Abstract
Increased nuclear protein O-linked β-N-acetylglucosamine glycosylation (O-GlcNAcylation) mediated by high glucose treatment or the hyperglycemia of diabetes mellitus contributes to cardiac myocyte dysfunction. However, whether mitochondrial proteins in cardiac myocytes are also submitted to O-GlcNAcylation or excessive O-GlcNAcylation alters mitochondrial function is unknown. In this study, we determined if mitochondrial proteins are O-GlcNAcylated and explored if increased O-GlcNAcylation is linked to high glucose-induced mitochondrial dysfunction in neonatal rat cardiomyocytes. By immunoprecipitation, we found that several mitochondrial proteins, which are members of complexes of the respiratory chain, like subunit NDUFA9 of complex I, subunits core 1 and core 2 of complex III, and the mitochondrial DNA-encoded subunit I of complex IV (COX I) are O-GlcNAcylated. By mass spectrometry, we identified that serine 156 on NDUFA9 is O-GlcNAcylated. High glucose treatment (30 mm glucose) increases mitochondrial protein O-GlcNAcylation, including those of COX I and NDUFA9 which are reduced by expression of O-GlcNAcase (GCA). Increased mitochondrial O-GlcNAcylation is associated with impaired activity of complex I, III, and IV in addition to lower mitochondrial calcium and cellular ATP content. When the excessive O-GlcNAc modification is reduced by GCA expression, mitochondrial function improves; the activity of complex I, III, and IV increases to normal and mitochondrial calcium and cellular ATP content are returned to control levels. From these results we conclude that specific mitochondrial proteins of cardiac myocytes are O-GlcNAcylated and that exposure to high glucose increases mitochondrial protein O-GlcNAcylation, which in turn contributes to impaired mitochondrial function.
Previous reports have demonstrated that exposure to high glucose impairs calcium flux in cardiac myocytes (1, 2) and that this change is mediated through increased O-linked β-N-acetylglucosamine glycosylation (O-GlcNAcylation) of the nuclear transcription factor Sp1 resulting in decreased sarco (endo) plasmic reticulum Ca2+-ATPase (SERCA2a) gene expression (2). Furthermore, we found that decreasing excessive protein O-GlcNAcylation in the diabetic mouse heart by expression of a transgene encoding β-N-acetyl-d-glucosaminidase (O-GlcNAcase or GCA, (3, 4)) improved calcium handling and contractile function (5). O-GlcNAcylation is an abundant post-translational protein modification occurring on serine and threonine residues (6-8). O-GlcNAcylation and phosphorylation are reversible and highly dynamic processes (9) influencing overlapping signaling cascades. More than 500 proteins have been identified to be O-GlcNAcylated (10) including transcriptional factors (11, 12), kinases (13), cell cycle molecules (14), and nuclear pore proteins (15).
In contrast to enzymatic glycosylation of nuclear and cytosolic proteins, O-GlcNAcylation of mitochondrial proteins has only been explored in a limited fashion. In a recent report a shorter splice variant of O-GlcNAc transferase (OGT)2 has been identified which exhibits preferred mitochondrial localization (16). Recombinant mOGT has O-GlcNAc transferase activity and substrate specificity, which is different from that of OGT (17). Despite these facts, it is unclear if mitochondrial proteins in cardiac myocytes are modified by O-GlcNAcylation, if high glucose exposure of cells modifies the extent of O-GlcNAcylation or if increased mitochondrial protein O-GlcNAcylation alters mitochondrial function. Further exploring the occurrence of O-GlcNAcylation on mitochondrial proteins in cardiac myocytes and determining its influence on mitochondrial function has relevance for diabetes induced cardiovascular disease.
Cardiovascular complications are the leading cause of diabetes-related morbidity and mortality (18) and include a diabetes mediated cardiomyopathy which occurs in the absence of coronary artery disease or hypertension (19, 20). Several factors have been implicated in the development of diabetic cardiomyopathy, including metabolic, biochemical, and ultrastructural changes within the cardiac myocyte (21-23). In particular, because of the primary role of mitochondria in ATP production, impairment of mitochondrial respiratory function is a key contributing factor to decreased contractile function of the diabetic heart. The mechanisms, which contribute to the diabetes-induced decrease in mitochondrial function, have been incompletely explored.
Our results show for the first time that specific mitochondrial proteins of cardiac myocytes are O-GlcNAcylated and that exposure to high glucose further enhances the O-GlcNAc modification. A mitochondrial DNA-encoded protein representing subunit I of complex IV is also O-GlcNAcylated, making it likely that O-GlcNAcylation is a modification occurring in mitochondria. In addition, we found that the high glucose induced excessive mitochondrial protein O-GlcNAcylation impairs mitochondrial respiratory complex function, which can be markedly improved by diminishing O-GlcNAcylation through O-GlcNAcase expression. This report demonstrates therefore that the repertoire of O-GlcNAcylation modified proteins includes mitochondrial proteins from cardiac myocytes and that excessive mitochondrial protein O-GlcNAcylation has significant negative effects on mitochondrial function.
EXPERIMENTAL PROCEDURES
Isolation and Culture of Neonatal Rat Cardiac Myocytes (NCM)—Primary cultures of neonatal rat cardiomyocytes were prepared as described previously (24). Cells (3 × 106 per 10-cm plate) were plated onto gelatin-coated culture dishes. Plating medium consisted of 4.25:1 Dulbecco's modified Eagle's medium: M199, 10% horse serum, 5% fetal bovine serum, 1% penicillin/streptomycin/fungizone, and 5.5 mmol of l,d-glucose. Cells were allowed to adhere to the plates for at least 24 h before different treatments. Cells were cultured in maintenance medium (4.5:1 Dulbecco's modified Eagle's medium: M199, 2% fetal bovine serum, 1% penicillin/streptomycin/fungizone) supplemented with either normal glucose (5.5 mm plus 25 mm mannitol) or high glucose (30 mm) concentrations, referred to as NG and HG, respectively. Cells exposed to high glucose were also treated with adenovirus (Adv) encoding human O-GlcNAcase (Adv-GCA) or vector control (Adv-CTR) (2), and are referred to as HGGCA or HGCTR. Cells were infected at a multiplicity of infection of 20/cell for both viruses. The culture medium was changed daily for 2 days until the cells were harvested.
Mitochondrial Preparation and Subfractionation—Fresh mouse hearts were washed and minced, then homogenized in 10 volumes (v/w) of buffer H (in mmol/l: 5 HEPES, 250 mannitol, 0.1 EDTA and 0.1% (w/v) bovine serum albumin, pH 7.5) with a Polytron homogenizer at medium speed, 3 times for 5 s. After centrifugation at 300 × g for 10 min the supernatant was collected, and the pellet was resuspended with the same volume of buffer H, then the homogenization and centrifugation procedure was repeated. Combined supernatants were centrifuged at 8000 × g for 15 min, and the supernatant was collected as the cytosolic protein fraction. The pellet was collected as the mitochondrial fraction. Mitochondrial extracts were further purified with a percoll density gradient as described previously (25, 26). Essentially, mitochondrial extract was dissolved in buffer H and loaded in 20 ml of buffer C (in mmol/l: 225 mannitol, 1 EGTA, 25 HEPES, pH 7.5, 0.1% (w/v) bovine serum albumin, 30% (v/v) percoll), and centrifuged at 95,000 × g for 1 h in a Beckman type 60Ti rotor. The lower condensed yellow-brown band was transferred into a new tube, and 4 volumes of buffer H was added, mixed, and spun at 6,500 × g for 10 min. After one wash with buffer H, the purified mitochondria pellet was used for Western blot and immunoprecipitation experiments. Mitochondrial inner membrane isolation was performed using a procedure described before (25).
To isolate protein fractions from treated NCM, the cells were harvested and homogenized with a Polytron homogenizer at full speed, 3 times for 5 s in mitochondria buffer (in mmol/l: 10 Tris, 250 sucrose, 2 EDTA, 0.025 PUGNAc, and protease inhibitor mixture (1:1,000, Sigma), pH 7.5). Centrifugation was performed at 1,500 × g for 15 min at 4 °C, and the pellet containing the nuclear fraction and cellular debris was discarded. The supernatant was centrifuged at 8,000 × g for 15 min. The supernatant was collected as cytosolic protein fraction. The pellet containing mitochondria was washed with mitochondria buffer and spun at 10,000 × g for 15 min. The washing step was repeated two times. The pellet was resuspended in lysis buffer (in mmol/l: 20 Tris, 20 NaCl, 0.1 EDTA, 0.025 PUGNAc, 1% Triton X-100, 1:1000 diluted protease inhibitor mixture, pH 7.0). After centrifugation at 16,000 × g for 10 min, the supernatant was collected for Western blot analysis. Protein concentrations were determined by Bradford assay.
Western Blotting and Immunoprecipitation—Cytosolic or mitochondrial samples were loaded on NuPAGE 4-12% Bis-Tris gels (Invitrogen). Separated proteins were transferred to nitrocellulose membranes. The membranes were blocked in 5% nonfat milk. RL2 (1:1000) antibody was used to detect O-GlcNAc modification (Anti-O-GlcNAc). Anti-GCA was a gift from Dr. Gerald W. Hart (Johns Hopkins University School of Medicine, Baltimore, Maryland). Anti-SERCA2a (1:2000, Santa Cruz Biotechnology) and anti-complex IV subunit I (Anti-COX I, 1:1000, Mitosciences) were also used as primary antibodies. Anti-mouse IgG-horseradish peroxidase (HRP)-conjugated (1:2000, Amersham) and anti-goat IgG-HRP-conjugated (1:5000, Pierce) were used as secondary antibodies.
Purified mitochondrial pellets isolated from mice hearts were resuspended in Nonidet P-40 buffer (in mmol/l: 20 Tris, 150 NaCl, 0.025 PUGNAc, 1% Nonidet P-40, pH 7.4). To precipitate subunit I of complex IV, around 3 mg of mitochondrial lysate was incubated with 3 μg anti-COX I antibody on a rotator for 3 h at 4 °C, then 30 μl of protein A/G beads (Santa Cruz Biotechnology) were added and incubated for 2 h at 4 °C. After incubation, the beads were spun down and washed with Nonidet P-40 buffer 3 times; then 20 μl of 2× sample buffer was added followed by boiling for 5 min. Beads were spun down, and 15 μl of supernatants were loaded on 4-12% gradient Bis-Tris gels for Western analysis. In addition, 25 μg of purified mitochondria protein was used as input control, and 25 μg of mitochondrial inner membrane was also loaded. Immunoprecipitation of subunits NDUFA9, core1, and core 2 was performed similarly but instead of using protein A/G beads, the immunoprecipitation kit ExactaCruz™ E (Santa Cruz Biotechnology) was used to diminish nonspecific IgG heavy and light chain bands. RL2 antibody was used to detect O-GlcNAc-modified proteins. Antibodies for subunits NDUFA9, core1, and core2 were purchased from Mitosciences, Oregon.
To analyze the O-GlcNAc modification level on COX I, NCMs were treated in NG, HG, HGCTR, and HGGCA as described above for 2 days. Following treatment, the cells were lysed in Nonidet P-40 buffer and COX I was immunoprecipitated with anti-COX I antibody. Immunoprecipitates were analyzed by Western blot using RL2 antibody first, then the membrane was stripped and reblotted with anti-COX I antibody. Band density was quantified with ImageJ software. Relative O-GlcNAc/COX I ratios for each COX I band were calculated.
Detection of O-GlcNAcylated Mitochondrial Proteins with Wheat Germ Agglutinin-conjugated (WGA) Beads—WGA beads were used to pull-down the O-GlcNAcylated mitochondrial proteins with the same procedure described (27). 100 μg of cell lysate proteins in 200 μl of lysis buffer as described above were used in each reaction. After incubation with wheat germ agglutinin-conjugated beads, the beads were spun down at 5000 rpm for 5 min, and the supernatant was collected. The pellet containing beads and pull-down proteins was washed three times with lysate buffer, and then mixed with 20 μl of 2× sample buffer, boiled for 5 min, and analyzed by Western blot. The same volume of total cell lysate and unbound proteins in the supernatant were also analyzed in the same Western blot. The experiment was repeated three times with three independent NCM preparations and treatments.
Mapping O-GlcNAc Modification Sites on Mitochondrial Proteins with ß-Elimination followed by Michael Addition with Dithiothreitol (BEMAD) and Tandem Mass Spectrometry—Immunoprecipitated mitochondrial protein NDUFA9 from mouse hearts was in-gel trypsin-digested, followed by performic acid oxidation, and then treated by alkaline phosphatase for 4 h at 37 °C. The peptides were treated with BEMAD and O-GlcNAc-modified peptides were enriched with thiol affinity chromatography performed sequentially as described previously (28, 29) The peptides were analyzed by reverse phase chromatography prior to mass spectrometry analysis using the following method. Nano-electrospray capillary column tips were made in house using a P-100 laser puller (Sutter Instruments). The columns were packed with Zorbax SB-C18 stationary phase (Agilent) purchased in bulk (5-μm particles, 15-cm length, 75-μm inner diameter). The reverse phase gradient separation was performed using water and acetonitrile (0.1% formic acid) as the mobile phases. The gradient starts at 5% acetonitrile and is ramped to 8% acetonitrile over 10 min. The acetonitrile is ramped to 35% acetonitrile over 20 min, then increased to 90% acetonitrile for another 20 min and maintained for another 10 min prior to re-equilibration to 5% acetonitrile. The data-dependent MS/MS data were obtained on the LTQ linear ion trap mass spectrometer using a home built nanoelectrospray source at 2 KV at the tip. The instrument was used in data-dependent MS/MS mode. One MS spectrum was followed by 4 MS/MS scans on the most abundant ions after the application of the dynamic exclusion list. Protein identification and the assignment of the covalent attachment of DTT on serine was performed using Mascot (Matrix Science, London, UK, version 2.1.04) with threshold scores set at the 95% confidence level and <1% false positive rate.
Spectrophotometric Assay to Measure Complex I Activity—After NCM were treated with different glucose concentrations and Adv-GCA or Adv-CTR, the complex I activity was spectrophotometrically measured according to the method described (30).
Polarographic Assay to Measure Complex III and Complex IV Activity—The activity of complex III and complex IV of the mitochondrial respiration chain was measured according to a previously described protocol (31, 32) with modification. After treatment described above, NCM were trypsinized and permeabilized by incubation with digitonin (10 μg/ml) in medium A (in mmol/l: 20 HEPES, 250 sucrose, 10 MgCl2, pH 7.1) for 10 min. Protein concentration in each sample was measured. Samples with equal amounts of protein were used for complex function measurement. Oxygen consumption was measured in permeabilized NCM using a Clark-type oxygen electrode (Yellow Spring Co.) in a thermoregulated chamber set at 37 °C. Complex III and complex IV activity were defined as the rate of oxygen consumption in the presence of the specific substrates (succinate and glycerol-3-phosphate for complex III, and ascorbate and N,N,N′,N′-tetramethyl-p-phenylenediamine for complex IV). Antimycin for complex III, and sodium cyanide for complex IV were used as inhibitors.
Cellular ATP Content Measurement—The cellular ATP concentration was measured by bioluminescent somatic cell assay kit (Sigma) according to the manufacturer's protocol with some modifications. After treatments, NCM were washed twice with phosphate-buffered saline. Cellular ATP was released by addition of equal volumes of phosphate-buffered saline and releasing reagent. After incubation at room temperature for 5 min, with gentle mixing, the supernatants were collected and used for ATP measurement. The cells in plates were lysed in buffer B as mentioned above and collected. After centrifugation at 14,000 rpm for 10 min, the supernatant was collected, and protein concentration was measured to normalize cellular ATP content.
Mitochondrial Ca2+ Measurement—Mitochondrial Ca2+ concentration was measured using ratiometric mitochondrial pericam as described before by us (33).
GCA Assay—O-GlcNAcase activity was measured as described by Dong and Hart (3) using p-nitrophenyl-N-acetyl-β-d-glucosaminide (pNP-O-GlcNAc) as substrate. Assays were performed in 100 μl of 50 mm sodium cacodylate, pH 7.0, 2 mm pNP-O-GlcNAc with 0.5 mg/ml of bovine serum albumin at 37 °C for 30 min. The reactions were stopped with addition of 0.9 ml of 0.5 m sodium carbonate, and absorbance was measured at 405 nm. Enzyme activity is expressed as the consumption of pNP-O-GlcNAc in mmol/min.
Data Collection and Statistical Analysis—Experimental treatments on neonatal rat myocytes were evaluated from at least three different isolated preparations and experiments. All data are presented as mean ± S.D. or mean ± S.E. as described. One-way ANOVA with Bonferroni's multiple comparison test among groups was calculated with GraphPad Prism 3.0 software package. p < 0.05 was considered to be statistically significant.
RESULTS
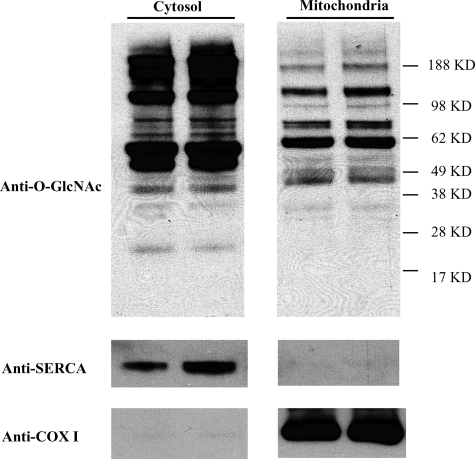
Mitochondrial Proteins Are O-GlcNAcylated—To determine if O-GlcNAc modification occurs on mitochondrial proteins mitochondrial and cytosolic proteins were isolated from wild-type mouse hearts and submitted to Western blotting analysis with the O-GlcNAc directed RL2 antibody (Fig. 1). O-GlcNAc modification was observed on several cytosolic and mitochondrial proteins. Compared with cytosolic proteins, mitochondrial proteins have overall less O-GlcNAc modification. Some mitochondrial proteins show marked O-GlcNAc modification, like the protein band in the 62-kDa molecular weight range. To ensure that the O-GlcNAc-modified proteins are derived from mitochondria and not from contamination by proteins from the cytosolic fraction, membranes were stripped and reblotted with an antibody against SERCA2a. As shown in Fig. 1, SERCA2a is not present in mitochondrial preparations. In contrast, the mitochondrial DNA encoded protein COX I has a strong and exclusive presence in the mitochondria-derived protein preparation.
FIGURE 1.
Overall protein O-GlcNAcylation in cytosolic and mitochondrial fractions. Cytosolic and mitochondrial proteins from mouse hearts were separated and analyzed by Western blot. 25 μg of cytosolic or 40 μg of mitochondrial proteins for each lane were loaded. The membrane was probed with anti-O-GlcNAc antibody (RL2), stripped and reblotted with anti-SERCA or an antibody for respiratory chain complex IV subunit I (anti-COX I).
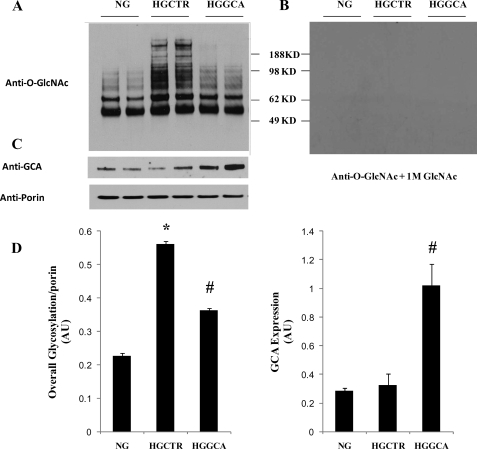
Mitochondrial Protein O-GlcNAc Modification Is Changed in Cardiac Myocytes by High Glucose Exposure and O-GlcNAcase Expression—NCM are exposed to normal glucose (5 mm glucose plus 25 mm mannitol to balance osmotic pressure) or high glucose (30 mm). In NCM exposed to normal glucose mitochondrial protein O-GlcNAcylation was similar to that observed in mitochondrial proteins isolated from wild-type mouse hearts (Fig. 2A). Exposure of NCM to high glucose with control adenovirus increased O-GlcNAcylation (Fig. 2, A and D). Our prior studies have shown that expression of an adenoviral vector-encoded GCA transgene in NCM markedly decreased O-GlcNAcylation of nuclear proteins (2). Here we tested if increasing GCA protein levels could also reduce the excessive mitochondrial protein O-GlcNAcylation occurring in NCM exposed to high glucose. The results show that mitochondrial O-GlcNAc modification is indeed markedly diminished by GCA expression. The same blots were stripped and incubated with same concentration of anti-O-GlcNAc antibody plus 1 m N-acetyl glucosamine (34), the blank Western blots prove the specificity of antibody to GlcNAc epitope (Fig. 2B). GCA expression was observed in the mitochondria and was increased 3-fold after exposure to Adv-GCA (Fig. 2, C and D). Anti-porin blots were done to check the protein loading (Fig. 2C). Relative overall mitochondrial protein O-GlcNAcylation from four experiments was analyzed with ImageJ software. The total O-GlcNAcylation signal was normalized with porin.
FIGURE 2.
Overall mitochondrial O-GlcNAcylation level increased with high glucose and was reduced with GCA overexpression. NCM were treated with normal glucose (NG), control adenovirus Adv-CTR (HGCTR) or Adv-GCA (HGGCA) for 48 h, then mitochondrial proteins were extracted separated on a 4-12% Bis-Tris gel. 40 μg of mitochondrial protein for each lane were loaded. The membrane was probed with anti-O-GlcNAc antibody (A), then stripped and reblotted with anti-O-GlcNAc antibody plus 1 m GlcNAc (B). Same blots were tested with anti-O-GlcNAcase and anti-porin antibody to ensure equal loading (C). Graphs showing summarized data of relative overall mitochondrial protein O-GlcNAcylation and GCA expression. Signal was normalized with porin (D). Results are presented as mean ± S.E., *, p < 0.05 versus NG; #, p < 0.05 versus HG, n = 4.
Reduction of Mitochondrial Protein O-GlcNAcylation by O-GlcNAcase Expression Is Accompanied by Improved Mitochondrial NADH Ubiquinone Oxidoreductase (Complex I), Ubiquinone Dehydrogenase (Complex III), and Cytochrome c Oxidase (Complex IV) Function Impaired by High Glucose—To evaluate if increased mitochondrial protein O-GlcNAcylation by high glucose exposure influences mitochondrial function, we determined mitochondrial respiratory complex function. The mitochondrial respiratory chain (RC) consists of five multi-subunit complexes which transport electrons and produce ATP. We determined the activities of complex I, III, IV, and V activities in NCM exposed to normal and high glucose.
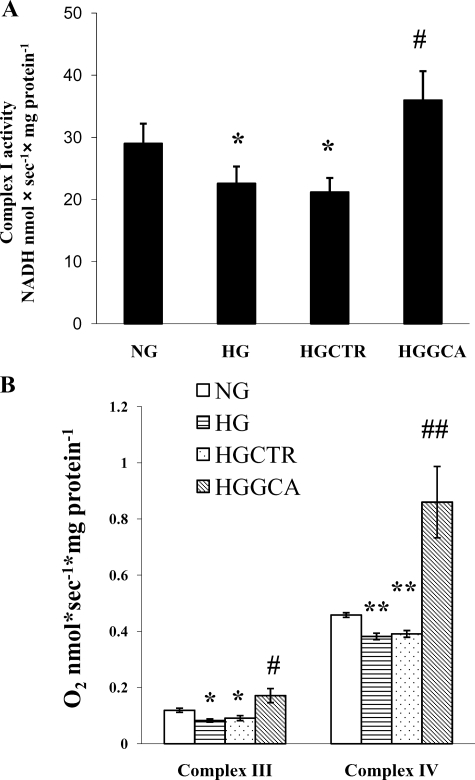
The activity of complex I is measured using a spectrometric method (30). As shown in Fig. 3A, high glucose treatment (HG) decreases complex I activity by 22.2% in comparison to normal glucose treatment (NG) (HG; 22.55 ± 2.77 versus NG; 28.99 ± 3.24, mean ± S.D., p < 0.05, n = 3). GCA expression in high glucose-treated cells (HGGCA) returned complex I activity to the control level (HGGCA; 35.96 ± 4.68 versus NG; 28.99 ± 3.24, mean ± S.D., p > 0.05, n = 3). Infection with a control adenovirus expressing no transgene had no effect on complex I activity (HGCTR; 21.17 ± 2.3; versus HG; 22.55 ± 2.8, mean ± S.D., p > 0.05, n = 3).
FIGURE 3.
The activities of mitochondrial respiratory chain complex I, III, and IV were impaired by high glucose treatment and increased by expression of O-GlcNAcase. NCM treatment conditions and abbreviations are the same as shown in Fig. 2. A, enzymatic activity of complex I. *, p < 0.05 for NG versus HG; #, p < 0.05 for HG versus HGGCA. The means and standard deviations are obtained from three independent experiments. B, activities of complex III and complex IV measured by polarographic assay. Open bars represent activities from cells in NG; light horizontal bars represent activities obtained from cells treated with HG; light downward diagonal bars represent activities obtained from cells treated with HGGCA. For complex III, *, p < 0.05 for HG versus NG; #, p < 0.05 for NG versus HGGCA. For complex IV, **, p < 0.05 for HG versus NG and ##, p < 0.001 for NG versus HGGCA. The means and S.D. are obtained from four independent experiments.
Similar results were found for complex III and IV activity measured by a polarographic method (31, 32). As shown in Fig. 3B, For complex III, high glucose exposure of NCM decreases the activity by 31% compared with normal glucose control (HG; 0.082 ± 0.006 versus NG; 0.119 ± 0.075, mean ± S.D., p > 0.05, n = 4). High glucose exposure decrease the activity of complex IV by 16% compared with normal glucose control (HG; 0.383 ± 0.012 versus NG; 0.458 ± 0.008, mean ± S.D., p > 0.05, n = 4). Surprisingly, GCA expression in NCM exposed to high glucose (HGGCA) significantly increased complex III and IV activities above control values: the activity of complex III increased by 44% compared with normal glucose control (HGGCA; 0.171 ± 0.025 versus NG; 0.119 ± 0.075, mean ± S.D., p > 0.05, n = 4), and the activity of complex IV increased by 88% compared with normal glucose (HGGCA; 0.860 ± 0.127 versus NG; 0.458 ± 0.008, mean ± S.D., p < 0.001, n = 4). Treatment with a control virus had no effect on complex III or IV activity (for complex III, HGCTR; 0.091 ± 0.009 versus HG; 0.082 ± 0.006, p > 0.05 and for complex IV, HGCTR; 0.391 ± 0.012 versus HG; 0.383 ± 0.012, mean ± S.D., p > 0.05, n = 4). Complex V activity was assessed by a spectrometric method, but did not change with high glucose exposure of NCM (data not shown).
O-GlcNAcase Expression Increases Cellular ATP Content—The high glucose treatment induced a decrease in mitochondrial complex activity, shown above, and is linked to diminished cellular ATP levels (Fig. 4). HG reduced whole cellular ATP content by 29% compared with normal (NG) (NG; 1 ± 0.12 versus HG; 0.79 ± 0.10, mean ± S.D., p < 0.05, n = 4). Expression of O-GlcNAcase increased cellular ATP levels in high glucose-treated cells (HGGCA; 1.23 ± 0.08 versus 0.79 ± 0.10, mean ± S.D., p < 0.05, n = 4). Treatment with the control virus did not change cellular ATP levels (HGCTR; 0.80 ± 0.09 versus HG; 0.79 ± 0.10, mean ± S.D., p < 0.05, n = 4), but treatment with GCA virus increased ATP content more than control virus treatment (HGGCA; 1.23 ± 0.08 versus NG; 1.12 ± 0.05, mean ± S.D., p < 0.05, n = 4).
FIGURE 4.
Cellular ATP content was decreased by high glucose and increased by expression of O-GlcNAcase. *, p < 0.05 for NG versus HG; #, p < 0.05 for HG versus HGGCA. The means and S.D. are obtained from four independent experiments.
Mitochondrial Calcium Levels Are Decreased by High Glucose Exposure and Normalized by O-GlcNAcase Expression—NCM were incubated in 5 mm or 25 mm glucose for 48 h followed by infection with Adv expressing the mitochondria directed green fluorescence based calcium indicator mPericam (35) and Adv expressing GCA or Adv not expressing a transgene. Adv-based transgene expression was maintained for an additional 48 h. At the end of this time period mitochondrial Ca2+ levels were determined. High glucose treatment decreased mitochondrial Ca2+ levels by 70%. Reduction of O-GlcNAcylation by O-GlcNAcase expression, in NCM treated with high glucose, increased mitochondrial calcium toward control levels (Fig. 5).
FIGURE 5.
Mitochondrial Ca2+ concentration in NCM treated with high glucose and O-GlcNAcase. High glucose decreased mitochondrial Ca2+ levels and expression of O-GlcNAcase increased mitochondrial Ca2+ toward control levels. Cells were maintained in 5 mm (NGCTR) or 25 mm (HGCTR, HGGCA) glucose for 48 h and then infected with Adv-mito-pericam and either Adv-CTR or Adv-GCA for 48 h. *, p < 0.05 for NGCTR versus HGCTR; **, p < 0.01 for HGGCA versus HGCTR. Values are mean ± S.E., n = 15 cells/group.
Identification of O-GlcNAcylated Protein Members in Specific Mitochondrial Respiratory Chain Complexes—To identify O-GlcNAc-modified protein members in mitochondrial RC complexes, specific protein members are immunoprecipitated using specific antibodies directed at them, followed by SDS-PAGE and Western blotting with the anti-O-GlcNAc antibody RL2. The same membranes are stripped and reblotted with subunit-specific antibodies to confirm the identity of the proteins. As shown in Fig. 6A, complex IV subunit I (COX I) was identified as an O-GlcNAcylated protein in the IP sample. COX I also appears as a lighter band in protein samples prepared from a whole lysate of purified mitochondria and protein samples derived from the inner mitochondrial membrane. As mentioned before COX I is encoded by mitochondrial DNA and resides in the mitochondrial inner membrane. Mitochondrial proteins encoded by genomic DNA such as subunit NDUFA9 of complex I, core 1, and core 2 of complex III are also O-GlcNAcylated (Fig. 6, B-D). Competition experiments with 1 m GlcNAc excluded nonspecific binding of the RL2 antibody (Fig. 6, B-D).
FIGURE 6.
Identified O-GlcNAc-modified subunits in respiratory chain complexes with immunoprecipitation and mass spectrometry. A, complex IV subunit I was immunoprecipitated from purified mitochondria. The membrane was first blotted with anti-O-GlcNAc antibody, stripped, and reblotted with antibody for complex IV subunit I (anti-COX I). Purified mitochondrial lysate (Mito) and inner mitochondrial membrane (IM) samples were also loaded. The arrow points to the band of O-GlcNAcylated COX I. Similar procedures were done for RC chain complex I subunit NDUFA9 (B), complex III core1 subunit (C), and core2 subunit (D). Input represents purified mitochondrial lysate used for immunoprecipitation. IP represents immunoprecipitated sample. E, mapping O-GlcNAc modification site (Ser-156) on NDUFA9 by mass spectrometry. O-GlcNAc is replaced by DTT through BEMAD. LC-MS/MS is used to detect the DTT-modified residue. MS/MS spectrum from collision-induced dissociation of a precursor ion selected at 2416.1554 (m/z 806.6 [M +3H]3+). F, summary of fragmentation patterns. Detected b and y ions are marked by rectangles. Serine 19 of the peptide fragment presented a mass of 223.1 because of DTT addition indicating the O-GlcNAc modification. The O-GlcNAc site is assigned to serine 156 of NDUFA9.
In addition to the immunoblotting method with antibody to detect O-GlcNAc-modified mitochondrial proteins (Figs. 1, 2, and 6, A-D), we performed site mapping of the O-GlcNAc sites on mitochondrial respiratory chain protein NDUFA9 with BEMAD treatment and MS/MS analysis as described under “Experimental Procedures.” Because of lability of O-GlcNAc modification for direct analysis by mass spectrometry, BEMAD treatment was used. With this technique O-GlcNAc-modified sites are replaced with DTT molecules, which are more stable. A differential mass increase of 136.2 Da to Ser or Thr indicated DTT-modified sites. Mapping of DTT sites assign the original O-GlcNAc modification sites on proteins. Using this method, we identified one O-GlcNAc-modified site (Ser-156) on NDUFA9 (Fig. 6, E and F, position 19 in the peptide).
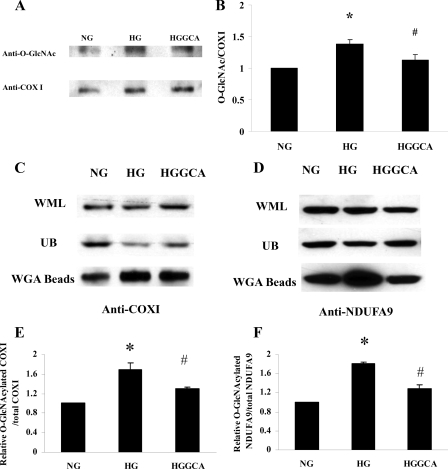
We also explored if high glucose treatment of NCM increases O-GlcNAcylation of COX I, which we found to be the case (Fig. 7A). In addition, we determined if increased O-GlcNAc modification of COX I induced by high glucose treatment can be diminished by GCA expression. For these experiments COX I protein is immunoprecipitated, followed by Western blot analysis with the anti-O-GlcNAc antibody (RL2). Densitometric analysis of COX I on Western blots probed with RL2 or anti-COX I antibodies was then performed. To quantitate O-GlcNAc modified COX I each sample is normalized for total COX I protein (Fig. 7B). The result shows that levels of O-GlcNAc modified COX I increased by 38.5% in high glucose-treated NCM compared with control (p < 0.01). The increased O-GlcNAcylation of COX I is returned to normal by O-GlcNAcase expression. Control adenovirus has no effect (data not shown).
FIGURE 7.
O-GlcNAcylation of COX I and NDUFA9 are increased with high glucose and decreased with expression of O-GlcNAcase. NCM were treated with normal glucose (NG), high glucose (HG), or Adv-GCA (HGGCA) for 48 h. After treatment, COX I was isolated by immunoprecipitation with anti-COX I antibody followed by Western blot analysis. The same membrane was first blotted with anti-O-GlcNAc antibody, and then stripped and reblotted with anti-COX I antibody. A, O-GlcNAcylated COX I (upper bands) and total COXI(bottom). B, relative COX I O-GlcNAcylation level. Values are mean ± S.D. obtained from three independent experiments. *, p < 0.001 for NG versus HG; #, p < 0.01 for HGGCA versus HG. Whole mitochondrial protein lysate (WML) were incubated with WGA beads to pull-down O-GlcNAcylated proteins. After the incubation, the beads were spun down, and the supernatants were proteins without O-GlcNAc modification (unbound, UB), the proteins on beads were O-GlcNAcylated proteins (WGA). Western blots with anti-COX I (C) and anti-NDUFA9 (D) were used to analyze the proteins in these three parts. Relative ratios of O-GlcNAcylated COX I to total COX I (E) or of O-GlcNAcylated NDUFA9 to total NDUFA9 (F) were compared. *, p < 0.05 for NG versus HG; #, p < 0.05 for HGGCA versus HG (E and F). Values are mean ± S.D. obtained from three independent experiments.
WGA binds to terminal GlcNAc residues and WGA-conjugated agarose beads bind to O-GlcNAc-modified proteins with high affinity, which has been used to pull-down O-GlcNAc-modified proteins (27, 36). Mitochondrial protein lysate from NCM treated with normal or high glucose or Adv-GCA was incubated with WGA beads to isolate the O-GlcNAc modified mitochondrial proteins. Western blots were used to measure the proportion of O-GlcNAc modified COXI or NDUFA9. As shown in Fig. 7, C and D, given that nearly the same amount of total COXI or NDUFA9 proteins under different treatments, in HG, more proteins are O-GlcNAcylated and bound to WGA beads and less proteins unbound and left in supernatant. GCA treatment reduced the proteins bound to WGA beads which suggested less O-GlcNAcylation on COXI or NDUFA9. Densitometric analysis of COX I or NDUFA9 on Western blots was performed. Fig. 7, E and F are relative proportions of O-GlcNAcylated COXI in total COXI (E), and O-GlcNAcylated NDUFA9 in total NDUFA9 (F). There are statistically significant differences between HG and NG and between HG and HGGCA (p < 0.05). The means and standard deviations were obtained from three independent experiments. Fig. 7, C and D show representative Western blots.
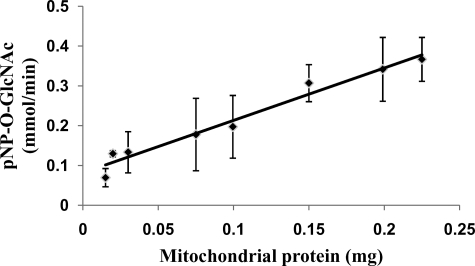
O-GlcNAcase Activity Is Detected in Mitochondria—Given that mitochondrial localized OGT has been reported (16) and mitochondrial GCA expression was detected here (Fig. 2C) we investigated if there is O-GlcNAcase activity within mitochondria. Therefore, we measured the O-GlcNAcase activity of purified mitochondria from mouse heart. The results show that there is O-GlcNAcase activity in mitochondria (Fig. 8).
FIGURE 8.
O-GlcNAcase activity in mitochondria from mouse heart. O-GlcNAcase activity assay was performed on different mitochondrial protein concentrations. Enzyme activity is expressed as the consumption of pNP-O-GlcNAc in mmol/min. Values are presented as mean ± S.D., n = 5.
DISCUSSION
In this report we demonstrate that in cardiac myocytes mitochondrial proteins are modified by O-GlcNAcylation and that the O-GlcNAcylation status influences mitochondrial function. We show that many mitochondrial proteins are O-GlcNAcylated including mitochondrial proteins of the respiratory chain complexes like NDUFA9 of complex I, core 1, and core 2 of complex III, and subunit I of complex IV (COX I). For the first time, we found the O-GlcNAc modification site on mitochondrial protein: serine-156 of NDUFA9 (Fig. 6, E and F), which directly proves that O-GlcNAc modification occurs in mitochondrial proteins. Moreover, In addition to these identified protein members of mitochondrial complexes, other currently unidentified mitochondrial proteins exhibit significant O-GlcNAcylation (Figs. 1 and 2). Several protein bands with marked O-GlcNAcylation are identifiable on Western blots of percoll gradient-purified mitochondrial proteins when probed with a specific antibody directed at O-linked N-acetylglucosamine (RL2 antibody). It should also be noted that the level of overall O-GlcNAc modification of cytosolic proteins is much higher than that of mitochondrial proteins (Fig. 1).
The O-GlcNAcylation of specific mitochondrial proteins presents, according to our findings, an enzymatic process occurring inside mitochondria. The O-GlcNAc-modified COX I subunit of complex IV is encoded by mitochondrial DNA making it highly likely that its O-GlcNAcylation occurs in the mitochondrial matrix. It should also be noted that a mitochondria directed shorter splice variant of OGT has been described, which has different substrate specificity from that of full-length OGT. In addition, we observed O-GlcNAcase expression and activity in the mitochondria. These results together reinforce the idea that O-GlcNAcylation takes place in the mitochondria as well. Prior studies in other cell types have reported the occurrence of mitochondrial protein O-GlcNAcylation, including on malate dehydrogenase (37). Here, we also identified that malate dehydrogenase, mitochondrial precursor (mouse) is O-GlcNAc modified at serine 51 (not shown). Aside from a few reports, mitochondrial protein O-GlcNAcylation was largely unexplored and its functional impact is unclear.
In this work, using mass spectrometry approach, we found that NDUFA9 can be GlcNAc modified at serine 156. This finding provided a direct and unambiguous evidence for O-GlcNAc modification on mitochondrial proteins. Unlike mapping phosphorylation sites, conventional mass spectrometry techniques are hardly successful to identify O-GlcNAc sites because the glycosylation suppresses signal and β-GlcNAc is extremely labile. BEMAD treatment uses base to beta-eliminate O-GlcNAc and replace it with a more stable DTT tag which can be enriched by thiol affinity chromatography. The DTT tag increases Ser or Thr mass by 136.2 daltons. Combining BEMAD and LC-MS/MS analysis it is possible to map O-GlcNAc sites on proteins (29, 34).
Prior reports have shown that exposure of cells to high glucose results in increased O-GlcNAcylation of nuclear and cytosolic proteins (2, 5) but its impact on O-GlcNAcylation of mitochondrial proteins has not been explored. We observed that high glucose exposure of cardiac myocytes results in increased O-GlcNAcylation of mitochondrial proteins. In particular, we analyzed the mitochondrial DNA encoded COX I protein of complex IV and nuclear DNA encoded NDUFA9 protein of complex I. We found that these proteins exhibit increased O-GlcNAcylation after treatment with high glucose. The high glucose-mediated increase in O-GlcNAcylation of mitochondrial proteins was accompanied by decreased mitochondrial complex function in addition to diminished cellular ATP levels and decreased mitochondrial calcium levels. Expression of a viral vector-encoded O-GlcNAcase transgene in cardiac myocytes exposed to high glucose markedly reduces mitochondrial protein O-GlcNAcylation. Reduction of mitochondrial protein O-GlcNAcylation was accompanied by increases in complex activity, ATP levels and normalization of mitochondrial calcium levels despite persistent exposure to high glucose. These findings imply that excessive O-GlcNAcylation of mitochondrial proteins has detrimental effects on mitochondrial function.
Currently, the molecular mechanisms by which protein O-GlcNAcylation influences protein function are actively explored (10). O-GlcNAcylation of proteins is a dynamic process resembling phosphorylation, and it is responsive to numerous stimuli. Post-translational protein modifications on OH- groups of serine and threonine by phosphorylation and O-GlcNAcylation can have reciprocal effects on protein function. Examples include modification of the same site by phosphorylation or O-GlcNAcylation; O-GlcNAc prevents the phosphorylation induced modification and its effect on protein function. In addition, competitive and alternate modification of adjacent sites occurs, as well as other interactions (10). Overall modification of protein function by O-GlcNAc derives from the interplay between protein modification by phosphorylation and O-GlcNAc. Our findings show a strong correlation between increased O-GlcNAcylation of specific proteins, for example, COX I protein of complex IV and decreased activity of complex IV. The detailed molecular mechanisms and a potential interplay between O-GlcNAcylation induced alterations of specific phosphorylation events remains currently unclear.
Hyperglycemia is a hallmark of diabetes mellitus and a diabetes mediated propensity for cardiovascular disease, including the development of heart failure, is well recognized (38). Decreased mitochondrial function in the diabetic cardiac myocyte is also well established (39) but the underlying mechanisms have not been completely explored. Based on our findings of increased O-GlcNAcylation of mitochondrial proteins in cardiac myocytes exposed to high glucose, one may speculate that excessive O-GlcNAcylation of mitochondrial proteins also occurs in the diabetic heart. In summary, our study demonstrated that O-GlcNAc modification occurs on mitochondrial proteins of cardiac myocytes and that excessive O-GlcNAc modification caused by high glucose contributes to impaired mitochondrial function.
This work was supported, in whole or in part, by NHLBI, National Institutes of Health Grant HL-66917 (to W. H. D.) and by Grant Number P60 MD00220, from the San Diego EXPORT Center, National Center of Minority Health and Health Disparities, National Institutes of Health. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: OGT, O-GlcNAc transferase; NCM, neonatal rat cardiac myocytes; GCA, O-GlcNAcase; pNP-O-GlcNAc, p-nitrophenyl-N-acetyl-β-d-glucosaminide; HG, high glucose; NG, normal glucose; DTT, dithiothreitol; WGA, wheat germ agglutinin.
References
- 1.Davidoff, A. J., and Ren, J. (1997) Am. J. Physiol. 272 H159-H167 [DOI] [PubMed] [Google Scholar]
- 2.Clark, R. J., McDonough, P. M., Swanson, E., Trost, S. U., Suzuki, M., Fukuda, M., and Dillmann, W. H. (2003) J. Biol. Chem. 278 44230-44237 [DOI] [PubMed] [Google Scholar]
- 3.Dong, D. L., and Hart, G. W. (1994) J. Biol. Chem. 269 19321-19330 [PubMed] [Google Scholar]
- 4.Gao, Y., Wells, L., Comer, F. I., Parker, G. J., and Hart, G. W. (2001) J. Biol. Chem. 276 9838-9845 [DOI] [PubMed] [Google Scholar]
- 5.Hu, Y., Belke, D., Suarez, J., Swanson, E., Clark, R., Hoshijima, M., and Dillmann, W. H. (2005) Circ. Res. 96 1006-1013 [DOI] [PubMed] [Google Scholar]
- 6.Vosseller, K., Sakabe, K., Wells, L., and Hart, G. W. (2002) Curr. Opin. Chem. Biol. 6 851-857 [DOI] [PubMed] [Google Scholar]
- 7.Wells, L., and Hart, G. W. (2003) FEBS Lett. 546 154-158 [DOI] [PubMed] [Google Scholar]
- 8.Zachara, N. E., and Hart, G. W. (2006) Biochim. Biophys. Acta 1761 599-617 [DOI] [PubMed] [Google Scholar]
- 9.Khidekel, N., Ficarro, S. B., Clark, P. M., Bryan, M. C., Swaney, D. L., Rexach, J. E., Sun, Y. E., Coon, J. J., Peters, E. C., and Hsieh-Wilson, L. C. (2007) Nat. Chem. Biol. 3 339-348 [DOI] [PubMed] [Google Scholar]
- 10.Hart, G. W., Housley, M. P., and Slawson, C. (2007) Nature 446 1017-1022 [DOI] [PubMed] [Google Scholar]
- 11.Gewinner, C., Hart, G., Zachara, N., Cole, R., Beisenherz-Huss, C., and Groner, B. (2004) J. Biol. Chem. 279 3563-3572 [DOI] [PubMed] [Google Scholar]
- 12.Roos, M. D., Su, K., Baker, J. R., and Kudlow, J. E. (1997) Mol. Cell. Biol. 17 6472-6480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parker, G. J., Lund, K. C., Taylor, R. P., and McClain, D. A. (2003) J. Biol. Chem. 278 10022-10027 [DOI] [PubMed] [Google Scholar]
- 14.Slawson, C., Shafii, S., Amburgey, J., and Potter, R. (2002) Biochim. Biophys. Acta 1573 121-129 [DOI] [PubMed] [Google Scholar]
- 15.Holt, G. D., and Hart, G. W. (1986) J. Biol. Chem. 261 8049-8057 [PubMed] [Google Scholar]
- 16.Love, D. C., Kochan, J., Cathey, R. L., Shin, S. H., and Hanover, J. A. (2003) J. Cell Sci. 116 647-654 [DOI] [PubMed] [Google Scholar]
- 17.Lazarus, B. D., Love, D. C., and Hanover, J. A. (2006) Glycobiology 16 415-421 [DOI] [PubMed] [Google Scholar]
- 18.Comer, F. I., Vosseller, K., Wells, L., Accavitti, M. A., and Hart, G. W. (2001) Anal. Biochem. 293 169-177 [DOI] [PubMed] [Google Scholar]
- 19.Holt, G., Snow, C., Senior, A., Haltiwanger, R., Gerace, L., and Hart, G. (1987) J. Cell Biol. 104 1157-1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Walgren, J. L., Vincent, T. S., Schey, K. L., and Buse, M. G. (2003) Am. J. Physiol. Endocrinol Metab 284 E424-E434 [DOI] [PubMed] [Google Scholar]
- 21.Belke, D. D., Larsen, T. S., Gibbs, E. M., and Severson, D. L. (2000) Am. J. Physiol. Endocrinol Metab 279 E1104-E1113 [DOI] [PubMed] [Google Scholar]
- 22.Bhimji, S., Godin, D. V., and McNeill, J. H. (1986) Acta Anat (Basel) 125 195-200 [DOI] [PubMed] [Google Scholar]
- 23.Rodrigues, B., Cam, M. C., and McNeill, J. H. (1998) Mol. Cell Biochem. 180 53-57 [PubMed] [Google Scholar]
- 24.Hartong, R., Villarreal, F. J., Giordano, F., Hilal-Dandan, R., McDonough, P. M., and Dillmann, W. H. (1996) J. Mol. Cell Cardiol. 28 2467-2477 [DOI] [PubMed] [Google Scholar]
- 25.Hovius, R., Lambrechts, H., Nicolay, K., and de Kruijff, B. (1990) Biochim. Biophys. Acta 1021 217-226 [DOI] [PubMed] [Google Scholar]
- 26.Turko, I. V., and Murad, F. (2003) J. Biol. Chem. 278 35844-35849 [DOI] [PubMed] [Google Scholar]
- 27.Gandy, J. C., Rountree, A. E., and Bijur, G. N. (2006) FEBS Lett. 580 3051-3058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Whelan, S. A., and Hart, G. W. (2006) Methods Enzymol. 415 113-133 [DOI] [PubMed] [Google Scholar]
- 29.Wells, L., Vosseller, K., Cole, R. N., Cronshaw, J. M., Matunis, M. J., and Hart, G. W. (2002) Mol. Cell Proteomics 1 791-804 [DOI] [PubMed] [Google Scholar]
- 30.Chretien, D., Benit, P., Chol, M., Lebon, S., Rotig, A., Munnich, A., and Rustin, P. (2003) Biochem. Biophys. Res. Commun. 301 222-224 [DOI] [PubMed] [Google Scholar]
- 31.Hofhaus, G., Shakeley, R. M., and Attardi, G. (1996) Methods Enzymol. 264 476-483 [DOI] [PubMed] [Google Scholar]
- 32.Hollander, J. M., Lin, K. M., Scott, B. T., and Dillmann, W. H. (2003) Free Radic Biol. Med 35 742-751 [DOI] [PubMed] [Google Scholar]
- 33.Belke, D. D., Swanson, E., Suarez, J., Scott, B. T., Stenbit, A. E., and Dillmann, W. H. (2007) Am. J. Physiol. Heart Circ. Physiol. 292 H1755-H1763 [DOI] [PubMed] [Google Scholar]
- 34.Wang, Z., Pandey, A., and Hart, G. W. (2007) Mol. Cell Proteomics 6 1365-1379 [DOI] [PubMed] [Google Scholar]
- 35.Nagai, T., Sawano, A., Park, E. S., and Miyawaki, A. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 3197-3202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roquemore, E. P., Chou, T. Y., and Hart, G. W. (1994) Methods Enzymol. 230 443-460 [DOI] [PubMed] [Google Scholar]
- 37.Cieniewski-Bernard, C., Bastide, B., Lefebvre, T., Lemoine, J., Mounier, Y., and Michalski, J. C. (2004) Mol. Cell Proteomics 3 577-585 [DOI] [PubMed] [Google Scholar]
- 38.Boudina, S., and Abel, E. D. (2007) Circulation 115 3213-3223 [DOI] [PubMed] [Google Scholar]
- 39.Tanaka, Y., Konno, N., and Kako, K. J. (1992) Cardiovasc Res. 26 409-414 [DOI] [PubMed] [Google Scholar]