029 [DOI] [PubMed] [Google Scholar]
- 2.Kobayashi, T., and Narumiya, S. (2002) Prostaglandins Other Lipid Mediat. 68 557-573 [DOI] [PubMed] [Google Scholar]
- 3.Tanioka, T., Nakatani, Y., Semmyo, N., Murakami, M., and Kudo, I. (2000) J. Biol. Chem. 275 32775-32782 [DOI] [PubMed] [Google Scholar]
- 4.Jakobsson, P. J., Thoren, S., Morgenstern, R., and Samuelsson, B. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 7220-7225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Watanabe, K., Kurihara, K., and Suzuki, T. (1999) Biochim. Biophys. Acta 1439 406-414 [DOI] [PubMed] [Google Scholar]
- 6.Murakami, M., Naraba, H., Tanioka, T., Semmyo, N., Nakatani, Y., Kojima, F., Ikeda, T., Fueki, M., Ueno, A., Oh, S., and Kudo, I. (2000) J. Biol. Chem. 275 32783-32792 [DOI] [PubMed] [Google Scholar]
- 7.Samuelsson, B., Morgenstern, R., and Jakobsson, P. J. (2007) Pharmacol. Rev. 59 207-224 [DOI] [PubMed] [Google Scholar]
- 8.Jakobsson, P. J., Morgenstern, R., Mancini, J., Ford-Hutchinson, A., and Persson, B. (2000) Am. J. Respir. Crit. Care Med. 161 S20-S24 [DOI] [PubMed] [Google Scholar]
- 9.Holm, P. J., Bhakat, P., Jegerschöld, C., Gyobu, N., Mitsuoka, K., Fujiyoshi, Y., Morgenstern, R., and Hebert, H. (2006) J. Mol. Biol. 360 934-945 [DOI] [PubMed] [Google Scholar]
- 10.Ferguson, A. D., McKeever, B. M., Xu, S., Wisniewski, D., Miller, D. K., Yamin, T. T., Spencer, R. H., Chu, L., Ujjainwalla, F., Cunningham, B. R., Evans, J. F., and Becker, J. W. (2007) Science 317 510-512 [DOI] [PubMed] [Google Scholar]
- 11.Ago, H., Kanaoka, Y., Irikura, D., Lam, B. K., Shimamura, T., Austen, K. F., and Miyano, M. (2007) Nature 448 609-612 [DOI] [PubMed] [Google Scholar]
- 12.Martinez Molina, D., Wetterholm, A., Kohl, A., McCarthy, A. A., Niegowski, D., Ohlson, E., Hammarberg, T., Eshaghi, S., Haeggström, J.Z., and Nordlund, P. (2007) Nature 448 613-616 [DOI] [PubMed] [Google Scholar]
- 13.Bubeck, P., Winkler, M., and Bautsch, W. (1993) Nucleic Acids Res. 21 3601-3602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martinez Molina, D., Eshaghi, S., and Nordlund, P. (2008) Curr. Opin. Struct. Biol. 4 442-449 [DOI] [PubMed] [Google Scholar]
- 15.Thoren, S., Weinander, R., Saha, S., Jegerschöld, C., Pettersson, P. L., Samuelsson, B., Hebert, H., Hamberg, M., Morgenstern, R., and Jakobsson, P. J. (2003) J. Biol. Chem. 278 22199-22209 [DOI] [PubMed] [Google Scholar]
- 16.Miyamoto, T., Yamamoto, S., and Hayaishi, O. (1974) Proc. Natl. Acad. Sci. U. S. A. 71 3645-3648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lands, W., Lee, R., and Smith, W. (1971) Ann. N. Y. Acad. Sci. 180 107-122 [DOI] [PubMed] [Google Scholar]
- 18.Burgess, J. R., and Reddy, C. C. (1997) Biochem. Mol. Biol. Int. 41 217-226 [DOI] [PubMed] [Google Scholar]
- 19.Burgess, J. R., Chow, N. W., Reddy, C. C., and Tu, C. P. (1989) Biochem. Biophys. Res. Commun. 158 497-502 [DOI] [PubMed] [Google Scholar]
- 20.Jegerschöld, C., Pawelzik, S. C., Purhonen, P., Bhakat, P., Gheorghe, K. R., Gyobu, N., Mitsuoka, K., Morgenstern, R., Jakobsson, P. J., and Hebert, H. (2008) Proc. Natl. Acad. Sci. U. S. A. 105 11110-11115 [DOI] [PMC free article] [PubMed] [Google Scholar]