Abstract
Pluripotent mesenchymal stem cells (MSCs) are bone marrow stromal progenitor cells that can differentiate into osteogenic, chondrogenic, adipogenic, and myogenic lineages. We previously demonstrated that bone morphogenetic protein (BMP) 9 is one of the most potent and yet least characterized BMPs that are able to induce osteogenic differentiation of MSCs both in vitro and in vivo. Here, we conducted gene expression-profiling analysis and identified that Hey1 of the hairy/Enhancer of split-related repressor protein basic helix-loop-helix family was among the most significantly up-regulated early targets in BMP9-stimulated MSCs. We demonstrated that Hey1 expression was up-regulated at the immediate early stage of BMP9-induced osteogenic differentiation. Chromatin immunoprecipitation analysis indicated that Hey1 may be a direct target of the BMP9-induced Smad signaling pathway. Silencing Hey1 expression diminished BMP9-induced osteogenic differentiation both in vitro and in vivo and led to chondrogenic differentiation. Likewise, constitutive Hey1 expression augmented BMP9-mediated bone matrix mineralization. Hey1 and Runx2 were shown to act synergistically in BMP9-induced osteogenic differentiation, and Runx2 expression significantly decreased in the absence of Hey1, suggesting that Runx2 may function downstream of Hey1. Accordingly, the defective osteogenic differentiation caused by Hey1 knockdown was rescued by exogenous Runx2 expression. Thus, our findings suggest that Hey1, through its interplay with Runx2, may play an important role in regulating BMP9-induced osteoblast lineage differentiation of MSCs.
Mesenchymal stem cells (MSCs)5 represent a very small fraction of the total population of nucleated cells in bone marrow (1) and are adherent multipotent marrow stromal cells (1-6). Although primarily located within the bone marrow compartment (5, 7, 8), MSCs have been isolated from periosteum, trabecular bone, adipose tissue, synovium, skeletal muscle, and deciduous teeth (9). As members of the transforming growth factor-β superfamily, BMPs play an important role in stem cell biology (10, 11) and regulate cell proliferation and differentiation during development (12, 13). Several BMPs have been shown to regulate osteoblast differentiation and subsequent bone formation (12-15). Genetic disruptions of BMPs result in various skeletal and extraskeletal abnormalities during development (14, 16). We have recently conducted a comprehensive analysis of the osteogenic activity of 14 human BMPs and demonstrated that BMP9 is one of the most potent BMPs promoting osteogenic differentiation of MSCs both in vitro and in vivo (17, 18). We also demonstrated that osteogenic BMPs regulate a distinct set of downstream targets in MSCs (6, 19-21).
BMP9 (also known as GDF2) was originally identified from fetal mouse liver cDNA libraries and is a relatively uncharacterized member of the BMP family. BMP9 is highly expressed in the developing mouse liver, and recombinant human BMP9 stimulates hepatocyte proliferation (22, 23). BMP9 has been shown to be a potent synergistic factor for hematopoietic progenitor-cell generation and colony formation and may play a role in the induction and maintenance of the neuronal cholinergic phenotype in the central nervous system (24). Although the recombinant human BMP9 protein was shown to exert little osteoinductive activity in vivo (22), we and others have demonstrated that exogenously expressed BMP9 is highly capable of inducing osteogenic differentiation (6, 17, 18, 25). However, the molecular mechanism behind BMP9-regulated MSC differentiation remains to be elucidated.
In this study, we investigated the mechanism underlying BMP9-regulated osteoblast lineage-specific differentiation of MSCs. Through gene expression-profiling analysis, we identified that Hey1 (also known as Hesr1, HRT1, CHF2, and HERP2), a transcription factor of the HERP family (26), is among the most significantly up-regulated targets in MSCs in response to BMP9. The HERP family of basic helix-loop-helix transcription factors is a direct target of the Notch signaling pathway (27), which is implicated in cell fate decision. Although Hey1 is up-regulated in early somitogenesis (26), the role of Hey1 in osteoblast differentiation is unclear. One study demonstrated that Hey1 is specifically up-regulated by BMP2 when MSCs differentiate to osteoblast lineage (28), whereas another study showed that Hey1 inhibited osteoblast maturation via interaction with Runx2, a known regulator of osteogenic differentiation (29). We sought to determine the functional role of Hey1 in BMP9-induced osteogenic differentiation.
We demonstrated that Hey1 is an important and direct target of BMP-9 signaling and is up-regulated at the immediate early stage of osteogenic differentiation. Silencing Hey1 expression reduced BMP9-stimulated osteogenic differentiation both in vitro and in vivo. Hey1 overexpression in MSCs increased BMP9-stimulated osteogenic differentiation in vitro, and enhanced mineralization of bone matrix in response to BMP9 in vivo. We further demonstrated that Hey1 and Runx2 act synergistically in BMP9-induced osteogenic differentiation and that exogenous expression of Runx2 provides a partial rescue of the Hey1 knockdown phenotype. Thus, our results suggest that Hey1 may play an important role in mediating BMP9-induced MSC differentiation.
EXPERIMENTAL PROCEDURES
Cell Culture and Chemicals—HEK293 and C3H10T1/2 lines were obtained from the ATCC (Manassas, VA) and maintained in complete Dulbecco's modified Eagle's medium and basal medium Eagle, respectively. Unless indicated, all chemicals were purchased from Sigma or Fisher Scientific.
Isolation of Mouse Embryo Fibroblasts—MEFs were isolated from post coitus day 13.5 mice, as previously described (30). Each embryo was dissected into 10-ml sterile phosphate-buffered saline, voided of its internal organs, and sheared through an 18-gauge syringe in the presence of 1 ml of 0.25% trypsin and 1 mm EDTA. After 15-min incubation with gentle shaking at 37 °C, Dulbecco's modified Eagle's medium with 10% fetal calf serum was added to inactivate trypsin. The cells were plated on 100-mm dishes and incubated for 24 h at 37 °C. Adherent cells were used as MEF cells. Aliquots were kept in a liquid nitrogen tank. All MEFs used in this study were within five passages.
Isolation of Total RNA—Subconfluent cells were seeded in 75-cm2 cell culture flasks in complete medium supplemented with 0.5% fetal calf serum and infected with a predetermined optimal titer of adenoviruses, such as AdBMP9 or AdGFP. At the indicated time after infection, total RNA was isolated using TRIzol reagents (Invitrogen), according to the manufacturer's instructions.
Microarray Analysis—Subconfluent C3H10T1/2 cells were maintained in basal medium Eagle containing 0.5% fetal calf serum and infected with AdBMP9 or AdGFP. At 30 h after infection, total RNA was isolated. The fully characterized RNA samples were used for target preparation and subjected to hybridizations to Affymetrix mouse gene chips 430A (containing ∼22,000 known genes and expressed sequence tags). The acquisition and initial quantitation of array images were performed using Affymetrix MAS 5.0 with the default parameters as previously described (19-21, 31). The acquired microarray raw data were further filtered and normalized to remove noise, whereas retaining true biological information by filtering out the genes with signal intensity in all samples <100 intensity units, and by removing the genes that received an “absent” call for all hybridizations. The clustering analysis was carried out by using the DNA-Chip Analyzer (dChip) software (51). Thresholds for selecting significant genes were set at a relative difference >2-fold, an absolute difference >100 signal intensity units, and a statistical difference at p < 0.05 (supplemental Table S1).
Establishment of Stable MSC Lines Expressing Hey1—We PCR-amplified and cloned the mouse Hey1 coding region into a retroviral vector that also conferred resistance to blasticidin, designated RV-Hey1. The cloning regions and PCR-amplified coding regions were verified by DNA sequencing. The RV-Hey1 vector was transfected into a retroviral packaging line (empty vector as a control), and the recombinant retrovirus was used to infect C3H10T1/2 cells followed by blasticidin selection. The resultant stable pooled clones were designated as C3H10-Hey1. Overexpression of Hey1 in this line was verified by qPCR. Construction details are available upon request.
Construction and Validation of siRNA Expression Vectors Targeting Mouse Hey1—We constructed and validated siRNA target sites (supplemental Table S2) for mouse Hey1 using our recently developed pSOS system (32). The selected target sites were first screened for their knockdown efficiency using our recently developed pSOS system (32). These vectors, RV-sim-Hey1, were transfected into a retroviral packing cell line, and the recombinant retrovirus was used to infect C3H10T1/2 cells followed by blasticidin selection. The resultant stable pooled clones were designated as C3H10-Hey1KD. The knockdown of Hey1 expression in this line was verified by qPCR. Construction details are available upon request.
Adenoviruses Expressing BMP9, Hey1, Runx2, and simHey1—Recombinant adenoviruses expressing BMP9 were generated as previously described (17, 18, 33). An analogous adenovirus expressing only GFP (AdGFP) was used as a control (34, 35). Adenoviruses expressing mouse Hey1 and Runx2 (AdHey1 and AdRunx2) were generated in the same fashion using the AdEasy technology as described (33, 36). The adenovirus expressing mouse Hey1 siRNA (Ad-simHey1) was generated using our recently developed pSES system (32). All PCR-amplified fragments and cloning junctions were verified by DNA sequencing.
qPCR Analysis—The qPCR was carried out as described (20, 21, 31). Ten micrograms of total RNA was used to generate cDNA templates by reverse transcription with hexamer and Superscript II reverse transcriptase (Invitrogen). The first strand cDNA products were further diluted 5- to 10-fold and used as qPCR templates. The qPCR primers (supplemental Table S2) were 18-mers, designed by using the Primer3 program, frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi, to amplify the 3′-end (∼120 bp) of the gene of interest. SYBR Green-based qPCR analysis was carried out using the Opticon DNA Engine (MJ Research). The specificity of each qPCR reaction was verified by melting curve analysis and further confirmed by resolving the PCR products on 1.5% agarose gels. pUC19, 5-fold serially diluted, was used as a standard. Duplicate reactions were carried out for each sample. All samples were normalized by the expression level of glyceraldehyde-3-phosphate dehydrogenase.
Measurement of ALP Activity—ALP activity was assessed by the colorimetric assay (using p-nitrophenyl phosphate as a substrate) and/or histochemical staining assay (using a mixture of 0.1 mg/ml napthol AS-MX phosphate and 0.6 mg/ml Fast Blue BB salt) as previously described (17-21, 31).
Stem Cell Implantation—The use and care of animals was approved by the Institutional Animal Care and Use Committee. Subconfluent C3H10T1/2 control line, C3H10-Hey1, and C3H10-Hey1KD cells were infected with AdBMP9, AdGFP, or co-infected with AdBMP9, AdRunx2, or AdHey1 for 15 h, and collected for subcutaneous injection (5 × 106 cells per injection) into the flanks of athymic nude (nu/nu) mice (four injections per group, 4- to 6-week-old, male, Harlan Sprague-Dawley). At 6 weeks after implantation, animals were sacrificed for MicroCT imaging, and the implantation sites were retrieved for histologic evaluation. A similar procedure was followed for the MEF implantation experiments, except that the MEFs were infected with Ad-simHey1 or AdHey1.
ChIP Analysis—Subconfluent C3H10T1/2 cells were infected with AdGFP or AdBMP9. At 30 h after infection, cells were cross-linked and subjected to ChIP analysis as previously described (31). Smad4 antibody, Smad1/5/8 antibody, or control IgG was used to pull down the protein-DNA complexes. The presence of Hey1 promoter sequence was detected by PCR using three pairs of primers corresponding to mouse Hey1 promoter region.
MicroCT Imaging Analysis—Animals were sacrificed at end-points and subjected to a high performance MicroCT imager that has a spatial resolution of 10-50 μm and a high contrast resolution. This unit provides quantitative measurements regarding the number and volume of each mass in each animal. MicroCT data were acquired and reconstructed into a three-dimensional image, and bone mass was quantified. To calculate the volume of each mass, the ImageJ program was used to determine the surface area of each planar slice of the MicroCT, the surface areas were summed, and volume was calculated as [(sum of the surface area of each slice) * 0.0543]. These volumes were averaged by dividing by the number of samples for each respective injection condition (n = 4).
Hematoxylin and Eosin, Trichrome, and Alcian Blue Staining—Retrieved tissues were fixed in 10% formalin overnight and embedded in paraffin. Serial sections of the embedded specimens were stained with hematoxylin and eosin. Trichrome and Alcian Blue stains were carried out as previously described (18, 37).
RESULTS
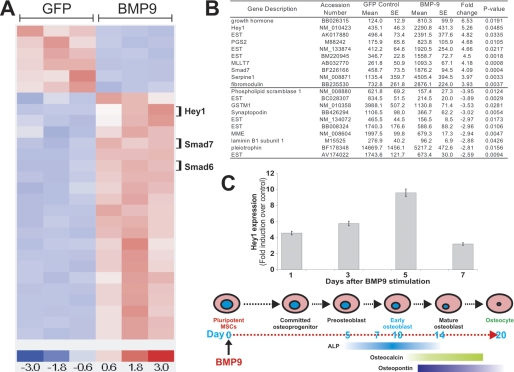
Hey1 Is Among the Most Significantly Up-regulated Targets of BMP9-induced Osteogenic Differentiation of MSCs—We recently evaluated the role of BMPs in regulating MSC differentiation and demonstrated that among the 14 BMPs, BMP9 is one of the most potent osteoinductive BMPs both in vitro and in vivo (17, 18). To gain further insight into the molecular basis of BMP9-mediated osteogenic differentiation of MSCs, we performed an expression profiling analysis of ∼22,000 genes in C3H10T1/2 MSCs stimulated with BMP9 or the GFP control. C3H10T1/2 cells are commonly used and well characterized MSCs (6, 20, 21). Using the dChip analysis under a high stringency (>2.0-fold and p < 0.05), we found that 40 genes were most significantly regulated in MSCs upon BMP9 stimulation; 30 genes were up-regulated and 10 were down-regulated (Fig. 1, A and B, and supplemental Table S1). This dChip analysis indicated that the Hey1 gene was among the most significantly up-regulated genes in response to BMP9 (Fig. 1B).
FIGURE 1.
Identification and verification of Hey1 as an early target of BMP9 signaling. A, dChip clustering analysis of the top 40 most significantly regulated genes by BMP9 in MSCs. B, list of the top 10 up-regulated and down-regulated target genes of BMP9 in MSCs. C, verification of BMP9-induced Hey1 expression in MSCs. Subconfluent C3H10T1/2 cells were infected with AdBMP9 or AdGFP. Total RNA was collected at the indicated times and subjected to qPCR analysis. The timeline of MSC differentiation leading to osteocytes is indicated at the bottom. All samples were normalized for glyceraldehyde-3-phosphate dehydrogenase expression. Reactions were done in duplicate. The fold induction was calculated by dividing the transcript level of BMP9-treated samples with that from GFP-treated samples.
To verify that Hey1 was induced by BMP9 in MSCs, C3H10T1/2 cells were infected with an adenovirus expressing BMP9 or GFP (AdBMP9 and AdGFP), and total RNA was collected at 1, 3, 5, and 7 days after infection. Using qPCR, we demonstrated that Hey1 expression was up-regulated by BMP9, and Hey1 expression level increased 5-fold, 6-fold, and 10-fold at days 1, 3, and 5, with a trend returning toward basal levels after 7 days. By day 7, BMP9-stimulated MSCs are usually committed to pre-osteoblast or early osteoblasts as indicated by the elevated ALP activity, a well established early osteogenic marker (Fig. 1C). Interestingly, although closely related, Hey2 gene expression was not significantly affected by BMP9 (data not shown). Thus, our results indicate that Hey1 expression is significantly induced at the immediate early stage of BMP9 stimulation in MSCs.
ChIP Analysis Indicates That Hey1 Is a Direct Target of BMP9-induced Smad Signaling—We next performed ChIP assay to determine whether Hey1 is a direct target of BMP9-induced Smad signaling pathway. We examined mouse Hey1 promoter sequence and found that several putative Smad binding sites were located in the-2.5-kb region. Accordingly, we designed three pairs of PCR oligonucleotides for ChIP analysis (Fig. 2A). For ChIP analysis, we infected subconfluent C3H10T1/2 with AdGFP or AdBMP9 for 30 h. Cells were cross-linked, and genomic DNA was sonicated, following immunoprecipitation with an anti-Smad4 antibody or IgG control. The retrieved genomic DNA fragments were subjected to PCR amplification using three pairs of primers, PP-1, PP-2, and PP-3. As shown in Fig. 2B, anti-Smad4 antibody, but not IgG, pulled down genomic fragments containing the mouse Hey1 promoter region. Smad4 binding to Hey1 promoter was seemingly BMP9-dependent, although there might be some basal binding of Smad4 to Hey1 promoter, as indicated by the primer pair PP-3. All ChIP assays had a similar input of genomic DNA (Fig. 2C). Similar results were obtained when Smad1/5/8 antibody was used for ChIP assays (Fig. 2D). These results strongly suggest that Hey1 may be a direct target of BMP9-induced Smad signaling in MSCs.
FIGURE 2.
ChIP analysis indicates that Hey1 is a direct target of BMP9-induced Smad signaling. A, schematic depiction of mouse Hey1 promoter region. The approximate locations of the three pairs of primers and their expected product sizes are indicated. B, subconfluent C3H10T1/2 cells were infected with AdGFP or AdBMP9 for 30 h. Cells were cross-linked. Genomic DNA was sonicated, following immunoprecipitation with anti-Smad4 or IgG. The retrieved genomic DNA was subjected to PCR using the three pairs of primers, PP-1, PP-2, and PP-3. The arrows indicate the locations of the expected products. C, control assays demonstrated that a similar amount of input materials was used for immunoprecipitation experiments. The arrows indicate the locations of the expected products. D, ChIP assays were carried out essentially the same as that described in B and C, except that Smad1/5/8 antibody was used for pulldown experiments. The arrows indicate the locations of the expected products. The ChIP analysis was performed in three independent experiments, and the representative results are shown.
Hey1 Knockdown Inhibits BMP9-stimulated Osteogenic Differentiation of MSCs—Recent studies reported conflicting results about the role of Hey1 in osteoblast differentiation (28, 29). Therefore, we sought to determine if Hey1 is critical to BMP9-mediated osteoblast differentiation via RNA interference-mediated knockdown of Hey1 expression. Hey1 silencing was achieved by establishing stable cell lines or a recombinant adenovirus that expressed siRNA targeting mouse Hey1. Using our recently developed pSOS retroviral vector system for simplified screening and validating of siRNA target sites (32), we evaluated the silencing efficiency of the four siRNA sites targeting the mouse Hey1 coding sequence. The top three sites were shown to effectively silence the GFP-Hey1 chimeric transcript (Fig. 3A). The retroviral vectors containing the top three siRNA sites were pooled and used to generate a retrovirus pool via co-transfection with the packaging plasmid. The resultant retrovirus was used to infect C3H10T1/2 cells and to establish stable cells using blasticidin selection. The pooled stable line was designated as C3H10-Hey1KD. As shown in Fig. 3B, Hey1 expression was reduced by 62% in the C3H10-Hey1KD cells.
FIGURE 3.
Inhibition of BMP9-induced osteogenic differentiation by RNA interference-mediated knockdown of Hey1 gene expression. A, selection and verification of siRNAs targeting mouse Hey1. The target sites were subcloned and tested using the pSOS system (32). The resultant vectors were transfected into 293 cells, and knockdown of chimeric GFP/Hey1 expression was recorded 5 days after transfection. B, verification of Hey1 knockdown in the stable line C3H10-Hey1KD. Total RNA was collected from subconfluent stable and control lines, and subjected to qPCR analysis using primers corresponding to the 3′-untranslated repeat of mouse Hey1 (pUC19 as a DNA quantitation standard). All samples were normalized for glyceraldehyde-3-phosphate dehydrogenase expression. C and D, inhibition of ALP activity by Hey1 gene knockdown in MSCs. RNA interference-mediated knockdown of the Hey1 expression reduced BMP9-stimulated ALP activity as compared with the control line (C3H10T1/2) demonstrated by colorimetric assays at days 7 and 10 (C) and by histochemical staining at day 10 (D). Representative results of three independent experiments are shown.
To evaluate the effect of Hey1 knockdown on BMP9-mediated osteogenic differentiation, we infected subconfluent C3H10-Hey1KD and C3H10T1/2 cells with either AdBMP9 or AdGFP and performed quantitative colorimetric analysis of ALP activity at days 7 and 10. BMP9 infection of C3H10T1/2 cells resulted in 18- and 67-fold increase in ALP activity at days 7 and 10, respectively (in comparison to the cells infected with AdGFP). However, knockdown of Hey1 expression led to a 94% and 88% decrease in ALP activity at days 7 and 10, respectively, in comparison to C3H10T1/2 cells (Fig. 3C). Similar results were obtained when ALP activity was evaluated qualitatively using a histochemical assay at day 10 (Fig. 3D). These results indicate that Hey1 is an important mediator of BMP9-induced osteogenic differentiation and that Hey1 knockdown may diminish BMP9-mediated bone formation.
Constitutive Overexpression of Hey1 Enhances BMP9-induced Osteogenic Differentiation of MSCs—We next examined the effect of continuous Hey1 expression on BMP9-induced osteogenic differentiation of MSCs. We constructed a retroviral vector expressing the mouse Hey1 gene to establish a stable cell pool of C3H10-Hey1 cells. Using qPCR analysis, we confirmed that Hey1 expression increased by ∼3-fold in C3H10-Hey1 cells when compared with the C3H10T1/2 control cells (Fig. 4A). We also constructed a recombinant adenovirus that constitutively expresses mouse Hey1 (also known as AdHey1). As shown in Fig. 4B, Hey1 overexpression synergized with BMP9-induced ALP activity in MSCs, especially at the early immediate stage of BMP9-induced osteogenic differentiation (see also supplemental Fig. S1A). Similar synergistic effects were observed by using histochemical staining of ALP activity (Fig. 4C). Taken together, these results indicate that Hey1, as an immediate early mediator of BMP9 signaling, acts synergistically with BMP9 in osteogenic differentiation of MSCs. Therefore, Hey1 overexpression may promote early osteogenic differentiation and hence accelerate bone formation.
FIGURE 4.
Constitutive Hey1 expression enhances BMP9-induced osteogenic differentiation of MSCs. A, confirmation of Hey1 expression in the C3H10-Hey1 stable line. Total RNA was collected from subconfluent cells and subjected to qPCR analysis. All samples were normalized for glyceraldehyde-3-phosphate dehydrogenase. B and C, synergistic induction of BMP9-induced ALP activity by Hey1 overexpression. C3H10-Hey1 and C3H10T1/2 control lines were infected with AdGFP or AdBMP9. Alternatively, C3H10T1/2 cells were infected with AdGFP, AdBMP9, and/or AdHey1. ALP activity was determined at indicated time points using colorimetric assays (B) or histochemical staining (C). Representative results of three independent experiments are shown.
Hey1 Knockdown Decreases BMP9-induced Bone Formation and Promotes Chondrogenesis, whereas Hey1 Overexpression Enhances Mineralization in BMP9-stimulated MSC Implantations in Vivo—Results from our in vitro knockdown and overexpression studies suggest that Hey1 may play an important role in BMP9-induced osteogenic differentiation of MSCs. We sought to determine the effect of Hey1 knockdown and overexpression on BMP9-mediated osteoblast differentiation in vivo using the stem cell implantation approach. Subconfluent C3H10T1/2 cells, C3H10-Hey1KD, and C3H10-Hey1 were infected with AdBMP9 or AdGFP. We also co-infected C3H10T1/2 cells with AdHey1 and AdBMP9, Ad-simHey1, or AdGFP. At 20 h after infection, cells were collected and injected subcutaneously into athymic nude mice. At 6 weeks following injection, animals were sacrificed and subjected to MicroCT imaging and histological evaluation. Ossification was readily detected by MicroCT imaging in C3H10T1/2 control cells stimulated with BMP9 (Fig. 5A), while groups receiving AdGFP or AdHey1 only did not exhibit any detectable ossification (data not shown). In comparison to the BMP9-transduced control C3H10T1/2 cell line, no significant ossification was detected in the mice injected with the BMP9-transduced C3H10-Hey1KD. Interestingly, Hey1 overexpression (C3H10-Hey1) reduced ectopic bone formation (Fig. 5A), but formed more mineralized bone as shown in MicroCT sectional images (supplemental Fig. S1C). Volumetric calculation of the bone masses indicated that BMP9-transduced C3H10T1/2 cells formed much larger bone masses than the BMP9-transduced C3H10-Hey1KD (7.16 mm3 versus 0.27 mm3, p < 0.05). BMP9-transduced C3H10-Hey1 also reduced ectopic bone formation as compared with the BMP9-stimulated C3H10T1/2 cells, but the difference was not statistically significant (3.5 mm3 versus 7.16 mm3, p > 0.05) (Fig. 5B).
FIGURE 5.
Critical role of Hey1 in BMP9-induced ectopic bone formation. C3H10-Hey1KD, C3H10-Hey1, and C3H10T1/2 control lines were transduced with AdBMP9 or AdGFP in vitro and implanted subcutaneously in athymic nude mice. At 6 weeks, animals were sacrificed and subjected to MicroCT imaging. A, MicroCT imaging analysis. Representative three-dimensional reconstructed images are shown. The injections sites are indicated by arrows. B, volumetric analysis of ectopic bone formation. Knockdown of Hey1 expression decreased ectopic bone formation (C3H10-Hey1KD = 0.27 mm3, C3H10T1/2 = 7.16 mm3, p = 0.02). Hey1 overexpression also reduced ectopic bone formation, although not statistically significant (C3H10-Hey1 = 3.5 mm3, C3H10T1/2 = 7.16 mm3, p = 0.08). C, histologic hematoxylin and eosin staining of retrieved samples. CC, chondrocyte; CM, chondroid matrix; MM, mineralized matrix; OB, osteoblast; OC, osteocyte; UD, undifferentiated MSCs. D, Trichrome (panels a and b) and Alcian Blue (panels c and d) staining of the samples retrieved AdBMP9 or AdBMP9 and Ad-simHey1 infected MEFs (3 weeks).
The implantation sites were retrieved and subjected to histologic analysis. Consistent with MicroCT imaging, no ossified tissues or cell masses were found in the mice injected with the GFP-transduced C3H10T1/2, C3H10-Hey1KD, or C3H10-Hey1 cells. The samples retrieved from the mice injected with BMP9-transduced C3H10T1/2 cells demonstrated undifferentiated stromal cells with a predominance of osteoid matrix and a shell-like rim of mature trabecular bone (Fig. 5C). The samples retrieved from the mice injected with BMP9-transduced C3H10-Hey1 cells exhibited the most mature bone of all the samples, yet the bone volume was smaller than the C3H10T1/2 control cells. This enhanced ossification phenotype was also observed in the mice injected with C3H10T1/2 cells co-infected with AdHey1 and AdBMP9 (supplemental Fig. S1D). Although MicroCT imaging failed to detect any bone formation, the samples retrieved from the BMP9-transduced C3H10-Hey1KD displayed smaller but readily detectable masses of undifferentiated stromal cells intermixed with chondroid matrix, chondroblasts, and chondrocytes, yet exhibited minimal mature trabecular bone (Fig. 5C). The decrease in BMP9-induced matrix mineralization was further confirmed by Trichrome staining assay. We found that adenovirus-mediated expression of mouse sim-Hey1 in MEFs remarkably reduced BMP9-induced matrix mineralization (Fig. 5D, panels a versus b). Conversely, knockdown of Hey1 expression increased the presence of chondroid matrix when the MEFs were stimulated with BMP9 (Fig. 5D, panels c versus d). Taken together, these results demonstrate that BMP9-induced osteogenic differentiation can be inhibited by silencing Hey1 expression and enhanced by Hey1 overexpression, suggesting that Hey1 may play an important role in promoting osteogenic differentiation and inhibiting chondrogenic differentiation.
Runx2 Can Partially Rescue the Hey1 Knockdown Phenotype—The absence of mature osteoblasts and ossification in BMP9-stimulated Hey1KD MSCs is reminiscent of the Runx2-deficient phenotype (38). Runx2 has been identified as a master regulator of osteoblast differentiation and chondrogenesis (38). Runx2 (also known as Cbfa1, Osf2, and AML3) is a member of the Runx class of transcription factors that contains a highly conserved 128-amino acid motif that functions in DNA binding, protein-protein interactions, and ATP binding (39). Runx2 is an essential regulator of osteoblast differentiation and skeletogenesis, because Runx2-deficient mice die shortly after birth and demonstrate a cartilaginous skeleton with complete absence of ossification (40). Despite the cartilaginous phenotype in the Runx2-null mice, histologic analysis demonstrated a delayed chondrocyte maturation phenotype, suggesting the importance of Runx2 in both chondrogenesis and osteogenesis (41).
Our in vivo studies suggest that Hey1 may promote osteogenic differentiation and inhibit chondrogenic differentiation. Therefore, we sought to test the functional relationship between Runx2 and Hey1. Similar to Hey1 overexpression, Runx2 overexpression in C3H10T1/2 MSCs was not sufficient to induce any detectable ALP activity in vitro and de novo bone formation in vivo (data not shown). These results suggest that Hey1 and Runx2 are not sufficient to initiate osteogenesis from MSCs and are only able to regulate osteogenic differentiation in committed osteoblast progenitors. Consistent with this possible role of Hey1 in the committed stage of osteogenesis, we demonstrated that the late osteogenic marker osteocalcin was up-regulated by BMP9 in the C3H10T1/2 cells, but not effectively induced by BMP9 in Hey1KD MSCs (Fig. 6A). Nonetheless, Runx2 acts synergistically with BMP9 in inducing osteogenic differentiation (i.e. ALP activity) in a Runx2 dosage-dependent fashion independent of Hey1 status (Fig. 6B and data not shown), suggesting that Runx2 may be regulated by BMP9 in a parallel pathway and/or that Runx2 may function downstream of Hey1 in BMP9-induced osteogenic differentiation.
FIGURE 6.
Inhibition of BMP9-mediated osteogenic differentiation due to Hey1 deficiency can be partially rescued by Runx2. A, Hey1 regulates expression of the late osteogenic marker. C3H10-Hey1KD and control lines were infected with AdBMP9 or AdGFP. Total RNA was collected at the indicated time points and subjected to qPCR analysis of osteocalcin (OC) expression. B, Runx2 and BMP9 act synergistically in the C3H10-Hey1KD line to increase osteogenic differentiation. Subconfluent C3H10-Hey1KD cells were infected with a low titer of AdBMP9 and increasing doses of AdRunx2. At indicated time points, cells were collected for colorimetric assays of ALP activity. C, Runx2 rescue on Hey1 knockdown was demonstrated in vivo by MicroCT imaging. D, Runx2 rescues the Hey1 knockdown phenotype. MicroCT imaging and volumetric data acquisition were carried out as described in Fig. 5B. E, histologic evidence of the Runx2 rescue effect on the Hey1KD MSCs. MM, mineralized matrix; OC, osteocyte; OB, osteoblast; UD, undifferentiated MSCs; magnification, 200×. F, regulation of Runx2 expression by Hey1. Total RNA was isolated from C3H10-Hey1KD and C3H10T1/2 control lines, and subjected to qPCR analysis for mouse Runx2 expression. See text for details.
Given the facts that Runx2 and BMP9 act synergistically to induce osteogenic differentiation of MSCs and Runx2 has been shown to play an important role in endochondral ossification (42, 43), we postulated that Runx2 may rescue the Hey1 knockdown phenotype. To test this possibility, we co-infected the C3H10-Hey1KD cells with AdRunx2 and AdBMP9 or AdGFP and implanted them subcutaneously into athymic nude mice. At 6 weeks after implantation, mice were sacrificed and ectopic ossification was evaluated with MicroCT imaging. Although no detectable ossification was observed in the mice injected with the C3H10-Hey1KD cells transduced with AdRunx2/AdGFP, co-expression of AdRunx2/AdBMP9 in C3H10-Hey1KD cells led to the formation of readily detectable ossified masses (Fig. 6C). Co-infection of AdBMP9 and AdRunx2 in the C3H10-Hey1KD cells resulted in over an increase of 20-fold in average bone volume in comparison to the C3H10-Hey1KD line stimulated by AdBMP9 alone. The average bone volumes in the BMP9- and Runx2-stimulated C3H10-Hey1KD cells was not statistically different from that of the BMP9-stimulated control C3H10T1/2 cells (5.49 mm3 versus 7.16 mm3, p = 0.53) (Fig. 6D). Histological evaluation of the retrieved samples revealed ossified masses with multiple foci of mature trabecular bone intermixed with osteoid matrix and undifferentiated MSCs (Fig. 6E and supplemental Fig. S1B). In comparison to the results shown in Fig. 5C, the addition of Runx2 resulted in a significant increase in mature trabecular bone.
The above results have demonstrated that exogenous expression of Runx2 partially rescued Hey1 knockdown-mediated inhibition of bone formation. One possible explanation could be that Runx2 may function downstream of Hey1 during BMP9-induced osteogenesis. To test whether Runx2 expression is dependent on Hey1, we conducted qPCR and demonstrated that Runx2 expression decreased by 79% in the Hey1 knockdown line C3H10-Hey1KD, in comparison to the control C3H10T1/2 cells (Fig. 6F). Thus, these results may provide an explanation regarding the ability of Runx2 to rescue the Hey1 knockdown phenotype in C3H10T1/2 cells.
DISCUSSION
Hey1 Is an Important and Direct Target of the BMP9 Osteogenic Signaling Pathway—We have recently identified BMP9 as one of the most potent osteogenic BMPs, and yet it remains one of the least characterized BMPs (6, 17, 18). To gain insights into the molecular basis of BMP9-mediated osteogenesis, we conducted an expression profiling analysis of genes regulated by BMP9 and found that Hey1 was among the most significantly up-regulated genes in MSCs upon BMP9 stimulation. We further investigated the functional role of Hey1 in BMP9-mediated osteogenic signaling in MSCs. We first confirmed that Hey1 expression was significantly up-regulated at the immediate early stage of BMP9 stimulation coinciding with the commitment of MSCs to osteoblast lineage. ChIP analysis indicated that Hey1 is a direct target of BMP9-induced Smad signaling. RNA interference-mediated knockdown of Hey1 expression diminished BMP9-stimulated osteogenic differentiation, and constitutive overexpression enhanced BMP9-induced differentiation in vitro and in vivo, suggesting Hey1 expression may play an important role in BMP9-induced osteogenic differentiation of MSCs. These results are in contrast to our previous studies of early targets, such as Id HLH and CTGF, in which their constitutive expression promotes proliferation and inhibits BMP-induced osteogenic differentiation of MSCs (6, 19-21).
Hey1 May Function Upstream of Runx2 during BMP9-induced Osteogenic Differentiation—Our in vivo results demonstrate that Hey1 may play an important role in promoting osteogenic differentiation and inhibiting chondrogenic differentiation. The absence of mature osteoblast and subsequent ossification in BMP9-stimulated Hey1KD MSCs is reminiscent of the Runx2-deficient phenotype (40, 44). Runx2 (runt-related gene 2; also known as, Cbfa1) a known target of Notch signaling, plays a critical role in osteogenesis (39). Furthermore, Runx2 is an essential transcription factor for osteoblast differentiation, and its expression can be-induced by both BMP2 and BMP7 (38, 44-46). Runx2 can directly stimulate expression of most of the well established bone markers, including ALP, osteopontin, and osteocalcin, in osteoblast progenitors (38, 47-49). Runx2 also plays an important role in chondrogenesis and endochondral ossification. Runx2 overexpression in chondrocytes via the chondrocyte-specific type II collagen promoter results in ectopic chondrocyte hypertrophy and endochondral ossification, thereby demonstrating the importance of Runx2 in controlling differentiation of both chondrocytes and osteoblasts (42, 43). It has been reported that Hey1 is induced by BMP2 in Runx2-deficient progenitors that failed to differentiate into bone-forming osteoblasts (50), and thus Runx2 may function downstream of Hey1 in osteogenesis. Consistent with this possibility are our findings, in which we have demonstrated that exogenous Runx2 could partially rescue the decreased osteogenic differentiation in Hey1KD MSCs both in vitro and in vivo.
Based on our findings, we propose a working model that depicts a possible mechanism of Hey1 function in BMP9-induced osteogenic differentiation. In normal MSCs, osteogenic BMP9 induces Hey1 expression at the immediate early stage, followed by Runx2 expression at the intermediate stage. BMP9-induced expression of Hey1 may promote osteogenic differentiation and inhibit chondrogenesis. Conversely, BMP9-stimulated expression of Runx2 stimulates both osteogenesis and chondrogenesis (42, 43). In the absence of differentiation stimuli, such as BMP9, expression of Hey1 and/or Runx2 is necessary but not sufficient for osteogenic differentiation of MSCs. Removal of Hey1 diminishes BMP9-induced osteogenic differentiation, which can be rescued by an exogenous expression of Runx2 that is functioning downstream or in parallel with Hey1. Consistent with our model are the reported findings in which Hey1 mRNA is up-regulated by BMP2 treatment when MSCs differentiate to osteoblast lineage and its expression pattern is overlapping with Runx2's (28). Furthermore, it has been shown that Hey1 mRNA is up-regulated in Runx2-deficient cells by BMP-2 stimulation, indirectly suggesting that Hey1 may function upstream of Runx2 during BMP-stimulated osteoblast differentiation (50).
The Exact Role of Hey1 Basic-Helix-Loop-Helix Protein in Osteogenesis Remains to Be Understood—Although our findings suggest that Hey1 may play an important role in BMP9-induced osteogenic differentiation of MSCs, the molecular mechanism behind the Hey1 functional role in osteogenesis remains to be defined. Interestingly, it has been reported that siRNA-mediated inhibition of Hey1 induction led to an increase in osteoblast matrix mineralization in vitro via inhibition of Runx2 transcriptional activity, suggesting that Hey1 may be a negative regulator of osteoblast maturation (29). Although we do not know the possible reasons for the discrepancy about the consequence of Hey1 knockdown, the experiments were conducted in the mature osteoblast line MC3T3, and it is unclear if the Hey1 knockdown-mediated enhancement of bone formation would be observed in vivo. In contrast to these findings reported by Zamurovic et al. (29), it has been demonstrated that Hes1, the binding partner of Hey1, functions to promote the transactivating ability of Runx2 (51). It is also conceivable that BMP9 may regulate Hey1 function distinctly from that of the BMP2 osteogenic mixture used in a previous study (29). However, as a target of both BMP2 and Notch signaling, Hey1 is clearly implicated in BMP- and Notch-mediated osteogenic differentiation (52, 53). Interestingly, Hey1-deficient mice exhibit behavioral alterations through the dopaminergic nervous system (54) and otherwise unremarkable phenotypic changes (55), although a combined loss of Hey1/HeyL or Hey1/Hey2 causes defects in heart development (56, 57). The bone and skeletal phenotypes of Hey1-deficient mice have not been extensively analyzed, and the osteoblast-specific Hey1 deletion has yet to be carried out.
In summary, we have demonstrated that Hey1 is an important mediator of BMP9-induced osteogenic differentiation of MSCs and that both Hey1 and Runx2 are involved in BMP9 osteogenic signaling, because Runx2 provides a partial rescue of the Hey1KD phenotype. Future studies should be directed at osteoblast-specific deletion of Hey1 in animal models, as well as further examination of the interaction between Hey1 and other regulatory factors such as Runx2 and Hes1 in BMP9-mediated osteogenic differentiation.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants CA106569, AR50142, and AR054381 (to R. C. H., T. C. H., and H. H. L.). This work was also supported by research grants from the American Cancer Society (to T. C. H. and H. H. L.), The Brinson Foundation (to T. C. H.), and the Orthopaedic Research and Education Foundation (OREF) (to R. C. H. and H. H. L.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2 and Table S1.
Footnotes
The abbreviations used are: MSC, mesenchymal stem cell; ALP, alkaline phosphatase; BMP, bone morphogenetic protein; ChIP, chromatin immunoprecipitation; GFP, green fluorescent protein; HES, hairy/Enhancer of split; HERP, HES-related repressor protein; MEF, mouse embryo fibroblast; qPCR, quantitative real-time PCR; MicroCT, microcomputed tomography; siRNA, small interference RNA; AdGFP, adenovirus expressing only GFP.
References
- 1.Pittenger, M. F., Mackay, A. M., Beck, S. C., Jaiswal, R. K., Douglas, R., Mosca, J. D., Moorman, M. A., Simonetti, D. W., Craig, S., and Marshak, D. R. (1999) Science 284 143-147 [DOI] [PubMed] [Google Scholar]
- 2.Prockop, D. J. (1997) Science 276 71-74 [DOI] [PubMed] [Google Scholar]
- 3.Aubin, J. E. (1998) J. Cell. Biochem. supplemental 31 73-82 [Google Scholar]
- 4.Ducy, P., Schinke, T., and Karsenty, G. (2000) Science 289 1501-1504 [DOI] [PubMed] [Google Scholar]
- 5.Caplan, A. I., and Bruder, S. P. (2001) Trends Mol. Med. 7 259-264 [DOI] [PubMed] [Google Scholar]
- 6.Luu, H. H., Song, W. X., Luo, X., Manning, D., Luo, J., Deng, Z. L., Sharff, K. A., Montag, A. G., Haydon, R. C., and He, T. C. (2007) J. Orthop. Res. 25 665-677 [DOI] [PubMed] [Google Scholar]
- 7.Friedenstein, A. J., Piatetzky, S., II, and Petrakova, K. V. (1966) J. Embryol. Exp. Morphol. 16 381-390 [PubMed] [Google Scholar]
- 8.Baksh, D., Song, L., and Tuan, R. S. (2004) J. Cell. Mol. Med. 8 301-316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barry, F. P., and Murphy, J. M. (2004) Int. J. Biochem. Cell Biol. 36 568-584 [DOI] [PubMed] [Google Scholar]
- 10.Varga, A. C., and Wrana, J. L. (2005) Oncogene 24 5713-5721 [DOI] [PubMed] [Google Scholar]
- 11.Zhang, J., and Li, L. (2005) Dev. Biol. 284 1-11 [DOI] [PubMed] [Google Scholar]
- 12.Shi, Y., and Massague, J. (2003) Cell 113 685-700 [DOI] [PubMed] [Google Scholar]
- 13.Attisano, L., and Wrana, J. L. (2002) Science 296 1646-1647 [DOI] [PubMed] [Google Scholar]
- 14.Hogan, B. L. (1996) Genes Dev. 10 1580-1594 [DOI] [PubMed] [Google Scholar]
- 15.Luo, J., Sun, M. H., Kang, Q., Peng, Y., Jiang, W., Luu, H. H., Luo, Q., Park, J. Y., Li, Y., Haydon, R. C., and He, T. C. (2005) Curr. Gene. Ther. 5 167-179 [DOI] [PubMed] [Google Scholar]
- 16.Zhao, G. Q. (2003) Genesis 35 43-56 [DOI] [PubMed] [Google Scholar]
- 17.Cheng, H., Jiang, W., Phillips, F. M., Haydon, R. C., Peng, Y., Zhou, L., Luu, H. H., An, N., Breyer, B., Vanichakarn, P., Szatkowski, J. P., Park, J. Y., and He, T. C. (2003) J. Bone Joint Surg. Am. 85A 1544-1552 [DOI] [PubMed] [Google Scholar]
- 18.Kang, Q., Sun, M. H., Cheng, H., Peng, Y., Montag, A. G., Deyrup, A. T., Jiang, W., Luu, H. H., Luo, J., Szatkowski, J. P., Vanichakarn, P., Park, J. Y., Li, Y., Haydon, R. C., and He, T. C. (2004) Gene Ther. 11 1312-1320 [DOI] [PubMed] [Google Scholar]
- 19.Peng, Y., Kang, Q., Cheng, H., Li, X., Sun, M. H., Jiang, W., Luu, H. H., Park, J. Y., Haydon, R. C., and He, T. C. (2003) J. Cell. Biochem. 90 1149-1165 [DOI] [PubMed] [Google Scholar]
- 20.Peng, Y., Kang, Q., Luo, Q., Jiang, W., Si, W., Liu, B. A., Luu, H. H., Park, J. K., Li, X., Luo, J., Montag, A. G., Haydon, R. C., and He, T. C. (2004) J. Biol. Chem. 279 32941-32949 [DOI] [PubMed] [Google Scholar]
- 21.Luo, Q., Kang, Q., Si, W., Jiang, W., Park, J. K., Peng, Y., Li, X., Luu, H. H., Luo, J., Montag, A. G., Haydon, R. C., and He, T. C. (2004) J. Biol. Chem. 279 55958-55968 [DOI] [PubMed] [Google Scholar]
- 22.Song, J. J., Celeste, A. J., Kong, F. M., Jirtle, R. L., Rosen, V., and Thies, R. S. (1995) Endocrinology 136 4293-4297 [DOI] [PubMed] [Google Scholar]
- 23.Miller, A. F., Harvey, S. A., Thies, R. S., and Olson, M. S. (2000) J. Biol. Chem. 275 17937-17945 [DOI] [PubMed] [Google Scholar]
- 24.Lopez-Coviella, I., Berse, B., Krauss, R., Thies, R. S., and Blusztajn, J. K. (2000) Science 289 313-316 [DOI] [PubMed] [Google Scholar]
- 25.Jane, J., Dunford, B., Kron, A., Pittman, D., Sasaki, T., Li, J., Li, H., Alden, T., Dayoub, H., Hankins, G., Kallmes, D., and Helm, G. (2002) Mol. Ther. 6 464. [DOI] [PubMed] [Google Scholar]
- 26.Leimeister, C., Externbrink, A., Klamt, B., and Gessler, M. (1999) Mech. Dev. 85 173-177 [DOI] [PubMed] [Google Scholar]
- 27.Iso, T., Kedes, L., and Hamamori, Y. (2003) J. Cell. Physiol. 194 237-255 [DOI] [PubMed] [Google Scholar]
- 28.de Jong, D. S., Vaes, B. L., Dechering, K. J., Feijen, A., Hendriks, J. M., Wehrens, R., Mummery, C. L., van Zoelen, E. J., Olijve, W., and Steegenga, W. T. (2004) J. Bone Miner Res. 19 947-958 [DOI] [PubMed] [Google Scholar]
- 29.Zamurovic, N., Cappellen, D., Rohner, D., and Susa, M. (2004) J. Biol. Chem. 279 37704-37715 [DOI] [PubMed] [Google Scholar]
- 30.Lengner, C. J., Lepper, C., van Wijnen, A. J., Stein, J. L., Stein, G. S., and Lian, J. B. (2004) J. Cell. Physiol. 200 327-333 [DOI] [PubMed] [Google Scholar]
- 31.Si, W., Kang, Q., Luu, H. H., Park, J. K., Luo, Q., Song, W. X., Jiang, W., Luo, X., Li, X., Yin, H., Montag, A. G., Haydon, R. C., and He, T. C. (2006) Mol. Cell. Biol. 26 2955-2964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luo, Q., Kang, Q., Song, W. X., Luu, H. H., Luo, X., An, N., Luo, J., Deng, Z. L., Jiang, W., Yin, H., Chen, J., Sharff, K. A., Tang, N., Bennett, E., Haydon, R. C., and He, T. C. (2007) Gene (Amst.) 395 160-169 [DOI] [PubMed] [Google Scholar]
- 33.He, T. C., Zhou, S., da Costa, L. T., Yu, J., Kinzler, K. W., and Vogelstein, B. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 2509-2514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.He, T. C., Sparks, A. B., Rago, C., Hermeking, H., Zawel, L., da Costa, L. T., Morin, P. J., Vogelstein, B., and Kinzler, K. W. (1998) Science 281 1509-1512 [DOI] [PubMed] [Google Scholar]
- 35.He, T. C., Chan, T. A., Vogelstein, B., and Kinzler, K. W. (1999) Cell 99 335-345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Luo, J., Deng, Z. L., Luo, X., Tang, N., Song, W. X., Chen, J., Sharff, K. A., Luu, H. H., Haydon, R. C., Kinzler, K. W., Vogelstein, B., and He, T. C. (2007) Nat. Protoc. 2 1236-1247 [DOI] [PubMed] [Google Scholar]
- 37.Kang, Q., Song, W. X., Luo, Q., Tang, N., Luo, J., Luo, X., Chen, J., Bi, Y., He, B. C., Park, J. K., Jiang, W., Tang, Y., Huang, J., Su, Y., Zhu, G. H., He, Y., Yin, H., Hu, Z., Wang, Y., Chen, L., Zuo, G. W., Pan, X., Shen, J., Vokes, T., Reid, R., Haydon, H. C., Luu, H. H., and He, T. C. (2008) Stem Cells Dev. doi: 10.1089/scd.2008.0130 [DOI] [PMC free article] [PubMed]
- 38.Ducy, P., Zhang, R., Geoffroy, V., Ridall, A. L., and Karsenty, G. (1997) Cell 89 747-754 [DOI] [PubMed] [Google Scholar]
- 39.Karsenty, G. (2000) Semin. Cell Dev. Biol. 11 343-346 [DOI] [PubMed] [Google Scholar]
- 40.Komori, T., Yagi, H., Nomura, S., Yamaguchi, A., Sasaki, K., Deguchi, K., Shimizu, Y., Bronson, R. T., Gao, Y. H., Inada, M., Sato, M., Okamoto, R., Kitamura, Y., Yoshiki, S., and Kishimoto, T. (1997) Cell 89 755-764 [DOI] [PubMed] [Google Scholar]
- 41.Inada, M., Yasui, T., Nomura, S., Miyake, S., Deguchi, K., Himeno, M., Sato, M., Yamagiwa, H., Kimura, T., Yasui, N., Ochi, T., Endo, N., Kitamura, Y., Kishimoto, T., and Komori, T. (1999) Dev. Dyn. 214 279-290 [DOI] [PubMed] [Google Scholar]
- 42.Ueta, C., Iwamoto, M., Kanatani, N., Yoshida, C., Liu, Y., Enomoto-Iwamoto, M., Ohmori, T., Enomoto, H., Nakata, K., Takada, K., Kurisu, K., and Komori, T. (2001) J. Cell Biol. 153 87-100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Takeda, S., Bonnamy, J. P., Owen, M. J., Ducy, P., and Karsenty, G. (2001) Genes Dev. 15 467-481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Otto, F., Thornell, A. P., Crompton, T., Denzel, A., Gilmour, K. C., Rosewell, I. R., Stamp, G. W., Beddington, R. S., Mundlos, S., Olsen, B. R., Selby, P. B., and Owen, M. J. (1997) Cell 89 765-771 [DOI] [PubMed] [Google Scholar]
- 45.Lee, K. S., Kim, H. J., Li, Q. L., Chi, X. Z., Ueta, C., Komori, T., Wozney, J. M., Kim, E. G., Choi, J. Y., Ryoo, H. M., and Bae, S. C. (2000) Mol. Cell. Biol. 20 8783-8792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gu, K., Zhang, L., Jin, T., and Rutherford, R. B. (2004) Cells Tissues Organs 176 28-40 [DOI] [PubMed] [Google Scholar]
- 47.Kern, B., Shen, J., Starbuck, M., and Karsenty, G. (2001) J. Biol. Chem. 276 7101-7107 [DOI] [PubMed] [Google Scholar]
- 48.Harada, H., Tagashira, S., Fujiwara, M., Ogawa, S., Katsumata, T., Yamaguchi, A., Komori, T., and Nakatsuka, M. (1999) J. Biol. Chem. 274 6972-6978 [DOI] [PubMed] [Google Scholar]
- 49.Javed, A., Barnes, G. L., Jasanya, B. O., Stein, J. L., Gerstenfeld, L., Lian, J. B., and Stein, G. S. (2001) Mol. Cell. Biol. 21 2891-2905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu, T., Gao, Y., Sakamoto, K., Minamizato, T., Furukawa, K., Tsukazaki, T., Shibata, Y., Bessho, K., Komori, T., and Yamaguchi, A. (2007) J. Cell. Physiol. 211 728-735 [DOI] [PubMed] [Google Scholar]
- 51.McLarren, K. W., Lo, R., Grbavec, D., Thirunavukkarasu, K., Karsenty, G., and Stifani, S. (2000) J. Biol. Chem. 275 530-538 [DOI] [PubMed] [Google Scholar]
- 52.Nobta, M., Tsukazaki, T., Shibata, Y., Xin, C., Moriishi, T., Sakano, S., Shindo, H., and Yamaguchi, A. (2005) J. Biol. Chem. 280 15842-15848 [DOI] [PubMed] [Google Scholar]
- 53.Minamizato, T., Sakamoto, K., Liu, T., Kokubo, H., Katsube, K., Perbal, B., Nakamura, S., and Yamaguchi, A. (2007) Biochem. Biophys. Res. Commun. 354 567-573 [DOI] [PubMed] [Google Scholar]
- 54.Fuke, S., Minami, N., Kokubo, H., Yoshikawa, A., Yasumatsu, H., Sasagawa, N., Saga, Y., Tsukahara, T., and Ishiura, S. (2006) J. Neurosci. Res. 84 1555-1563 [DOI] [PubMed] [Google Scholar]
- 55.Fischer, A., Schumacher, N., Maier, M., Sendtner, M., and Gessler, M. (2004) Genes Dev. 18 901-911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fischer, A., Steidl, C., Wagner, T. U., Lang, E., Jakob, P. M., Friedl, P., Knobeloch, K. P., and Gessler, M. (2007) Circ. Res. 100 764-765 [DOI] [PubMed] [Google Scholar]
- 57.Fischer, A., Klattig, J., Kneitz, B., Diez, H., Maier, M., Holtmann, B., Englert, C., and Gessler, M. (2005) Mol. Cell. Biol. 25 8960-8970 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.