Abstract
Glutathione S-transferase Pi (GSTπ) is a marker protein in many cancers and high levels are linked to drug resistance, even when the selecting drug is not a substrate. S-Glutathionylation of proteins is critical to cellular stress response, but characteristics of the forward reaction are not known. Our results show that GSTπ potentiates S-glutathionylation reactions following oxidative and nitrosative stress in vitro and in vivo. Mutational analysis indicated that the catalytic activity of GST is required. GSTπ is itself redox-regulated. S-Glutathionylation on Cys47 and Cys101 autoregulates GSTπ, breaks ligand binding interactions with c-Jun NH2-terminal kinase (JNK), and causes GSTπ multimer formation, all critical to stress response. Catalysis of S-glutathionylation at low pK cysteines in proteins is a novel property for GSTπ and may be a cause for its abundance in tumors and cells resistant to a range of mechanistically unrelated anticancer drugs.
Glutathione S-transferases (GSTs)2 are classified as a family of Phase II detoxification enzymes that have classically been described as catalyzing the conjugation of glutathione (GSH) to electrophilic compounds through thioether linkages (1). The Pi class (GSTπ) is present at high levels in many solid tumors (particularly ovarian, non-small cell lung, breast, liver, pancreas, colon, and lymphomas) and has been indicated in many reports to be overexpressed in drug-resistant tumors (2, 3). Although its increased expression was frequently linked with enhancement of drug detoxification, in most instances the selecting drugs were not substrates of GSTπ. This ambiguity and the high prevalence of GSTπ in tumors have intimated cellular functions for the protein that are unrelated to catalytic detoxification. Recently GSTπ has been identified as an endogenous protein binding partner and regulator of c-Jun NH2-terminal kinase (JNK) and peroxiredoxin VI (1-cysPrx) (4–6). Moreover, oxidative stress causes increased GSTπ expression, the regulation of which has been identified as a downstream event linked to wild-type p53 function (7).
Cellular response to oxidative or nitrosative stress includes S-glutathionylation, a post-translational modification characterized by conjugation of glutathione to low pK cysteine sulfhydryl or sulfenic acid moieties in target proteins. This adds a three-amino acid side chain and introduces a net negative charge (as a consequence of glutamic acid) to the protein (8). Consequently, protection from further oxidative damage and/or alteration of protein conformation affecting function and/or cellular localization occurs. Proteins so far identified that are susceptible to S-glutathionylation can be categorized into six distinct clusters: cytoskeletal, glycolysis/energy metabolism, kinases and signaling pathways, calcium homeostasis, antioxidant enzymes, and protein folding (9). Reversibility of S-glutathionylation spontaneously by GSH or catalytically by glutaredoxin or sulfiredoxin (8, 10)3 provides the cell with a dynamic cycle of regulatory events. A plausible link between GSTπ and such regulation was provided by the observation that GSTπ could conjugate glutathione to a member of the peroxiredoxin family, peroxiredoxin VI, a non-selenoglutathione-dependent peroxidase that converts lipids and other hydroperoxides to corresponding alcohols (6, 12). The catalytically important cysteine residue on peroxiredoxin VI is sterically inaccessible for GSH within the globular homodimeric complex and GSTπ facilitated transfer of GS- to this site. The resultant activation of the enzyme served a cellular regulatory role through maintaining intracellular H2O2 levels (13), particularly with respect to antioxidant protection of cell membranes. Moreover, S-glutathionylation of protein-tyrosine phosphatase (PTP1B) or protein-disulfide isomerase inactivates these enzymes and can have impact on kinase-mediated proliferative events and protein folding (14, 15).3
Both endogenous and exogenous reactive oxygen and nitrogen species generation can cause S-glutathionylation of certain targeted proteins. For the present studies, we developed multiple model systems of oxidative and nitrosative stress to investigate the role of GSTs in S-glutathionylation reactions, using oxidized glutathione (GSSG) and two agents, NOV-002 and PABA/NO. NOV-002 is a mimetic of oxidized glutathione and causes oxidative stress induction (16). PABA/NO (O2-{2,4-dinitro-5-[4-(N-methylamino)benzoyloxy]phenyl}1-(N,N-dimethylamino)diazen-1-ium-1,2-diolate) (17) is a prodrug, which releases nitric oxide (NO) and activates stress response pathways that involve JNK, eventually leading to apoptosis (14, 18). Elevated levels of NO provide the primary source of reactive nitrogen species and can alter protein function directly through post-translational modifications (nitration/nitrosylation) or indirectly through interactions with oxygen, superoxide, thiols, and heavy metals, the products of which can lead to S-glutathionylation.
Recognition that S-glutathionylation of proteins and its cycling is important in regulating critical cellular events means that there is a need to define both the forward and reverse components of the cycle. Until this time, the forward reaction of S-glutathionylation has been tacitly assumed to occur spontaneously. Our present study shows for the first time that GSTπ catalyzes protein S-glutathionylation in vivo and is contributory to its high expression levels in many drug-resistant tumors.
EXPERIMENTAL PROCEDURES
Materials and Cell Lines—Generation of GSTπ wild-type and knock-out mice (19), extraction of the mouse embryo fibroblasts (MEF) cells, and details of the transfected NIH3T3 cell lines were described previously (20, 21). Cell lines were maintained in Dulbecco's modified Eagle's medium containing 10% fetal calf serum, 100 μg/ml streptomycin, 100 units/ml penicillin, and 2 mm l-glutamine at 5% CO2 and 37 °C. GSH (reduced) and GSSG (oxidized) were obtained from MP Biomedicals (Aurora, OH). PABA/NO was prepared as previously described (14). NOV-002 was provided by Novelos Therapeutics, Inc. (Newton, MA).
Human GSTπ Mutagenesis—Human GSTπ was amplified using PCR from a cDNA prepared from HEK293 cells using the following oligonucleotides: 5′-GCGCGAATTCATGCCGCCCTACACCGTGGT-3′ and 5′-GCGCAAGCTTTCACTGTTTCCCGTTGCCAT-3′. The resulting PCR fragments were cloned into the EcoRI/HindIII recognition sites of pCMV2b FLAG tag vector for mammalian expression and into the NheI/BamHI recognition sites of pET28b for expression in Escherichia coli. Cysteines 47 and 101 were changed to alanine by site-directed mutagenesis using the kit supplied by Stratagene (La Jolla, CA). The catalytically inactive mutant was generated by a single mutation (Y7F) by site-directed mutagenesis. The integrity of the inserts was confirmed by sequence analysis.
Transfected Cell Lines—HEK293 cells were transfected with pCMV2b vector containing human wild-type (HEK293-WT), C47A mutation (HEK-Cys47), C101A mutation (HEK-Cys101), or the catalytically impaired mutant GSTπ (HEK-Y7F) using the transfection agent, Lipofectamine (Sigma). Mock transfectants (HEK-VA) were generated by transfecting HEK293 cells with the pCMV2b empty vector.
GSTπ Activity Assays—The specific enzyme activity was evaluated with the 1-chloro-2,4-dinitrobenzene (CDNB) substrate as previously described (22). 48 h after transfection, HEK293 cells were lysed and 100 μg of protein per reaction assayed. The reaction mixture was carried out in 100 mm PBS with 1.0 mm EDTA, pH 6.5, containing 1 mm GSH and CDNB. The absorbance at 340 nm was monitored every 30 s for 2 min in a Bio-Rad Benchmark Plus Microplate Spectrophotometer. The rate of spontaneous conjugation of GSH to CDNB was subtracted from the rates of GSTπ-catalyzed reactions. The extinction coefficient (5.3 m-1 cm-1) for the CDNB conjugate at 340 nm in 96-well plates was used to calculate the specific activity.
Protein Preparation—Cells (5 × 105 per treatment group) were collected and washed with PBS. Cell pellets were suspended in lysis buffer containing 20 mm Tris-HCl, pH 7.5, 15 mm NaCl, 1 mm EDTA, 1 mm EGTA, 1% Triton X-100, 2.5 mm sodium pyrophosphate, and 1 mm glycerophosphate with freshly added phosphatase inhibitors (5 mm sodium fluoride and 1 mm Na3VO4) and a protease inhibitor mixture (Sigma). Cells were incubated for 30 min on ice, disrupted by sonication (3 times for 10 s), and centrifuged for 30 min at 10,000 × g (4 °C). Protein concentrations in the supernatant were determined with the Bradford method (Bio-Rad) using IgG as a standard.
Immunoblot Analysis—Equivalent amounts of protein were electrophoretically resolved under non-reducing conditions on 10% SDS-polyacrylamide gels; unmodified proteins were separated under reducing conditions. Proteins were transferred onto nitrocellulose membranes (Bio-Rad). Nonspecific binding was reduced by incubating the membrane in blocking buffer (20 mm Tris-HCl, pH 7.5, 150 mm NaCl, 0.1% Tween 20, 1 μm protease inhibitors, 5 mm sodium fluoride, and 1 mm Na3VO4) containing 10% nonfat dried milk for 1 h. Membranes were incubated with the indicated antibody (blocking buffer containing 5% nonfat dried milk) overnight at 4 °C, washed 3 times with PBS for 15 min, and incubated with the appropriate secondary antibody conjugated to horseradish peroxidase for 1 h. The membranes were washed 3 times and developed with enhanced chemiluminescence detection reagents (Bio-Rad). The blots were scanned with a Bio-Rad ChemiDoc system and visualized with a transilluminator. The images were stored in a TIFF format. The relative intensity of bands was evaluated using Quantity One software (version 4.5.2; Bio-Rad) and plotted as arbitrary units in relation to actin.
An infrared excited dual color (red and green) fluorescent imaging system (Odyssey LI-COR, NE) was used to detect S-glutathionylation of GSTπ (GST-SSG) in HEK293 cells after treatment with increasing amounts of PABA/NO. Proteins from cell lysates were resolved electrophoretically and transferred to polyvinylidene difluoride membranes as described above. Membranes were treated with proprietary blocking buffer (LI-COR, NE) and incubated with both polyclonal anti-GSTπ (1:1,000 dilution, Medical & Biological Laboratories Co., Ltd. (MDL), Nagoya, Japan) and monoclonal anti-glutathionylated protein antibodies (1:1,000 dilution, Virogen, Watertown, MA) as described above. After washing 3 times with PBS an immunoblot was developed with anti-rabbit IRDye®800CW (green) and anti-mouse IRDye®680 (red) secondary antibodies simultaneously (1:1,000 dilution, both from LI-COR, NE) according to the manufacturer's protocol. Finally, immunoblots were imaged with Odyssey imaging system (LI-COR) and quantified with standard software (LI-COR, NE).
In Vivo Models—GSTπ wild-type (GSTP1P2+/+) and knock-out (GSTP1P2-/-) mice (4 and 12 weeks old) were housed in the Animal Resource Facility of the Medical University of South Carolina. Animal care was provided in accordance with the procedures outlined in the Guide for the Care and Use of Laboratory Animals (NIH Publication 86-23, 1985). Animals were treated with 5 mg/kg PABA/NO or 25 mg/kg NOV-002 by tail vein injection. Blood was collected in heparin-coated tubes by orbital bleed at various time points. The liver was harvested and protein lysates were prepared and immediately assayed for cysteine modification.
Identification of S-Glutathionylated Serum Proteins—100 μg of control and drug-treated serum was separated by two-dimensional SDS-PAGE under non-reducing conditions. Proteins were resuspended in 8 m urea with 0.5% carrier ampholytes and run on immobilized pH gradients covering exponentially the pH range from 3 to 10. For the second dimension, the immobilized pH gradient strips were separated on 10% polyacrylamide gels. Protein spots of interest were excised and subject to trypsin digestion. Peptide mass was analyzed by MALDI-TOF mass spectrometry at the Proteomics Core Facility of the Medical University of South Carolina. Protein identification was performed using software from the National Center for Biotechnology Information protein data base.
Detection of Protein Sulfhydryls in Liver Homogenates of GSTπ+/+ and GSTπ-/- Mice—The supernatants of liver homogenate (100 μl) were passed through a BioSpin6 (Bio-Rad) micro-column to eliminate both GSH and GSSG. Then 20 μl of eluent was added to 2 ml of 40 mm PB, pH 7.4, in a quartz cuvette of PTI QM-8 spectrofluorometer (PTI Inc., Birmingham, NJ) under constant stirring at 37 °C. The emission (513 nm, excitation at 379 nm) of each sample was recorded for 1 min (background) before and 2 min after an addition of 5 μm (final concentration) ThioGlo-1 (Calbiochem). Each sample fluorescence saturation value corresponds to a concentration of free sulfhydryls. At the end of each experiment 1 μm reduced glutathione was added to the sample to ensure that saturation was not associated with the concentration of ThioGlo-1. The data represent mean ± S.E. (SigmaStat 10, SyStat, MA) of at least three independent measurements per condition.
Spectroscopic Analysis of GSTπ in Vitro—The effect of GSTπ S-glutathionylation on enzyme secondary structure was examined by circular dichroism (CD) where measurements were carried out on a 202 AVIV Associates (Lakewood, NJ) using a semimicro quartz rectangular 1 × 10 × 40-mm cuvette. GSTπ samples (>95% homogeneous, 40 μm in 20 mm PB, pH 7.4) were maintained at 22 °C using a Pelletier element. Spectra were recorded while scanning in the far-ultraviolet region (190–260 nm), with bandwidth of 1.0 nm, step size of 1.0 nm, integration time of 30 s, and three repeats. The output of the CD spectrometer was recalculated according to the protein concentration, amino acid content, and cuvette thickness into molecular ellipticity units (degrees/cm2/dmol).
Protein tryptophan fluorescence was recorded on an F2500 spectrofluorometer (Hitachi) using 10 × 10 × 40-mm quartz cuvette, excitation and emission slits were 2.5 and 5.0 nm, respectively. GSTπ samples (>95% homogeneous, 1.0 μm in 20 mm PB, pH 7.4) before and after treatment with PABA/NO and GSH were used. The excitation wavelength was 295 nm to minimize an effect of protein tyrosines and phenylalanines. Background spectra were subtracted from final emission of the protein. GSTπ (∼2 mg/ml) was S-glutathionylated by treatment with 25 μm PABA/NO and 1 mm GSH for 10 min in 40 mm PB, pH 7.4, at room temperature. Excess PABA/NO and GSH were eliminated by triplicate washing through Biospin-6 (Bio-Rad) SEC micro-spin columns with 20 mm PBS. Protein concentration was determined by the Bradford method (Bio-Rad).
Data Analysis—Mean ± S.D. for at least three experiments were calculated for each treatment group. The immunoblots shown are representative of at least three essentially identical results. Data were analyzed for statistically significant differences between the control and experimental/treated groups with Student's paired t test using SigmaStat 3.5 (Systat Software Inc., MA).
RESULTS
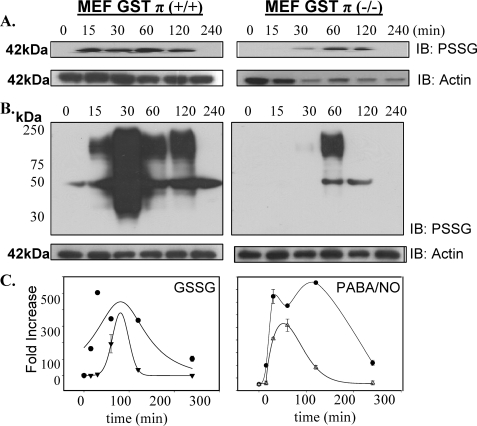
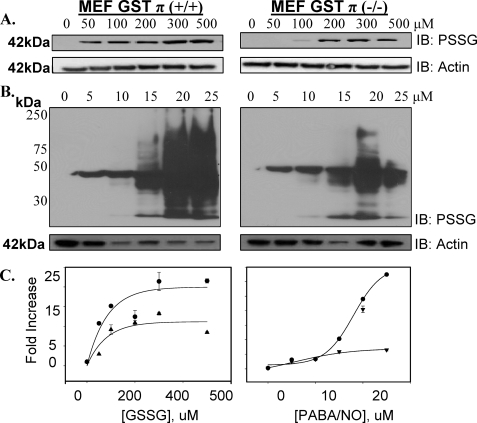
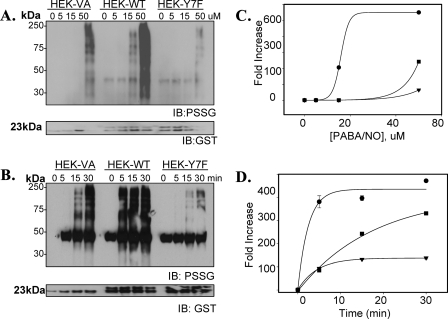
Impact of GSTπ Phenotype on Protein S-Glutathionylation following Oxidative and Nitrosative Stress—The physiological importance of S-glutathionylation is established and becomes critical to cellular stress responses (for review see Refs. 15). The mechanism of this reaction has not been defined as spontaneous or enzymatically catalyzed. In the first model system, MEF derived from GSTπ wild-type (GSTP1P2+/+) and null (GSTP1/P2-/-) mice were used to evaluate the dynamics of S-glutathionylation following exposure to agents that induce either oxidative (e.g. GSSG) or nitrosative stresses (e.g. PABA/NO). Using monoclonal antibodies directed toward the S-glutathionylated moiety (Virogen, Watertown, MA), we observed a time- (Fig. 1) and dose- (Fig. 2) dependent increase in protein modification (PSSG) following oxidative (GSSG) and nitrosative (PABA/NO) stress in MEF-GSTπ+/+ cells. Under basal conditions, S-glutathionylated proteins were essentially undetectable in either MEF-GSTπ+/+ or MEF-GSTπ-/- cells. The quantification of experimental data from 3 immunoblots was averaged and the exponential rise to maximal was statistically analyzed using SigmaStat. The dose dependence of PABA/NO-induced S-glutathionylation elicits a sigmoidal shape only for GSTπ+/+ indicative of an enzyme-controlled process (Fig. 2, panel C). The lag period may be explained by the accumulation of PABA/NO active metabolite(s). The time-dependent kinetics of PABA/NO-induced S-glutathionylation are described by similar bell-shaped curves for both GSTπ+/+ and GSTπ-/- cells (Fig. 1, panel C), perhaps corresponding to the dynamic, reversible origin of S-glutathionylation. The S-glutathionylation of cellular proteins in GSTπ+/+ cells is higher, starts earlier, and lasts longer when compared with the GSTπ-/- cells, p < 0.01. These data suggest that whereas basal levels of S-glutathionylation may be spontaneous or catalyzed by other GSTs, GSTπ contributes significantly to its facilitation and amplification. The dose dependence of GSSG-induced S-glutathionylation shows the expected rise-to-maximum shape indicative of a direct effect of this drug. The presence of GSTπ results in significant amplification of this effect (Fig. 2, panel C). The kinetics of GSSG-induced S-glutathionylation was similar to those for PABA/NO (Fig. 1, panel C). A major difference is that the shorter period of maximal basal S-glutathionylation may correspond to a direct effect of oxidant stress or a direct involvement of glutathione reductase.
FIGURE 1.
GSTπ expression enhances time-dependent S-glutathionylation of proteins. Mouse embryo fibroblast cells derived from GSTπ wild-type or knock-out animals were treated with 200 μm GSSG (A) or 25 μm PABA/NO for 0 to 240 min (B). Proteins were separated by non-reducing SDS-PAGE and S-glutathionylation evaluated by immunoblot (IB) with anti-glutathionylated protein monoclonal antibody (PSSG, Virogen, n = 4). Even loading of protein was confirmed by stripping the membrane and re-probing for actin. The kinetics of the S-glutathionylation reaction were analyzed using a standard 2 parameter exponential rise to maximum fitting procedure (Sigma Plot 10, SyStat, MA) for time dependence following GSSG and PABA/NO (C) for GSTπ+/+ (•) and GST-/- MEF (▴). Data represent mean ± variance, p < 0.01.
FIGURE 2.
GSTπ expression enhances dose-dependent S-glutathionylation of proteins. Mouse embryo fibroblast cells derived from GSTπ+/+ or GSTπ-/- animals were treated with various concentrations of GSSG (A) or PABA/NO (B) for 1 h. Proteins were resolved by non-reducing SDS-PAGE and S-glutathionylation evaluated by immunoblot (IB) with PSSG antibody (n = 4). The kinetics of the S-glutathionylation reaction were analyzed using a standard 2-parameter exponential rise to maximum fitting procedure (Sigma Plot 10, SyStat, MA) for dose dependence following GSSG or PABA/NO (C) for GSTπ+/+ (•) and GST-/- MEF (▴). Data represent mean ± variance; p < 0.01.
Nitrosative stress induced by PABA/NO treatment leads to S-glutathionylation of multiple proteins, 10 of which were identified in a prior report (14). Total protein S-glutathionylation (e.g. proteins ranging from ∼30 to ∼200 kDa) was quantitated following a 25 μm PABA/NO treatment where there were 7-fold more in MEF-GSTπ+/+ compared with GSTπ-/- cells (307 ± 72 versus 44 ± 6 arbitrary units, p < 0.01) (Fig. 2). Maximal PABA/NO-induced S-glutathionylation of proteins in MEF-GSTπ+/+ cells was evident at 30 min, whereas MEF-GSTπ-/- cells had no detectable levels of modified proteins at this time (Fig. 1).
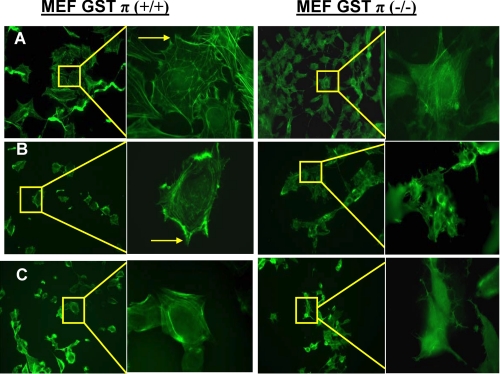
Oxidative stress induced by GSSG treatment produced a dose- and time-dependent S-glutathionylation of a single 42-kDa protein that we previously identified as actin (14). Maximal S-glutathionylation of actin was detected at 300 μm GSSG (Fig. 2), was 1.8-fold higher in MEF-GSTπ+/+ (p < 0.01), and occurred maximally at 15 min compared with 1 h for MEF-GSTπ-/- cells. Individual globular actin (G-actin) subunits polymerize to form filamentous actin (F-actin). S-Glutathionylation of G-actin subunits prevents its polymerization, and decreases both stress fiber and focal contact formation. We examined the morphological effects of GSSG-induced S-glutathionylated actin in MEF-GSTπ+/+ or MEF-GSTπ-/- cells. Phalloidin staining shows that in MEF-GSTπ+/+ there is a decrease in formation of both stress fibers and focal contacts (Fig. 3) compared with MEF-GSTπ-/- cells. These morphological data are quite consistent with enhanced S-glutathionylation of cellular G-actin as a consequence of GSTπ.
FIGURE 3.
S-Glutathionylation of actin is elevated in MEF-GST+/+ cells. MEF-GST+/+ and MEF-GST-/- cells were seeded on glass coverslips and treated with: A, vehicle; B, 300 μm GSSG; C, 15 μm PABA/NO for 1 h and stained with phalloidin to visualize actin polymerization/stress fiber formation. Arrow highlights focal contacts.
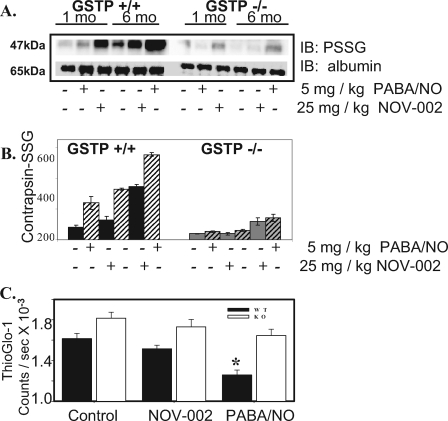
GSTπ-/- Mice Have Reduced Levels of an S-Glutathionylated Serum Protein, Contrapsin, following Drug Treatment—One- and three-month-old male GSTP1P2+/+ and GST1P2-/- mice were used as an in vivo model to evaluate GST-mediated S-glutathionylation of serum and liver proteins. Mice were treated with a single dose of either 5 mg/kg PABA/NO or 25 mg/kg NOV-002 (Fig. 4, panel A). NOV-002 treatment leads to enhanced S-glutathionylation of contrapsin in serum in a dose- and time-dependent manner. S-Glutathionylation of contrapsin was evaluated in serum 1 h following treatment. The averaged relative density of S-glutathionylated contrapsin bands (Fig. 4, panel B) following PABA/NO and NOV-002 treatment at 1 h was significantly higher in GSTP1P2+/+ mice compared with GSTP1P2-/- (n = 6, p < 0.01).
FIGURE 4.
S-Glutathionylation of serum and liver proteins in GSTπ WT and KO mice. Male GSTπ wild-type (GSTP1P2+/+) and knock-out (GSTP1P2-/-) mice (ages 1 and 3 months) were treated with an intravenous bolus of the oxidized glutathione mimetic, NOV-002 at 25 mg/kg or PABA/NO at 5 mg/kg. Blood was collected at various time points via orbital bleed. The plasma proteins (A) were separated by non-reducing SDS-PAGE and S-glutathionylated contrapsin was evaluated by immunoblot (IB) with PSSG antibody. The ratios of S-glutathionylated contrapsin to albumin were plotted in arbitrary units (B), solid, 1-month-old; hatched, 3-month-old animals (6 animals per treatment group). Free protein sulfhydryl content was measured in the liver homogenates of 3-month-old GSTπ wild-type (+/+) and knock-out (-/-) mice (C) using the fluorescent sulfhydryl-specific probe ThioGlo-1. The ThioGlo-1 emission (at 513 nm) for each treatment group was averaged and plotted as mean ± S.D. (n = 3); solid bars represent GSTπ WT (+/+) and open bars represent KO (-/-) animals. The difference between WT and KO treated and untreated animals is statistically significant (*, p ≤ 0.001).
Analysis of Cysteine Modification in Liver Tissue—The fluorescent probe, ThioGlo-1 specifically reacts with sulfhydryls (e.g. unmodified cysteine residues) and was used to assess hepatic proteins following PABA/NO and NOV-002 treatment in 3-month-old GSTP1P2+/+ and GST1P2-/- mice (Fig. 4, panel C). Reduced ThioGlo-1 emission corresponding with decreased protein sulfhydryl concentration (e.g. cysteine modification) was observed following PABA/NO and NOV-002 treatment in the liver homogenates of GSTP1P2+/+ mice (control, 16,152 ± 509; NOV-002, 15,155 ± 352; PABA/NO, 12,583 ± 476, p < 0.001). Protein sulfhydryl modification in the liver was greater for the PABA/NO-treated (∼22%) versus NOV-002-treated (∼6%) animals (n = 3, p < 0.001). These data are consistent with immunoblot analysis showing a large number of S-glutathionylated cellular proteins following PABA/NO treatment, whereas oxidized glutathione treatment led to S-glutathionylation of actin. However, no statistical differences were detected in the liver lysate from GSTP1P2-/- mice following treatment. Our data indicate that GSTπ expression quantitatively influences oxidative/nitrosative stress-mediated protein S-glutathionylation in vivo.
The Enzymatic Activity of GSTπ Is Crucial for Protein S-Glutathionylation in Vivo—The third model system was designed to test if the enzymatic activity of GSTπ is required for S-glutathionylation reactions. HEK293 cells were transfected with either wild-type (HEK-WT) or a catalytically inactive mutant (23) (HEK-Y7F) of GSTπ. To validate the model system, a general GST substrate (CDNB) was used to measure the specific activity (Table 1). Mock-transfected cells (HEK-VA) had GST specific activity of 59 ± 3 units/mg, whereas HEK-WT cells were significantly higher (75 ± 2 units/mg; p < 0.01). The specific activity of GST was significantly reduced in HEK-Y7F cells (12 ± 5 units/mg; p < 0.01), presumably as a consequence of a dominant negative effect (the catalytic activity of GST is attributed to the dimeric form). Residual activity can be attributed to other isoforms of GST present in HEK293 cells. This model system was then used to demonstrate that GSTπ activity caused the observed dose- and time-dependent increase in protein S-glutathionylation (Fig. 5). The dose-dependent studies demonstrated that at 15 μm PABA/NO, the amount of S-glutathionylated proteins following treatment was substantially higher in HEK-WT cells as compared with untreated controls (p < 0.05); whereas HEK-VA and HEK-Y7F were equivalent to untreated controls. The dose dependence of PABA/NO-induced S-glutathionylation in this model system (Fig. 5, panel C) shows similar kinetics as in MEF cells. The presence of enzymatically active GSTπ dramatically amplified S-glutathionylation. Temporal differences in S-glutathionylation patterns were also observed following a single dose of 50 μm PABA/NO (Fig. 5, panel B). Within 5 min, the relative levels of S-glutathionylation were saturated in HEK-WT cells (389 ± 12 arbitrary units, p < 0.001), whereas HEK-VA and HEK-Y7F cells were essentially equivalent to untreated controls (82 ± 9 and 63 ± 9 arbitrary units, p > 0.05). The HEK-Y7F cells (catalytically impaired GSTπ) had S-glutathionylation patterns that were below HEK-VA, suggesting a dominant negative effect. Kinetic differences of the HEK-293 model system (Fig. 5, panel C) compared with that of the MEFs (Fig. 2, panel C) may be a specific property attributable to basal S-glutathionylation in HEK293 cells.
TABLE 1.
Effect of S-glutathionylation on GST enzymatic activity The values represent the mean of 3 independent experiments ± S.D.
| GST specific activity | |
|---|---|
| μmol/mg protein/min | |
| Cells | |
| HEK-VA | 59.3 ± 3.2 |
| HEK-WT | 75.3 ± 2.1a |
| HEK-Y7F | 12.4 ± 5.1a |
| HEK-C47A | 12.8 ± 1.8a |
| HEK-C101A | 13.8 ± 0.8a |
| Purified protein | |
| GST | 53.2 ± 1.6 |
| GST + NOV-002 | 49.1 ± 2.3 |
| GST + PABA/NO | 13.0 ± 0.9a |
| GST(Y7F) | 4.5 ± 0.1a |
| GST(C47A) | 5.3 ± 2.5a |
| GST(C101A) | 10.1 ± 0.9a |
p < 0.01
FIGURE 5.
GSTπ enzymatic activity is crucial for protein S-glutathionylation. HEK293 cells were transiently transfected with vector (HEK-VA), wild-type GSTπ (HEK-WT), or an enzymatically inactive mutant form of GSTπ (HEK-Y7F). Concentration (A) and time dependence (B) effects following PABA/NO treatment illustrate that increased ectopic expression of GSTπ stimulates, whereas mutation of the catalytic tyrosine in the enzyme active site diminishes S-glutathionylation (PSSG). The corresponding relative abundance of modified proteins was plotted as the fold increase compared with untreated control for concentration (C) and time (D) dependence. Experimental data were fitted with standard sigmoid, 2-parameter exponential rise to maximum (Sigma Plot 10, SyStat). HEK-WT cells, •; HEK-VA cells, ▪; and HEK-Y7F cells, ▴; n = 3; p < 0.01. IB, immunoblot.
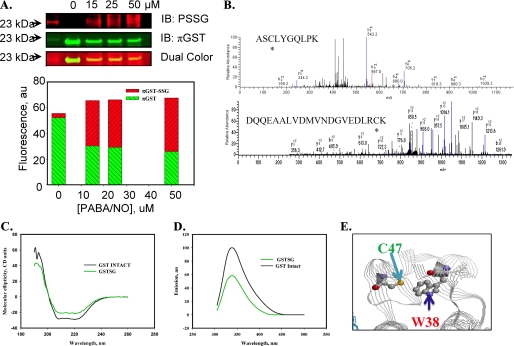
Autoregulation of GSTπ through S-Glutathionylation in Vitro and in Cells—Prior studies have shown that oxidative stress induces multimerization and inactivation of GSTπ (4). In determining the kinetics of GSTπ-catalyzed S-glutathionylation reactions, we discovered that GSTπ itself becomes S-glutathionylated under nitrosative stress conditions and this modification leads to its inactivation and multimerization. GSTπ is S-glutathionylated in HEK293-WT cells treated with 15–50 μm PABA/NO for 1 h (Fig. 6, panel A). Our data show that a band in lysates of PABA/NO-treated cells contains both intact and S-glutathionylated GSTπ (GST-SSG). In our present study we used polyclonal and monoclonal primary antibodies for GSTπ and S-glutathionylated proteins, respectively. The green and red fluorescently labeled secondary anti-rabbit and anti-mouse antibodies allowed dual-color infrared-excited fluorescence detection of co-localized red (PSSG; Fig. 6, panel A, upper image) and green (GSTπ; Fig. 6, panel A, middle image) fluorescence in one band (Fig. 6, panel A, lower image). The yellow band representing S-glutathionylated GSTπ has a lower intensity when compared with unmodified GST (green, middle image). These data indicate that only a fraction of GSTπ becomes S-glutathionylated under our experimental conditions. The dual colored image was quantified using standard Odyssey software (LI-COR, NE). The bar diagram represents the relative input of red and green fluorescence in each band. The relative portion of red fluorescence (associated with GST-SSG) is increased and green fluorescence (associated with unmodified GSTπ) is decreased proportional to PABA/NO treatment. In addition, S-glutathionylation of GSTπ is lower in the cells carrying the catalytically inactive mutant (Y7F, not shown), suggesting that GSTπ can catalytically mediate self-S-glutathionylation resulting in autoregulation of this enzyme.
FIGURE 6.
Autoregulation of GSTπ occurs through the Cys47 and Cys101S-glutathionylation. HEK293-WT cells were treated with 0–50 μm PABA/NO for 1 h (A). The proteins were separated by non-reducing SDS-PAGE and S-glutathionylation was evaluated by immunoblot (IB) with PSSG monoclonal primary antibody and GSTπ polyclonal primary antibody and detected simultaneously with both red (anti-mouse) and green (anti-rabbit) fluorescent secondary antibodies. The dual colored image was quantified using standard Odyssey (LI-COR, NE) software. The bars represent a relative input of red and green fluorescence in each band. MALDI-MS analysis of purified, expressed GSTπ treated with 50 μm PABA/NO and 0.5 mm GSH showed that peptides containing Cys47 and Cys101 are S-glutathionylated (B). S-Glutathionylation of Cys47 and Cys101 on GSTπ alters structure. Spectroscopic analysis of native (black) and PABA/NO + GSH-treated (green) GSTπ in vitro was performed using CD (C) and tryptophanyl fluorescence (D) of purified protein. According to the published crystal structure (31), the relative positions of Cys47 and Trp38 for GSTπ are depicted using RasMol 2.7.4.2 (E).
S-Glutathionylation of Cys47 and Cys101 of GSTπ Alters Protein Structure/Function—Tandem mass spectroscopy of S-glutathionylated GSTπ determined that Cys47 and Cys101 were modified by addition of GSH (∼305 Da) (Fig. 6, panel B). The effects of S-glutathionylation on the catalytic activity of GSTπ were evaluated in HEK293 cells and in vitro using purified protein (Table 1). The GST specific activity of either drug-treated recombinant GSTπ or C47A or C101A mutants of GSTπ (4.5–13 μmol/mg protein/min) was significantly diminished compared with wild-type (53.2 ± 1.6 μmol/mg of protein/min, Table 1). The mutated cysteine residues are not in the catalytic site and their modification may affect enzymatic activity through changes in protein structure. Therefore, we evaluated the effects of S-glutathionylation of GSTπ on its secondary structure. The CD spectrum (190–260 nm) of S-glutathionylated GSTπ was similar to native protein, and consistent with a small decrease in the α-helical content (206–220 nm) of the protein (Fig. 6, panel C). Based on the published crystal structure of GSTπ (11), the sulfhydryl of Cys47 is proximal to Trp38 (∼3 Å). Consequently, consistent with the known quenching effect of the disulfide, the intrinsic fluorescence of GSTπ was substantially decreased upon its S-glutathionylation (Fig. 6, panel D). These data indicate that disulfide bonds of the S-glutathionylated protein remain in proximity to Trp38, suggesting small structural changes associated with S-glutathionylation. Simultaneously, a hypsochromic shift of protein tryptophanyl emission maximum ∼4 nm after protein S-glutathionylation indicates some shielding of Trp38 from a polar environment through Cys47 S-glutathionylation (Fig. 6, panel E). This result is consistent with some structural changes that decrease enzymatic activity. Thus, our spectroscopic analysis shows an effect of GSTπ S-glutathionylation on its tertiary (quaternary) structure.
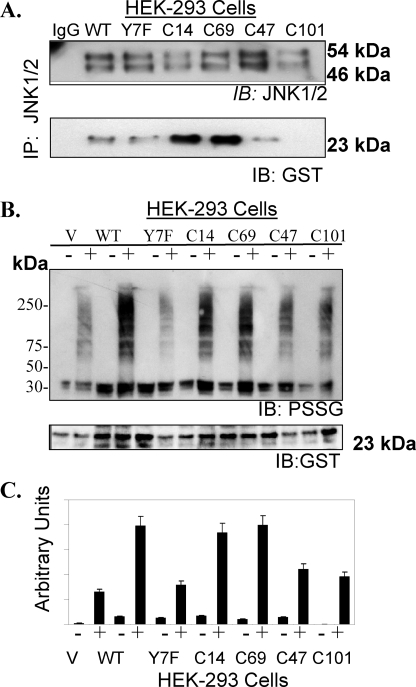
The cellular effects (e.g. catalytic activity and substrate binding) of nitrosative stress-induced S-glutathionylation or site-specific mutation of Cys47 and Cys101 were evaluated in HEK293 cells. The activity of GSTπ (CDNB, substrate) in the lysate of cells transfected with C47A, C101A mutant vectors was ∼15% of that for HEK-WT cells (Table 1). These data are consistent with decreased activity observed for S-glutathionylated-GSTπ. Figs. 1, 2, 3, 4, 5, 6 demonstrate that the catalytic activity of GSTπ in concert with oxidative or nitrosative stress are required for S-glutathionylation of multiple target proteins. Following 30 μm PABA/NO, HEK293-WT cells had ∼2-fold higher levels of S-glutathionylated proteins than HEK293-C47A or -C101A cells (Fig. 7, panel B).
FIGURE 7.
Protein-protein interactions of GSTπ with JNK were evaluated in HEK293 cells transfected with GSTπ (WT, Y7F, C47A, and C101A mutants). JNK1/2 was immunoprecipitated from HEK293 cells and separated by SDS-PAGE followed by immunoblot for GSTπ and JNK1/2 (A). Protein S-glutathionylation (PSSG antibody) patterns were evaluated in transfected HEK293 cells treated with vehicle or 30 μm PABA/NO for 1 h (B). The membranes were stripped and reprobed for GSTπ. The relative densities of S-glutathionylated proteins bands were plotted as mean ± S.D., of at least three independent experiments (C). IB, immunoblot.
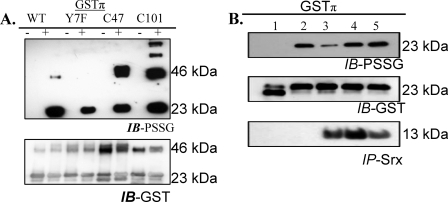
In non-stressed cells, protein-protein interactions between GSTπ and JNK are important and maintain both in an inactive state. However, oxidative stress leads to dissociation of the complex and activation of both proteins (4, 5). Immunoprecipitation of JNK1/2 in HEK293 cells demonstrated that Cys101 in GSTπ is critical for these protein-protein interactions (Fig. 7, panel A). Glutaredoxin and sulfiredoxin can mediate the deglutathionylation of proteins (10, 24, 25). PABA/NO induced S-glutathionylation of purified, recombinant GSTπ in vitro (Fig. 8, panel B). When recombinant, catalytically active hSrx1 was added to the extent that the modification was decreased. However, heat-inactivated hSrx1 did not cause this effect. Mutation of the only cysteine residue in sulfiredoxin (hSrx-C99A) resulted in failure to deglutathionylate GSTπ. Our data suggest that S-glutathionylation of Cys47 and/or Cys101 of GSTπ interferes with complex formation with other proteins. Where this impacts on the JNK-GST interaction, the result may be an alteration in cellular JNK signaling events.
FIGURE 8.
S-Glutathionylation of GSTπ in vitro. Recombinant human purified GSTπ (WT, Y7F, C47A, and C101A) was treated with 50 μm PABA/NO and 0.5 mm GSH for 1 h (A). The proteins were separated by non-reducing SDS-PAGE and S-glutathionylation was evaluated by immunoblot (IB) with PSSG antibody. Membranes were stripped and reprobed for GSTπ. Deglutathionylation of GSTπ by hSrx1 is shown in B. GSTπ was treated with: 1, control; 2, 50 μm PABA/NO and 0.5 mm GSH for 1 h; 3, 50 μm PABA/NO and 0.5 mm GSH for 30 min followed by addition of hSrx1 and further incubation 30 min; 0–50 μm PABA/NO and 0.5 mm GSH for 30 min followed by addition of heat-inactivated hSrx1 and further incubation 30 min; 5–50 μm PABA/NO and 0.5 mm GSH for 30 min followed by the addition of C99A mutant of hSrx1 and further incubation 30 min. IP, immunoprecipitation.
DISCUSSION
Despite the evidence that GSTπ is one of the more prevalent cytosolic proteins in many tumors (in particular ovarian, non-small cell lung, breast, colon, pancreas and lymphomas (26)), assigning functional role(s) for the protein has proved elusive. Our present results are the first to show that it is a catalytic determinant of the forward reaction of S-glutathionylation of proteins, contributing to the regulation of a functional S-glutathionylation cycle. High levels of GSTπ have been associated with carcinogen-induced transformation of such diverse tissues as liver and tongue (27) and GST can be regarded as a tumor marker. Although GSTπ knock-out mice are viable, they are more susceptible to certain carcinogens (19) and GSTπ knock-out mouse embryo fibroblasts have faster growth rates than their wild-type counterparts (21). The association of GSTπ with proliferative pathways is not explained by the detoxification properties of the protein. In general, the catalytic activity of GSTπ is modest for any of a number of identified substrates, particularly anticancer drugs. The overexpression of GSTπ frequently accompanies selection for drug resistance (2), even when the selecting drug is not a substrate for catalyzed thioether bond formation with GSH. Reports of protein-protein interactions between GSTπ and regulatory kinases (4) have identified a role for GSTπ in stress response pathways and perhaps provide a partial explanation for its high levels. Our present data are novel in that they provide a further important cellular process to which GSTπ contributes. The S-glutathionylation of target proteins is critical, both in protecting cells against oxidative stress and in altering structure/function and cellular localization of modified proteins. S-Glutathionylation does occur in the absence of GSTπ (in the null background) and may be regarded as a spontaneous reaction. However, the rate of S-glutathionylation is significantly enhanced in the presence of GSTπ. Our data show that GSTπ deletion results in enhanced stress fibers and focal adhesion formation under oxidative and nitrosative stress for mouse embryonic fibroblasts in tissue culture (Fig. 3). This is likely to influence transmembrane signal transduction to the cell surface. Because the rapidity of response to stress conditions is likely to be an important determinant of cell survival, such a rapid response may be critical.
The catalytic site of GSTπ is required for the S-glutathionylation reaction to occur. For the catalytic mechanism of GSTπ, the nucleophilicity of the sulfydryl group of GSH is enhanced through abstraction of a proton by Tyr7 (23) and this facilitates the formation of the thioether bond with electrophilic sites on putative substrates. Mutation of the Tyr7 residue interfered with the S-glutathionylation reaction. In this regard, the formation of a disulfide bond between the thiolate anion of GSH, bound to GSTπ, and the sulfhydryl (sulfenic acid) of specific cysteine(s) of the target protein appears to be a heretofore unreported physiologically relevant activity of GSTπ. The data suggest that the cysteine residues of GSTπ are of less consequence to the S-glutathionylation reaction, but are involved in protein complex formation with JNK and perhaps reflect the functional importance of different domains for the protein active conformation.
The two drugs used in these experiments have distinct mechanisms. NOV-002 provides a stable formulation of oxidized glutathione (GSSG) and can alter the intracellular redox balance through modification of GSH:GSSG ratios (16). PABA/NO is a GSTπ-activated prodrug that releases nitric oxide (28). The convergence of their pharmacology is the production of reactive oxygen species and subsequent alteration in redox balance resulting in a stress-oriented response. NOV-002 is presently under clinical development as a chemoprotectant that stimulates myeloproliferation. This physiological effect is reasonable given that mild oxidative stress is linked with increases in cell proliferation or differentiation. Although some pathways involving fyn kinase and c-Cbl have been implicated as broad determinants in the reactive oxygen species-driven division/differentiation of some progenitor cells of oligodendrocytes (29), the generality of these pathways in controlling cell proliferation is yet to be determined.
There are a number of instances where sulfur and phosphorus biochemistry may coalesce. For example, S-glutathionylation of a number of phosphatases that are critical to reversing kinase effects have been reported (30). Indeed, a number of proteins that are S-glutathionylated are frequently involved in growth regulatory pathways (15), further emphasizing the potential importance of GSTπ in mediating the forward reaction. The S-glutathionylation cycle may prove to be as critical in determining cellular regulatory/stress responses as the phosphorylation cascades governed by kinases or phosphatases.
The S-glutathionylation of GSTπ resulted in changes in its secondary structure (Fig. 6). These can be interpreted as a decrease of α-helical content in secondary structure as a consequence of a decreased number of disulfide bonds in the S-glutathionylated protein with consequent impact on tertiary and quaternary structure. GSTπ has only two tryptophan residues: Trp28 and Trp38. The Trp38 is ∼3.2 Å from the sulfhydryl of Cys47, where S-glutathionylation can quench tryptophan fluorescence (25). The treatment of GSTπ with PABA/NO and GSH resulted in a substantial decrease (∼50%) in fluorescence (Fig. 6, panel D), which correlates with site-specific S-glutathionylation of Cys47. This Cys47 residue of GSTπ could become oxidized to sulfenic or sulfinic acid, neither of which would influence Trp38 fluorescence. Presumably the thiolate or sulfenic states of Cys47 are most susceptible to S-glutathionylation. Thus, our data show that purified GSTπ used in this study is active, corresponding with our enzymatic analysis (Fig. 5). In addition, the tryptophan fluorescence of S-glutathionylated GSTπ shows a shift of emission maximum (∼4 nm) to a short wavelength, indicating a less polar environment around this residue. Thus, S-glutathionylation causes substantial changes in GSTπ tertiary and quaternary structure and suggests specific interactions with the target protein for catalytic S-glutathionylation.
In summary, in response to reactive oxygen/nitrogen species generated either by non-toxic (NOV-002) or toxic (PABA/NO) drugs, cells evoke a rapid and robust S-glutathionylation of target proteins. The rate of protein S-glutathionylation is significantly enhanced by the presence of a catalytically active GSTπ. Catalysis of this post-translational modification by a proximal donor of GS- and the low pK cysteine thiol of target proteins is a novel property for GSTπ and is an important forward reaction in the S-glutathionylation cycle. In context with the pleiotropic functions now ascribed to GSTπ, high expression levels in tumor cells may be a reflection of the aberrant redox pathways that can be cause/effect related to the transformation process.
Acknowledgments
We thank the Drug Metabolism and Pharmacokinetics, X-ray Crystallography and Proteomics Core Facilities at the Medical University of South Carolina. This work was conducted in a facility constructed with support by National Institutes of Health Grant C06 RR015455 from the Extramural Research Facilities Program of the National Center for Research Resources.
This work was supported, in whole or in part, by National Institutes of Health Grants CA08660 and CA117259. This work was also supported by the South Carolina Centers of Excellence program. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: GST, glutathione S-transferase; JNK, c-Jun NH2-terminal kinase; MALDI-TOF, matrix-assisted laser desorption ionization time-of-flight; NO, nitric oxide; MEF, mouse embryo fibroblasts; HEK, human embryonic kidney; CDNB, 1-chloro-2,4-dinitrobenzene; PBS, phosphate-buffered saline; WT, wild type; CD, circular dichroism; PABA, O2-{2,4-dinitro-5-[4-(N-methylamino)benzoyloxy]phenyl}1-(N,N-dimethylamino)diazen-1-ium-1,2-diolate.
Findlay, V. J., Townsend, D. M., Morris, T. E., Fraser, J. P., He, L., and Tew, K. D. (2006) Cancer Res. 66, 6800–6806
References
- 1.Boyland, E., and Chasseaud, L. F. (1967) Biochem. J. 104 95-102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tew, K. D. (1994) Cancer Res. 54 4313-4320 [PubMed] [Google Scholar]
- 3.Hayes, J. D., and Pulford, D. J. (1995) Crit. Rev. Biochem. Mol. Biol. 30 445-600 [DOI] [PubMed] [Google Scholar]
- 4.Adler, V., Yin, Z., Fuchs, S. Y., Benezra, M., Rosario, L., Tew, K. D., Pincus, M. R., Sardana, M., Henderson, C. J., Wolf, C. R., Davis, R. J., and Ronai, Z. (1999) EMBO J. 18 1321-1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang, T., Arifoglu, P., Ronai, Z., and Tew, K. D. (2001) J. Biol. Chem. 276 20999-21003 [DOI] [PubMed] [Google Scholar]
- 6.Manevich, Y., Feinstein, S. I., and Fisher, A. B. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 3780-3785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lo, H. W., Pleasants, L., Cao, X., Milas, M., Pollock, R., and Ali-Osman, F. (2008) Mol. Cancer Res. 6 848-850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shelton, M. D., Chock, P. B., and Mieyal, J. J. (2005) Antioxid. Redox Signal. 7 348-366 [DOI] [PubMed] [Google Scholar]
- 9.Mack, J. T., Beljanski, V., Soulika, A. M., Townsend, D. M., Brown, C. B., Davis, W., and Tew, K. D. (2007) Mol. Cell. Biol. 27 44-53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beer, S. M., Taylor, E. R., Brown, S. E., Dahm, C. C., Costa, N. J., Runswick, M. J., and Murphy, M. P. (2004) J. Biol. Chem. 279 47939-47951 [DOI] [PubMed] [Google Scholar]
- 11.Ji, X., Tordova, M., O'Donnell, R., Parsons, J. F., Hayden, J. B., Gilliland, G. L., and Zimniak, P. (1997) Biochemistry 36 9690-9702 [DOI] [PubMed] [Google Scholar]
- 12.Ralat, L. A., Manevich, Y., Fisher, A. B., and Colman, R. F. (2006) Biochemistry 45 360-372 [DOI] [PubMed] [Google Scholar]
- 13.Rhee, S. G. (2006) Science 312 1882-1883 [DOI] [PubMed] [Google Scholar]
- 14.Townsend, D. M., Findlay, V. J., Fazilev, F., Ogle, M., Fraser, J., Saavedra, J. E., Ji, X., Keefer, L. K., and Tew, K. D. (2006) Mol. Pharmacol 69 501-508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Townsend, D. M. (2007) Mol. Interv. 7 313-324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Townsend, D. M., He, L., Hutchens, S., VandenBerg, T. E., Pazoles, C. J., and Tew, K. D. (2008) Cancer Res. 68 2870-2877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saavedra, J. E., Srinivasan, A., Buzard, G. S., Davies, K. M., Waterhouse, D. J., Inami, K., Wilde, T. C., Citro, M. L., Cuellar, M., Deschamps, J. R., Parrish, D., Shami, P. J., Findlay, V. J., Townsend, D. M., Tew, K. D., Singh, S., Jia, L., Ji, X., and Keefer, L. K. (2006) J. Med. Chem. 49 1157-1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Findlay, V. J., Townsend, D. M., Saavedra, J. E., Buzard, G. S., Citro, M. L., Keefer, L. K., Ji, X., and Tew, K. D. (2004) Mol. Pharmacol 65 1070-1079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Henderson, C. J., Smith, A. G., Ure, J., Brown, K., Bacon, E. J., and Wolf, C. R. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 5275-5280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rosario, L. A., O'Brien, M. L., Henderson, C. J., Wolf, C. R., and Tew, K. D. (2000) Mol. Pharmacol 58 167-174 [DOI] [PubMed] [Google Scholar]
- 21.Ruscoe, J. E., Rosario, L. A., Wang, T., Gate, L., Arifoglu, P., Wolf, C. R., Henderson, C. J., Ronai, Z., and Tew, K. D. (2001) J. Pharmacol. Exp. Ther. 298 339-345 [PubMed] [Google Scholar]
- 22.Boyland, E., and Chasseaud, L. F. (1969) Adv. Enzymol. Relat. Areas Mol. Biol. 32 173-219 [DOI] [PubMed] [Google Scholar]
- 23.Dirr, H., Reinemer, P., and Huber, R. (1994) Eur. J. Biochem. 220 645-661 [DOI] [PubMed] [Google Scholar]
- 24.Gallogly, M. M., and Meyal, J. J. (2007) Curr. Opin. Pharmacol. 7 381-391 [DOI] [PubMed] [Google Scholar]
- 25.Peltoniemi, M. J., Karala, A. R., Jurvansuu, J. K., Kinnula, V. L., and Ruddock, L. W. (2006) J. Biol. Chem. 281 33107-33114 [DOI] [PubMed] [Google Scholar]
- 26.Tew, K. D. (2007) Biochem. Pharmacol 73 1257-1269 [DOI] [PubMed] [Google Scholar]
- 27.Tanaka, T., Makita, H., Kawabata, K., Mori, H., and El-Bayoumy, K. (1997) Cancer Res. 57 3644-3648 [PubMed] [Google Scholar]
- 28.Keefer, L. K. (2003) Annu. Rev. Pharmacol. Toxicol. 43 585-607 [DOI] [PubMed] [Google Scholar]
- 29.Li, Z., Dong, T., Proschel, C., and Noble, M. (2007) PLoS Biol. 5 e35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barrett, W. C., DeGnore, J. P., Konig, S., Fales, H. M., Keng, Y. F., Zhang, Z. Y., Yim, M. B., and Chock, P. B. (1999) Biochemistry 38 6699-6705 [DOI] [PubMed] [Google Scholar]