Abstract
Strains of uropathogenic Escherichia coli (UPEC) encode filamentous adhesive organelles called type 1 pili that promote bacterial colonization and invasion of the bladder epithelium. Type 1 pilus-mediated interactions with host receptors, including α3β1 integrin, trigger localized actin rearrangements that lead to internalization of adherent bacteria via a zipper-like mechanism. Here we report that type 1 pilus-mediated bacterial invasion of bladder cells also requires input from host microtubules and histone deacetylase 6 (HDAC6), a cytosolic enzyme that, by deacetylating α-tubulin, can alter the stability of microtubules along with the recruitment and directional trafficking of the kinesin-1 motor complex. We found that disruption of microtubules by nocodazole or vinblastine treatment, as well as microtubule stabilization by taxol, inhibited host cell invasion by UPEC, as did silencing of HDAC6 expression or pharmacological inhibition of HDAC6 activity. Invasion did not require two alternate HDAC6 substrates, Hsp90 and cortactin, but was dependent upon the kinesin-1 light chain KLC2 and an upstream activator of HDAC6, aurora A kinase. These results indicate that HDAC6 and microtubules act as vital regulatory elements during the invasion process, possibly via indirect effects on kinesin-1 and associated cargos.
Invasion of epithelial cells and other nonprofessional phagocytes facilitates the dissemination, growth, and persistence of many bacterial pathogens within their hosts. The invasion process requires either direct or indirect manipulation of host cytoskeletal dynamics by the incoming pathogens (1). Some bacteria, like the enteric pathogens Salmonella enterica and Shigella flexneri, inject effector molecules into target host cells in order to directly modulate actin polymerization and dynamics. The resultant cytoskeletal rearrangements trigger intense ruffling of the host plasma membrane and subsequent internalization of adherent bacteria. Alternately, invasive bacteria like Listeria monocytogenes and Yersinia pseudotuberculosis express so-called invasin proteins that bind host cell receptors such as integrins, promoting receptor clustering and stimulating localized actin rearrangements. This results in the formation of host plasma membrane extensions that zipper around and engulf the bound bacteria. Invasion of host cells via a zipper-like mechanism requires that the pathogen tap into host signaling networks that are normally activated during host cell adhesion, polarization, or migration.
Among bacteria that enter host cells by a zipper-like mechanism are strains of uropathogenic E. coli (UPEC)2 (2–4). These microbes are the major cause of urinary tract infections (UTIs), accounting for greater than 80% of cases worldwide (5, 6). Traditionally, UPEC strains were considered strictly extracellular pathogens. However, more recent work has demonstrated that UPEC can invade the epithelial cells that line the lumenal surfaces of the urinary tract (7). In mouse UTI models, intracellular UPEC have a notable survival advantage over their extracellular counterparts, and are able to persevere within the urinary tract for extended periods in the face of robust innate host defenses and even antibiotic treatments (3, 8–13). It has been speculated that these intracellular reservoirs of UPEC can act as a source for the recurrent and chronic UTIs that afflict many individuals throughout their lives (3, 9, 14).
To gain entry into uroepithelial cells, UPEC utilize peritrichously expressed filamentous adhesive organelles known as type 1 pili (2). These are composite fibers made up of a 7-nm thick rod comprised of repeating FimA subunits with a distally attached 3-nm-wide tip fibrillum structure consisting of two adaptor molecules, FimF and FimG, and the adhesin FimH (15, 16). The FimH adhesin binds mannose-containing glycoprotein receptors and is sufficient to stimulate bacterial uptake by bladder epithelial cells (2). Interactions between FimH and host receptors like α3β1 integrin activate a signaling cascade involving focal adhesion, Src, and phosphoinositide 3-kinases, as well as Rho-family GTPases and the actin bundling and adaptor proteins α-actinin and vinculin (2, 17, 18). The coordinated activation of these host factors drives localized rearrangements of the actin cytoskeleton, leading to the envelopment and internalization of bound UPEC. This is a cholesterol-dependent process that also requires clathrin, the canonical clathrin adaptor AP-2, and at least three alternate endocytic adaptors, Numb, ARH, and Dab2 (19).
A growing number of studies have shown that the dynamics of the actin cytoskeleton can be modulated by microtubules (20–22). For example, microtubule depolymerization can stimulate the formation of actin stress fibers and focal adhesions, as observed at sites of integrin attachment to the extracellular matrix (23, 24), while the polymerization of microtubules toward focal adhesions correlates with focal adhesion disassembly (25). This latter process is proposed to involve the delivery of so-called “relaxing” factor(s) to focal adhesions via microtubule-associated kinesin motor proteins (25, 26). The potential for cross-talk between the actin and microtubule cytoskeletal networks led us to ask if microtubules affect the integrin-mediated actin-dependent entry of type 1-piliated UPEC into host bladder epithelial cells. Our results indicate that microtubules, along with the microtubule-associated enzyme histone deacetylase 6 (HDAC6), the HDAC6 activator Aurora A kinase, and the kinesin-1 motor protein complex, have crucial regulatory roles during the invasion process.
EXPERIMENTAL PROCEDURES
Bacteria, Cell Culture, and Drugs—The UPEC cystitis isolate UTI89 was grown in static Luria-Bertani (LB) broth (Difco) at 37 °C for 48 h to induce type 1 pili expression prior to use in infection assays (9, 27). S. flexneri (ATCC 12022) and S. enterica serovar Typhimurium (SL1344), kindly provided by Dr. O. Steele-Mortimer (Rocky Mountain Labs), were grown shaking in LB broth to mid-log as previously described (28), while HB101/pRI203 was similarly grown to stationary phase (29). The human bladder epithelial cancer cell lines designated 5637 (ATCC HTB-9) and T24 (ATCC HTB-4) were maintained in RPMI 1640 and McCoy's 5A medium, respectively, supplemented with 10% heat inactivated fetal bovine serum (Hyclone). Normal primary human bladder epithelial cells were purchased from Lonza and maintained in Keratinocyte Growth Medium-2 (KGM-2) supplemented with a Single-Quots® Kit containing epidermal growth factor, bovine pituitary extract, insulin, hydrocortisone, epinephrine, and transferrin (Lonza). Paclitaxel/taxol, vinblastine sulfate, trichostatin A (TSA), sodium butyrate (NaB), nicotinamide (NA), and sodium orthovanadate were obtained from Sigma-Aldrich. Nocodazole and 17-AAG were purchased from Biomol, while aurintricarboxylic acid (ATA) and Aurora Kinase Inhibitor II (AKI II) were from Calbiochem. Tubacin and niltubacin were kind gifts from Drs. S. Schreiber and R. Mazitschek (Broad Institute of Harvard University and MIT).
Expression Plasmids—KLC2 cDNA was amplified from a human cDNA library using forward primers Klc2-F-Flag (5′-ctgaggatccatggattacaaggatgacgacgataagatggccatgatggtgtttcc-3′) or Klc2-F-HA (5′-ctgaggatccatgtacccatacgatgttccagattacgctatggccatgatggtgtttc-3′), and the reverse primer Klc2-R (5′-acgatgatatcttagcccaccagggagcttc-3′). Klc2-F-Flag and Klc2-F-HA were used to append sequences encoding N-terminal FLAG and HA tags, respectively. PCR products were digested using BamHI and EcoRV restriction enzymes and ligated into pcDNA3.1 (+) (Invitrogen) to create pKLC2_FLAG and pKLC2_HA. The sequence encoding HDAC6 was amplified by PCR from pBJ5-HDAC6 (kindly provided by S. Schreiber, Harvard University) using primer sets HDAC6-F (5′-attgcagatctatgacctcaaccggccaggattc-3′) and HDAC6-R (5′-ataatcgtcgactggtgtgggtggggcatatcct-3′). The PCR product was then digested using BglII and SalI restriction enzymes and ligated into pEGFP-N1 (Clontech) to generate pEGFP_HDAC6. All constructs were verified by sequencing.
Transfection and KLC2 Pull-down Assays—5637 cells were transfected with pKLC2_FLAG or pKLC2_HA using FuGENE® 6 Transfection Reagent (Roche Applied Science). 16–18 h later the transfected cells were treated with DMSO or TSA (300 nm) for 3 h and lysed in MT polymerization buffer (50 mm HEPES pH 7.4, 2.5 mm KCl, 2.5 mm MgCl2, 160 mm NaCl, 1.5 mm CaCl2, 1 mm phenylmethylsulfonyl fluoride, 1× protease inhibitor mixture (Roche Applied Science), and 1% Triton X-100) supplemented with DMSO or TSA. Lysates were centrifuged at 18,000 × g for 10 min at 4 °C to remove nuclei, unbroken cells, and large debris. Tubulin within the supernatants was polymerized by addition of 20 μm taxol and 1 mm GTP (Sigma-Aldrich) for 1 h at 33 °C. Polymerized microtubules and microtubule-associated proteins were pelleted by centrifugation at room temperature for 45 min at 18,000 × g. Pellets were then resuspended in MT polymerization buffer, boiled in 1× sample buffer, sonicated, and resolved by SDS-PAGE using 12.5% polyacrylamide gels prior to transfer to polyvinylidene difluoride membranes for Western blot analysis.
Protein Analysis—Western blots were performed and visualized using either enhanced chemiluminescence or an Odyssey Infrared Imaging System (LI-COR Biosciences) as previously described (18). Primary antibodies used include rabbit anti-HA (Bethyl Laboratories), rabbit anti-α-tubulin (Rockland), mouse anti-acetylated tubulin (clone 6-11B-1), and anti-FLAG M2 (Sigma-Aldrich), rabbit anti-cortactin (H222), anti-Aurora A (1G4), and anti-GEF-H1 (55B6, Cell Signaling), rabbit anti-HDAC6 (Santa Cruz Biotechnology), mouse anti-pan-actin, and anti-β-actin antibodies (Abcam).
Bacterial Invasion and Cell Association Assays—Gentamicin protection bacterial invasion and cell association assays were performed as described (18). 5637 cells were seeded into 24-well tissue culture plates and grown for 18–24 h to confluency. Two sets of triplicate wells of bladder cells were then treated with the indicated drugs or with carrier alone for the times specified prior to infection with UTI89 using a multiplicity of infection (MOI) of about 15 bacteria per host cell. Plates were spun at 600 × g for 5 min to expedite and synchronize bacterial contact with the host cell monolayers. After a 2-h incubation at 37 °C in the continued presence of drug or carrier, one set of wells was washed four times with PBS with added Mg+2 and Ca+2 (PBS-Mg+2/Ca+2) before being lysed in the same buffer containing 0.3% Triton X-100. Bacteria present in these lysates, representing the total number of host cell-associated bacteria, were enumerated by plating serial dilutions on LB agar plates. To determine numbers of internalized bacteria, after the initial 2-h incubation, the second set of triplicate wells was incubated for another 2 h in medium containing 100 μg/ml of gentamicin (Sigma-Aldrich) to kill any extracellular bacteria. Cells were then washed four times with PBS-Mg+2/Ca+2, lysed, and plated on LB-agar plates. In experiments using S. enterica, gentamicin was added for 1 h following an initial 30-min infection period. Levels of cell-associated and intracellular bacteria are expressed relative to untreated controls. All assays were repeated three or more times in triplicate.
Gene Silencing—Expression of cortactin (human CTTN), KLC2, and Rho GEF-H1 (human ARHGEF2) was silenced using Dharmacon ON-TARGET plus SMART pool siRNA oligos specific for each gene (Dharmacon). Oligos used to silence HDAC6 and Aurora A kinase have been described previously (30, 31), and were also purchased from Dharmacon. Nonspecific siRNA oligos were used as controls. 5637 bladder cells, grown in T25 flasks to a confluency of 70–80%, were transfected with oligos using DharmaFECT 1 reagent (Dharmacon). The cell culture medium was replaced 10–24 h post-transfection, and 24 h later the cells were seeded into 24-well plates for use in invasion and cell-association assays. Knockdown of Aurora A kinase, HDAC6, cortactin, and Rho GEF-H1 was assessed by Western blot analysis, while knockdown of KLC2 was verified by semi-quantitative RT-PCR using primers KLC2_for (5′-gaacatcctggcactggtct-3′) and KLC2_rev (5′-gccacatctgggtgaaactt-3′). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH), used as an internal control for RT-PCR, was amplified using primers GAPDH_for (5′-gagtcaacggatttggtcgt-3′) and GAPDH_rev (5′-ttgattttggagggatctcg-3′).
Microscopy—Bladder cells grown on 12-mm-diameter glass coverslips were washed with warm PBS-Mg+2/Ca+2 and fixed for 30 min at 37 °C in 3.7% paraformaldehyde/PBS following infection and/or treatment with the specified drugs or carrier alone. After 3 × 5-min washes with PBS, fixed samples were incubated for 30 min with blocking buffer (1% powder milk, 3% bovine serum albumin (BSA), and 0.1% Triton X-100 in PBS). Samples were stained for 1 h at room temperature with mouse anti-β tubulin (clone 2-28-33; Sigma-Aldrich) and rabbit or goat anti-E. coli (Biodesign International) in PBS containing 1% powdered milk and 3% BSA. Following 3 × 5 min washes in PBS, samples were incubated with secondary antibodies conjugated to Alexa488, -546, or -647 dyes (Molecular Probes) for 30 min. F-actin was detected using Alexa546-conjugated phalloidin (1:40, Molecular Probes). All samples were mounted, following final washes in PBS, using Fluorsave reagent (Calbiochem). Samples were visualized using either an Olympus IX70 FV300 or FV1000 confocal microscope equipped with ×60 and ×100 oil immersion objectives (Olympus PlanApo ×60 NA 1.42 and UPlanSApo ×100 NA 1.40 Oil immersion TIRFM) and argon and helium-neon (HeNe) lasers providing excitation energy at 488, 543, and 633 nm.
RhoA Activation Assays—Levels of RhoA activation in drug-treated and untreated 5637 cells were determined using a RhoA activation assay Biochem kit according to the manufacturer's instructions (Cytoskeleton, Inc.).
RESULTS
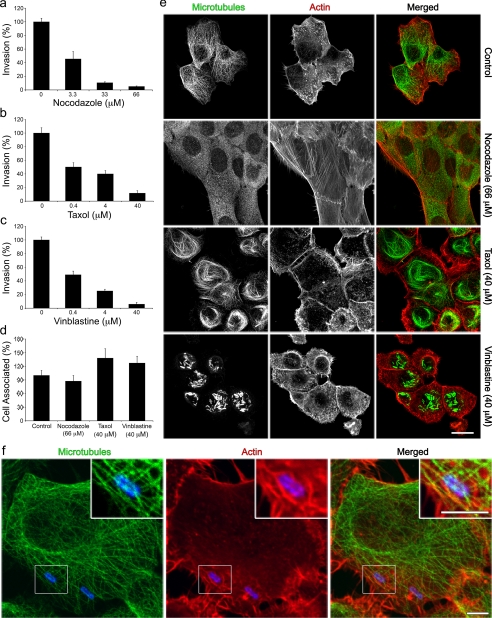
Microtubules Are Required for UPEC Invasion of Host Cells— A role for microtubules, which are made up of α- and β-tubulin heterodimers, in UPEC invasion of bladder epithelial cells was tested using three specific microtubule inhibitors: nocodazole, taxol, and vinblastine. Nocodazole binds β-tubulin and thereby stimulates microtubule depolymerization, while vinblastine induces depolymerization as well as aggregation of tubulin subunits (32, 33). Taxol, on the other hand, promotes the assembly and stabilization of microtubule filaments (34). Human bladder epithelial cells (ATCC 5637 cells) were incubated with increasing concentrations of each drug for 60 min prior to infection with the well-characterized type 1-piliated human cystitis isolate UTI89 (9, 27). Host cell invasion by UTI89 requires expression of type 1 pili, and specifically the FimH adhesin (2, 18). As determined by using standard gentamicin protection-based invasion assays, all three drugs were found to inhibit UTI89 invasion in a dose-dependent fashion (Fig. 1, a–c). None of the drugs affected bacterial viability or growth3 and none had any inhibitory effects on bacterial adherence to the bladder cells (Fig. 1d). Removal of the drugs just prior to infection made the bladder cells susceptible once again to invasion by UTI89, demonstrating that the effects of all three drugs were reversible (supplemental Fig. S1). Confocal microscopy verified that the microtubule filaments seen in control cells treated with carrier (DMSO) alone were completely disrupted by nocodazole treatment (Fig. 1e). In contrast, taxol caused the formation of elongated stabilized microtubules that arced through the cell interior beneath the cortical actin cytoskeleton; while vinblastine treatment led to the formation of perinuclear tubulin aggregates.
FIGURE 1.
Microtubule-dependent invasion of bladder cells by UPEC. 5637 bladder cell monolayers were treated with the indicated concentrations of (a) nocodazole, (b) taxol, or (c) vinblastine for 1 h prior to infection with the UPEC isolate UTI89. After a 2-h infection in the continued presence of drugs, intracellular (a–c) and total cell-associated (d) bacterial titers were determined. Data are expressed relative to untreated controls as the means ± S.E. of at least three independent experiments carried out in triplicate. e, confocal micrographs of bladder cells treated for 1 h with the indicated drugs or (f) infected for 30 min with UTI89 prior to staining to visualize microtubules (green), F-actin (red), and UPEC (blue). The boxed area in f was enlarged to show detail. Scale bars:(e) 20 μm and (f) 5 μm.
At low concentrations (≤300 nm), nocodazole can effectively perturb microtubule dynamics without causing substantial disruption of microtubule filaments (36). The decreasing effectiveness of low doses of nocodazole in our invasion assays (Fig. 1a) suggested that dynamic rearrangement of microtubules was not critical for UPEC invasion of host cells. Imaging of infected bladder cells, in the absence of drugs, supported this conclusion. Although we saw microtubules crisscrossing sites of UPEC entry, we could discern no specific recruitment of these filaments toward incoming bacteria (Fig. 1f).
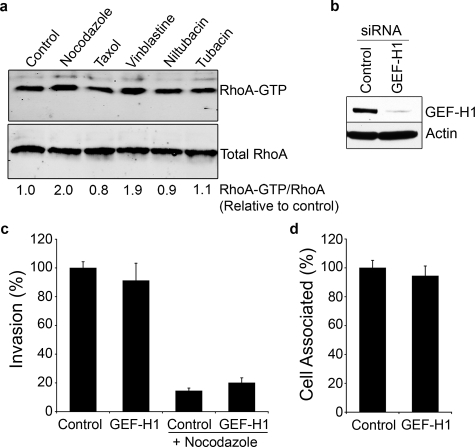
GEF-H1 Does Not Affect Host Cell Invasion by UPEC—In our assays we noted that nocodazole-induced disassembly of microtubules stimulated the formation of prominent actin stress fibers (Fig. 1e). These were rarely seen in untreated control bladder cells and were not obvious in cells treated with either taxol or vinblastine. The formation of actin stress fibers following treatment of other cell lines with nocodazole has been attributed to enhanced stimulation of Rho GTPases downstream of the microtubule-associated guanine nucleotide exchange factor GEF-H1, which can be activated following microtubule disassembly (24, 37–39). Relative to control and taxol-treated bladder cells, both nocodazole and vinblastine caused modest increases in the levels of activated RhoA-GTP, as determined using pull-down assays with the Rho binding domain of the RhoA effector rhotekin (Fig. 2a). These results suggested that aberrant activation of Rho GTPases downstream of GEF-H1 could be responsible for attenuating UPEC invasion following disruption of host microtubules. However, this does not appear to be the case since silencing of GEF-H1 expression using short interfering RNA (siRNA) had no discernable effect on the ability of UPEC to bind or invade bladder epithelial cells, regardless of nocodazole treatment (Fig. 2, b–d).
FIGURE 2.
Rho GEF-H1 activity is not necessary during UPEC entry. a, levels of activated RhoA-GTP, as determined by pull-down assays using the Rho binding domain of Rhotekin, were quantified relative to total RhoA in 5637 bladder cells following 30 min treatments with carrier alone (control), nocodazole (66 μm), taxol (40 μm), or vinblastine (40 μm), or 3 h treatments with 20 μm of either niltubacin or tubacin. b, Western blot showing levels of GEF-H1 in bladder cells 72 h after transfection with either control nonspecific siRNA or GEF-H1-specific siRNA. Blots were also probed using an anti-actin antibody to verify equal protein loading. Quantification of (c) internalized and (d) total cell-associated bacteria following infection of GEF-H1-silenced bladder cells by UTI89, with or without 33 μm nocodazole treatment. Data are expressed relative to control nonspecific siRNA-transfected samples as the means ± S.E. of at least three independent experiments carried out in triplicate.
HDAC6-mediated Invasion of Bladder Cells by UPEC—Microtubules are subject to a number of post-translational modifications, including acetylation, polyglutamylation, polyglycylation, detyrosination, and phosphorylation (40). Acetylation of the ε-amino group of a conserved lysine residue (Lys-40) within the N terminus of α-tubulin is associated with stabilized microtubules (41–43). While an acetyltransferase that modifies α-tubulin has not yet been identified, the cytoplasmically localized class IIb histone deacetylase HDAC6 has been shown to deacetylate this tubulin subunit (31, 44–46). Unlike other HDACs, HDAC6 has two functional deacetylase domains, one of which (TDAC) acts on α-tubulin (31, 47). Cycles of acetylation and deacetylation of α-tubulin are coupled with changes in the functionality and dynamic instability of microtubules and, consequently, may indirectly help regulate and coordinate actin-based cell functions, including the uptake of invading microbes (41, 48–52).
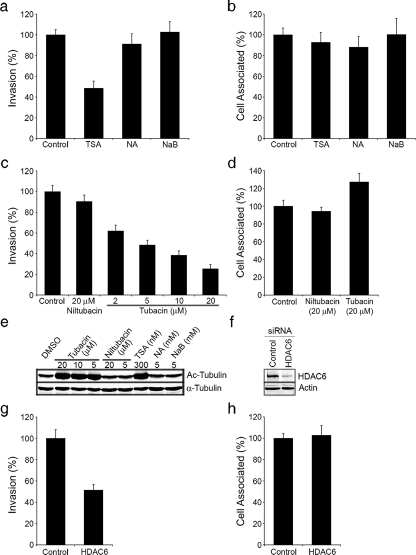
To address this possibility, we first employed a panel of HDAC inhibitors in bacterial invasion and cell association assays. The 18 known HDACs encoded by human cells are divided into four classes based on sequence homologies and domain organization (47). Trichostatin A (TSA) is a potent inhibitor of class I and class II HDACs, including HDAC6 (31). The ability of UTI89 to invade TSA-treated bladder cells was substantially reduced relative to cells treated with only the carrier DMSO (Fig. 3a). In contrast, the short-chain fatty acid sodium butyrate (NaB), a general HDAC inhibitor that does not affect the TDAC domain of HDAC6 (31, 53), had no effect on UTI89 invasion frequencies. The use of nicotinamide (NA) to inhibit Sirt2, a cytosolic protein that was reported to have tubulin deacetylase activity like HDAC6 (54), also failed to reduce UTI89 entry into bladder cells (Fig. 3a). Of note, the inhibitory effect of TSA on UTI89 internalization was not unique to 5637 cells. TSA treatment of either T24 human bladder cancer epithelial cells or primary normal human bladder epithelial cells (BdEC) significantly inhibited UTI89 invasion without affecting bacterial adherence, as seen with 5637 cells (supplemental Fig. S2).
FIGURE 3.
HDAC6 promotes UPEC entry into host bladder cells. 5637 bladder cells were pretreated for 3 h with (a and b) 300 nm TSA, 5 mm NA, or 5 mm NaB, or with (c and d) the indicated concentrations of tubacin and niltubacin. Cells were then infected with UTI89 for 2 h and (a and c) intracellular and (b and d) total cell-associated bacterial titers were determined relative to controls treated with carrier alone. e, Western blot analysis of bladder cells treated with the indicated inhibitors shows levels of both total and acetylated (Ac) α-tubulin. f, levels of HDAC6 in bladder cells 72 h after transfection with either control nonspecific siRNA or HDAC6-specific siRNA, as detected by Western blot. Blots were also probed using an anti-actin antibody to verify equal protein loading. Following a 2 h infection with UTI89, (g) intracellular and (h) total cell-associated bacterial titers recovered from siRNA-transfected cells were determined. Data in the graphs are expressed relative to appropriate controls as the means ± S.E. of at least three independent experiments carried out in triplicate.
These results implicate the TDAC domain of HDAC6 as a positive regulator of bladder cell invasion by UPEC. To further test this possibility we used tubacin, a specific small molecule inhibitor of the TDAC domain of HDAC6 (45). Tubacin blocked UTI89 invasion of 5637 bladder epithelial cells in a dose-dependent fashion, whereas cells treated with either carrier alone or with an inactive carboxylate analog of tubacin (niltubacin) had no effect (Fig. 3c). None of the drugs used in these assays affected UTI89 viability (data not shown) or adherence to the host bladder cells (Fig. 2, b and d). By Western blot analysis, only TSA and tubacin treatments increased levels of acetylated α-tubulin in the bladder epithelial cells (Fig. 3e), verifying the activity and specificity of the tested drugs. Notably, neither tubacin nor niltubicin affected RhoA activation (Fig. 2a), indicating that HDAC6 inhibition is likely not interfering with UPEC uptake via any sort of feedback effect on Rho GTPases.
The involvement of HDAC6 in the uptake of UPEC by bladder cells was further confirmed using siRNA. The knockdown of HDAC6 expression with HDAC6-specific siRNA substantially inhibited UTI89 entry into the bladder cells, but had no effect on bacterial association with the host cells (Fig. 3, f–h). Overexpression of HDAC6 had a similar, although less pronounced, inhibitory effect on invasion (supplemental Fig. S3). Taken together, these data demonstrate a key role for the TDAC domain of HDAC6 during UPEC entry into bladder epithelial cells.
By confocal microscopy, we observed that HDAC6 is primarily perinuclear, but also localizes at the periphery of 5637 bladder cells. We detected no significant alteration in either the overall localization or appearance of actin filaments or microtubules due to HDAC6 inhibition or knockdown (data not shown).
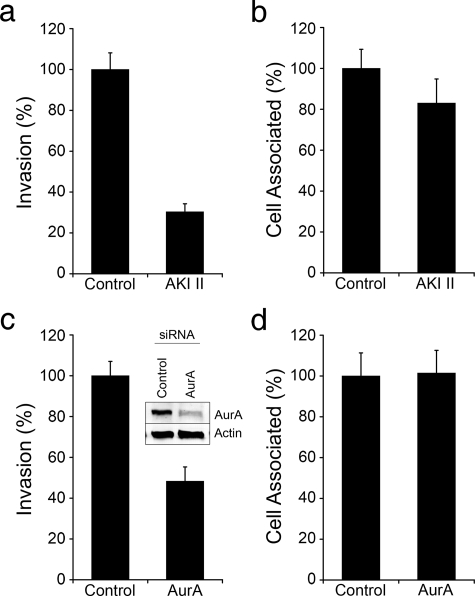
Aurora A Kinase Modulates UPEC Entry into Bladder Cells— Recently, it was reported that the mitotic serine/threonine kinase Aurora A (AurA) can phosphorylate and activate HDAC6 within human retinal pigment epithelial cells, thereby stimulating tubulin deacetylation and subsequent resorption of cilia (55). We found that treatment of 5637 bladder cells with the aurora kinase inhibitor AKI II substantially hindered UTI89 invasion, but had no significant effect on bacterial adherence (Fig. 4, a and b). Likewise, silencing of AurA expression using siRNA also inhibited UPEC invasion, without affecting bacterial association with the host cells (Fig. 4, c and d). These results indicate that AurA can promote UPEC entry into bladder cells, possibly via effects on HDAC6 activation.
FIGURE 4.
Aurora A kinase-dependent invasion of host cells by UPEC. a and b, 5637 bladder cells were pretreated for 3 h with 10 μm of AKI II or carrier alone. Cells were then infected with UTI89 for 2 h in the continued presence of drug or carrier after which (a) intracellular and (b) total cell-associated bacterial titers were determined. Similarly, numbers of (c) intracellular and (d) total host cell-associated bacteria were calculated 2-h post-infection of AurA-silenced and control siRNA-treated bladder cells. Inset in c shows levels of AurA and actin in bladder cells transfected with either AurA-specific or control non-specific siRNA. Data are expressed relative to controls as the means ± S.E. of at least three independent experiments carried out in triplicate.
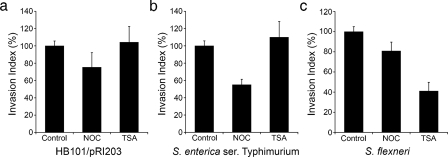
Alternate HDAC6 Substrates and Bacterial Invasion—We next asked if HDAC6 was a general requirement for bacterial entry into bladder epithelial cells. Host cells were treated with or without nocodazole or TSA prior to infection with the recombinant E. coli K12 strain HB101/pRI203, S. enterica serovar Typhimurium, or S. flexneri. HB101/pRI203 expresses the invasin protein from Y. pseudotuberculosis and is able to enter host cells via a zipper-like mechanism by binding integrin receptors (56). On the other hand, the enteric pathogens S. enterica and S. flexneri utilize type III secretion systems and injected effector molecules to trigger host membrane ruffling and bacterial uptake (1). Nocodazole treatment had a notable inhibitory effect only on host cell invasion by S. enterica, while TSA significantly inhibited only S. flexneri (Fig. 5). These results indicate that HDAC6 inhibition does not negatively affect all bacterial entry pathways.
FIGURE 5.
Differential requirements for HDAC6 by other invasive bacterial pathogens. Bladder epithelial cells were treated with carrier alone (control), 33 μm nocodazole, or 300 nm TSA prior to infection with the indicated pathogens. Invasion indices (calculated by dividing the number of gentamicin-protected intracellular bacteria by the total number of cell-associated bacteria) are presented relative to controls as the means ± S.E. of at least three independent experiments carried out in triplicate.
The differential effects of TSA on host cell invasion by S. enterica and S. flexneri may be explained by recent work showing that the host F-actin binding protein cortactin, like α-tubulin, is also a substrate for HDAC6 (57). Cortactin is necessary for S. flexneri entry into host cells, but is not required by S. enterica (58, 59). Hyperacetylation of lysine residues within cortactin in the absence of HDAC6 activity can interfere with the ability of cortactin to bind F-actin (57), and this may account for the inhibitory effect of TSA on host cell invasion by S. flexneri, but not S. enterica. In addition to cortactin, the cytosolic chaperone Hsp90 was also recently identified as an HDAC6 substrate (60, 61). Impaired activity of cortactin or Hsp90 as a consequence of HDAC6 inhibition or silencing could conceivably interfere with UPEC entry into bladder epithelial cells. However, neither silencing of cortactin expression using siRNA nor inhibition of Hsp90 using the geldanamycin analog 17-AAG had any inhibitory effect on host cell invasion by UTI89 (supplemental Fig. S4).
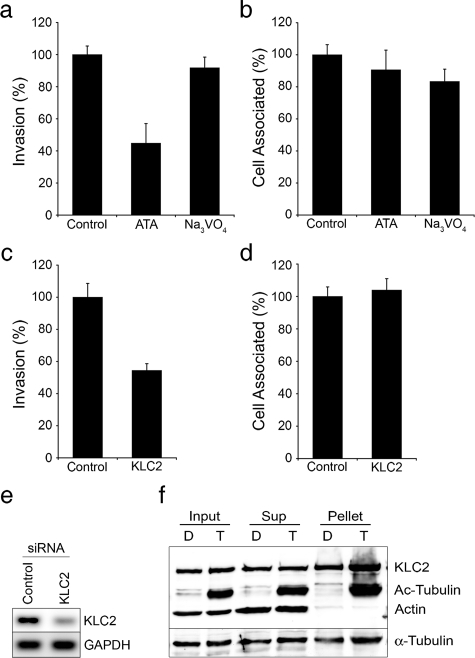
Kinesin-1 Facilitates UPEC Invasion of Bladder Cells—In addition to affecting microtubule dynamics, HDAC6 can also influence the recruitment of kinesin and dynein motor complexes. Specifically, HDAC6-mediated deacetylation of microtubules can alter the affinity and directional trafficking of these motor proteins (49, 66), and HDAC6 itself can act as an adaptor between dynein motors and aggregated protein cargos destined for aggresomes (63). These observations suggested that HDAC6 might modulate UPEC entry into host cells via indirect effects on either dynein or kinesin motor proteins. In support of this possibility, we found that inhibition of plus-end-directed kinesin motors in bladder epithelial cells by use of aurintricarboxylic acid (ATA) (64) markedly attenuated UPEC invasion, without affecting bacterial adherence to the host cells. In contrast, inhibition of the minus-end-directed motor dynein, by use of sodium orthovanadate (Na3VO4) (65), had no effect on UPEC entry (Fig. 6, a and b). Although these drugs have known pleiotropic effects, their use nonetheless suggested involvement of a kinesin motor in the invasion process. Use of siRNA verified this contention, showing that knockdown of a component of the conventional kinesin-1 motor complex (kinesin light chain-2, KLC2) significantly inhibits UPEC entry into host bladder cells without affecting bacterial adherence (Fig. 6, c–e). Similar to previous findings in neuronal cells (66), we found that increased acetylation of α-tubulin as a result of HDAC6 inhibition by TSA enhanced the recruitment of KLC2 to microtubules 2–5-fold in a concentration-dependent manner, as determined using pulldown assays (Fig. 6f and data not shown). These findings support the notion that HDAC6 can regulate UPEC entry into host cells by affecting the recruitment of kinesin motor proteins and associated cargoes.
FIGURE 6.
Kinesin-1 promotes host cell invasion by UPEC. Bladder cells were pre-treated with aurintricarboxylic acid (ATA, 50 μm) for 3 h or with sodium orthovanadate (Na3VO4, 100 μm) for 1 h prior to infection with UTI89. After 2 h in the continued presence of drugs (a) intracellular and (b) total cell-associated bacterial titers were calculated and are expressed relative to controls treated with only carrier. Quantification of (c) internalized and (d) total cell-associated bacteria following infection of KLC2-silenced bladder cells by UTI89. e, semi-quantitative RT-PCR showing KLC2 and GAPDH message levels in bladder cells 72 h after transfection with either control nonspecific siRNA or KLC2-specific siRNA. Data in graphs are expressed relative to appropriate controls as the means ± S.E. of at least three independent experiments carried out in triplicate. f, 5637 bladder cells, following transfection with pKLC2_FLAG, were treated with either DMSO alone (D) or 300 nm TSA (T) for 3 h prior to lysis. The Western blot shows levels of FLAG-tagged KLC2, α-tubulin, acetylated tubulin, and actin present in total cell lysates (input) or in supernatants (Sup) recovered after polymerizing and spinning out microtubules (pellets). Similar results were obtained using hemagglutinin-tagged KLC2.
DISCUSSION
While dynamic rearrangement of the actin cytoskeleton is necessary for nearly all invasive bacterial pathogens to gain entry into their target host cells, at least a few of these invasive microbes, including Haemophilus influenzae, Klebsiella pneumoniae, Mycoplasma penetrans, Porphyromonas gingivalis, and Grimontia (Vibrio) hollisae, also require a functional host microtubule network (67). In most cases, the mechanism by which microtubules affect the invasion process remains enigmatic. Here we found that the actin-dependent entry of type 1-piliated UPEC into bladder epithelial cells requires both microtubules and the TDAC activity of HDAC6.
By deacetylating α-tubulin, HDAC6 can enhance the instability of microtubules, which in turn can potentially alter the actin cytoskeleton via effects on GEF-H1 and Rho GTPases (39). In our assays we could not detect any overt rearrangement of microtubules at sites of bacterial entry and the inhibition of UPEC invasion by nocodazole, vinblastine, taxol, or TSA treatments was not attributable to aberrant GEF-H1 activation and subsequent stimulation of Rho GTPases. We also found that the HDAC6 substrates cortactin and Hsp90 were dispensable for UPEC entry. Moreover, knockdown or inhibition of HDAC6 did not cause any notable changes in the overall appearance or localization of either actin filaments or microtubules as determined by confocal microscopy. Additional results from the use of low concentrations of nocodazole indicate that microtubule dynamics, influenced by HDAC6 or by other means, are not strictly necessary for UPEC entry into bladder cells. Rather, we suggest that microtubules, HDAC6, and the upstream activator of HDAC6, AurA, function indirectly as essential regulatory elements affecting actin and membrane dynamics during the invasion process.
Previous studies have implicated HDAC6 in HIV-1 entry (51), the assembly of immune synapses in T cells (52), and the dynamic alteration of focal adhesions (50). Notably, the engagement of α3β1 integrin receptors by type 1 piliated UPEC stimulates the assembly and activation of signaling complexes similar to those associated with focal adhesions and less developed focal contacts (18). Kinesin motor proteins have been implicated in the relaxation and disassembly of focal adhesions, presumably by facilitating the delivery of key cytoskeletal regulatory or signaling factor(s) to the cell surface (25, 26). Although these factors have not yet been definitively identified, it is notable that kinesin-1 can bind and transport the actin nucleating protein WAVE1, which in turn can alter cortical actin cytoskeletal and plasma membrane dynamics (68, 69). Interestingly, we have found that a related actin nucleating protein, WAVE2, is critical for UPEC entry into bladder epithelial cells.3
By affecting the acetylation status of α-tubulin, HDAC6 can modulate the recruitment and directional trafficking of kinesin-1 and any associated cargos (see Fig. 6f and (49, 66)). Ultimately, the kinesin-dependent delivery of cargo like WAVE2 to the host cell periphery may account for the dependence of type 1 pili-mediated bacterial invasion on host microtubules, AurA, and HDAC6. However, we cannot rule out the possibility that other as-yet undefined targets of HDAC6, in addition to microtubules, may also influence UPEC invasion. Similarly, AurA may also affect the invasion process by modifying cellular targets other than HDAC6. For example, the Drosophila AurA homologue has recently been shown to modulate the localization of the alternate endocytic clathrin adaptor Numb, a protein that we identified previously as a key regulator of integrin-mediated, clathrin-dependent entry of UPEC into bladder cells (19, 70). Nevertheless, our finding that HDAC6 and AurA promote the clathrin-dependent uptake of UPEC downstream of integrin receptors suggests a broader role for both HDAC6 and AurA in endocytic pathways, plasma membrane dynamics, and possibly other cellular processes, including the invasive migration of cancer cells as mediated by integrin attachment and recycling.
Supplementary Material
Acknowledgments
We thank O. Steele-Mortimer (Rocky Mountain Laboratories) for Salmonella strain SL1344 and the University of Utah School of Medicine Cell Imaging Facility for help with the microscopy. We are also grateful to S. Schreiber and R. Mazitschek (Broad Institute of Harvard University and MIT) and the Initiative for Chemical Genetics (ICG, National Cancer Institute) for providing the FLAG-tagged HDAC6 construct, tubacin and niltubacin.
This work was supported, in whole or in part, by National Institutes of Health Grant DK068585. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
Footnotes
The abbreviations used are: UPEC, uropathogenic E. coli; ATA, aurintricarboxylic acid; AurA, aurora A kinase; AKI II, Aurora Kinase Inhibitor II; BSA, bovine serum albumin; GEF, guanine nucleotide exchange factor; DMSO, dimethyl sulfoxide; HDAC, histone deacetylase; NA, nicotinamide; NaB, sodium butyrate; PI, phosphoinositide; TDAC, tubacin deacetylase; TSA, trichostatin A; UTI, urinary tract infection; PBS, phosphate-buffered saline.
M. A. Mulvey, unpublished data.
References
- 1.Rottner, K., Stradal, T. E., and Wehland, J. (2005) Dev. Cell 9 3-17 [DOI] [PubMed] [Google Scholar]
- 2.Martinez, J. J., Mulvey, M. A., Schilling, J. D., Pinkner, J. S., and Hultgren, S. J. (2000) EMBO J. 19 2803-2812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mulvey, M. A., Lopez-Boado, Y. S., Wilson, C. L., Roth, R., Parks, W. C., Heuser, J., and Hultgren, S. J. (1998) Science 282 1494-1497 [DOI] [PubMed] [Google Scholar]
- 4.Wang, H., Liang, F. X., and Kong, X. P. (2008) J Histochem. Cytochem. 56 597-604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Foxman, B. (2002) Am. J. Med. 113 Suppl. 1A, 5S-13S [DOI] [PubMed] [Google Scholar]
- 6.Ronald, A. (2002) Am. J. Med. 113 Suppl. 1A, 14S-19S [DOI] [PubMed] [Google Scholar]
- 7.Bower, J. M., Eto, D. S., and Mulvey, M. A. (2005) Traffic 6 18-31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mulvey, M. A., Schilling, J. D., Martinez, J. J., and Hultgren, S. J. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 8829-8835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mulvey, M. A., Schilling, J. D., and Hultgren, S. J. (2001) Infect. Immun. 69 4572-4579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schilling, J. D., Lorenz, R. G., and Hultgren, S. J. (2002) Infect. Immun. 70 7042-7049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hvidberg, H., Struve, C., Krogfelt, K. A., Christensen, N., Rasmussen, S. N., and Frimodt-Moller, N. (2000) Antimicrob. Agents Chemother. 44 156-163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kerrn, M. B., Struve, C., Blom, J., Frimodt-Moller, N., and Krogfelt, K. A. (2005) J. Antimicrob. Chemother. 55 383-386 [DOI] [PubMed] [Google Scholar]
- 13.Justice, S. S., Hung, C., Theriot, J. A., Fletcher, D. A., Anderson, G. G., Footer, M. J., and Hultgren, S. J. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 1333-1338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eto, D. S., Sundsbak, J. L., and Mulvey, M. A. (2006) Cell Microbiol. 8 704-717 [DOI] [PubMed] [Google Scholar]
- 15.Russell, P. W., and Orndorff, P. E. (1992) J. Bacteriol. 174 5923-5935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jones, C. H., Pinkner, J. S., Roth, R., Heuser, J., Nicholes, A. V., Abraham, S. N., and Hultgren, S. J. (1995) Proc. Natl. Acad. Sci. U. S. A. 92 2081-2085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martinez, J. J., and Hultgren, S. J. (2002) Cell Microbiol. 4 19-28 [DOI] [PubMed] [Google Scholar]
- 18.Eto, D. S., Jones, T. A., Sundsbak, J. L., and Mulvey, M. A. (2007) PLoS Pathog. 3 e100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eto, D. S., Gordon, H. B., Dhakal, B. K., Jones, T. A., and Mulvey, M. A. (2008) Cell Microbiol. 10 2553-2567 [DOI] [PubMed] [Google Scholar]
- 20.Basu, R., and Chang, F. (2007) Curr. Opin. Cell. Biol. 19 88-94 [DOI] [PubMed] [Google Scholar]
- 21.Rodriguez, O. C., Schaefer, A. W., Mandato, C. A., Forscher, P., Bement, W. M., and Waterman-Storer, C. M. (2003) Nat. Cell Biol. 5 599-609 [DOI] [PubMed] [Google Scholar]
- 22.Bellestram, C., Magid, N., Zonis, J., Shtutman, M., and Bershadsky, A. (2004) in Cell Motility: From Molecules to Organisms (Ridley, A., Peckham, M., and Clark, P. eds), John Wiley & Sons, Ltd., Hoboken, NJ
- 23.Danowski, B. A. (1989) J. Cell Sci. 93 255-266 [DOI] [PubMed] [Google Scholar]
- 24.Liu, B. P., Chrzanowska-Wodnicka, M., and Burridge, K. (1998) Cell Adhes. Commun. 5 249-255 [DOI] [PubMed] [Google Scholar]
- 25.Kaverina, I., Krylyshkina, O., and Small, J. V. (1999) J. Cell Biol. 146 1033-1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Palazzo, A. F., and Gundersen, G. G. (2002) Sci STKE 2002 PE31. [DOI] [PubMed] [Google Scholar]
- 27.Chen, S. L., Hung, C. S., Xu, J., Reigstad, C. S., Magrini, V., Sabo, A., Blasiar, D., Bieri, T., Meyer, R. R., Ozersky, P., Armstrong, J. R., Fulton, R. S., Latreille, J. P., Spieth, J., Hooton, T. M., Mardis, E. R., Hultgren, S. J., and Gordon, J. I. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 5977-5982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Knodler, L. A., Finlay, B. B., and Steele-Mortimer, O. (2005) J. Biol. Chem. 280 9058-9064 [DOI] [PubMed] [Google Scholar]
- 29.Isberg, R. R., Voorhis, D. L., and Falkow, S. (1987) Cell 50 769-778 [DOI] [PubMed] [Google Scholar]
- 30.Kufer, T. A., Sillje, H. H., Korner, R., Gruss, O. J., Meraldi, P., and Nigg, E. A. (2002) J. Cell Biol. 158 617-623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hubbert, C., Guardiola, A., Shao, R., Kawaguchi, Y., Ito, A., Nixon, A., Yoshida, M., Wang, X. F., and Yao, T. P. (2002) Nature 417 455-458 [DOI] [PubMed] [Google Scholar]
- 32.Hoebeke, J., Van Nijen, G., and De Brabander, M. (1976) Biochem. Biophys. Res. Commun. 69 319-324 [DOI] [PubMed] [Google Scholar]
- 33.De Brabander, M., De May, J., Joniau, M., and Geuens, G. (1977) Cell Biol. Int. Rep. 1 177-183 [DOI] [PubMed] [Google Scholar]
- 34.Jordan, M. A. (2002) Curr. Med. Chem. Anticancer Agents 2 1-17 [DOI] [PubMed] [Google Scholar]
- 35.Deleted in proof
- 36.Liao, G., Nagasaki, T., and Gundersen, G. G. (1995) J. Cell Sci. 108 3473-3483 [DOI] [PubMed] [Google Scholar]
- 37.Krendel, M., Zenke, F. T., and Bokoch, G. M. (2002) Nat Cell Biol. 4 294-301 [DOI] [PubMed] [Google Scholar]
- 38.Enomoto, T. (1996) Cell Struct. Funct. 21 317-326 [DOI] [PubMed] [Google Scholar]
- 39.Birkenfeld, J., Nalbant, P., Yoon, S. H., and Bokoch, G. M. (2008) Trends Cell Biol. 18 210-219 [DOI] [PubMed] [Google Scholar]
- 40.MacRae, T. H. (1997) Eur J Biochem. 244 265-278 [DOI] [PubMed] [Google Scholar]
- 41.Westermann, S., and Weber, K. (2003) Nat. Rev. Mol. Cell. Biol. 4 938-947 [DOI] [PubMed] [Google Scholar]
- 42.L'Hernault, S. W., and Rosenbaum, J. L. (1985) Biochemistry 24 473-478 [DOI] [PubMed] [Google Scholar]
- 43.Piperno, G., LeDizet, M., and Chang, X. J. (1987) J. Cell Biol. 104 289-302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Matsuyama, A., Shimazu, T., Sumida, Y., Saito, A., Yoshimatsu, Y., Seigneurin-Berny, D., Osada, H., Komatsu, Y., Nishino, N., Khochbin, S., Horinouchi, S., and Yoshida, M. (2002) EMBO J. 21 6820-6831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Haggarty, S. J., Koeller, K. M., Wong, J. C., Grozinger, C. M., and Schreiber, S. L. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 4389-4394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang, Y., Li, N., Caron, C., Matthias, G., Hess, D., Khochbin, S., and Matthias, P. (2003) EMBO J. 22 1168-1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang, X. J., and Gregoire, S. (2005) Mol. Cell. Biol. 25 2873-2884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Destaing, O., Saltel, F., Gilquin, B., Chabadel, A., Khochbin, S., Ory, S., and Jurdic, P. (2005) J. Cell Sci. 118 2901-2911 [DOI] [PubMed] [Google Scholar]
- 49.Reed, N. A., Cai, D., Blasius, T. L., Jih, G. T., Meyhofer, E., Gaertig, J., and Verhey, K. J. (2006) Curr. Biol. 16 2166-2172 [DOI] [PubMed] [Google Scholar]
- 50.Tran, A. D., Marmo, T. P., Salam, A. A., Che, S., Finkelstein, E., Kabarriti, R., Xenias, H. S., Mazitschek, R., Hubbert, C., Kawaguchi, Y., Sheetz, M. P., Yao, T. P., and Bulinski, J. C. (2007) J. Cell Sci. 120 1469-1479 [DOI] [PubMed] [Google Scholar]
- 51.Valenzuela-Fernandez, A., Alvarez, S., Gordon-Alonso, M., Barrero, M., Ursa, A., Cabrero, J. R., Fernandez, G., Naranjo-Suarez, S., Yanez-Mo, M., Serrador, J. M., Munoz-Fernandez, M. A., and Sanchez-Madrid, F. (2005) Mol. Biol. Cell 16 5445-5454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Serrador, J. M., Cabrero, J. R., Sancho, D., Mittelbrunn, M., Urzainqui, A., and Sanchez-Madrid, F. (2004) Immunity 20 417-428 [DOI] [PubMed] [Google Scholar]
- 53.Guardiola, A. R., and Yao, T. P. (2002) J. Biol. Chem. 277 3350-3356 [DOI] [PubMed] [Google Scholar]
- 54.North, B. J., Marshall, B. L., Borra, M. T., Denu, J. M., and Verdin, E. (2003) Mol Cell 11 437-444 [DOI] [PubMed] [Google Scholar]
- 55.Pugacheva, E. N., Jablonski, S. A., Hartman, T. R., Henske, E. P., and Golemis, E. A. (2007) Cell 129 1351-1363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Isberg, R. R., and Leong, J. M. (1990) Cell 60 861-871 [DOI] [PubMed] [Google Scholar]
- 57.Zhang, X., Yuan, Z., Zhang, Y., Yong, S., Salas-Burgos, A., Koomen, J., Olashaw, N., Parsons, J. T., Yang, X. J., Dent, S. R., Yao, T. P., Lane, W. S., and Seto, E. (2007) Mol. Cell 27 197-213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Unsworth, K. E., Way, M., McNiven, M., Machesky, L., and Holden, D. W. (2004) Cell Microbiol. 6 1041-1055 [DOI] [PubMed] [Google Scholar]
- 59.Bougneres, L., Girardin, S. E., Weed, S. A., Karginov, A. V., Olivo-Marin, J. C., Parsons, J. T., Sansonetti, P. J., and Van Nhieu, G. T. (2004) J. Cell Biol. 166 225-235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kovacs, J. J., Murphy, P. J., Gaillard, S., Zhao, X., Wu, J. T., Nicchitta, C. V., Yoshida, M., Toft, D. O., Pratt, W. B., and Yao, T. P. (2005) Mol. Cell 18 601-607 [DOI] [PubMed] [Google Scholar]
- 61.Bali, P., Pranpat, M., Bradner, J., Balasis, M., Fiskus, W., Guo, F., Rocha, K., Kumaraswamy, S., Boyapalle, S., Atadja, P., Seto, E., and Bhalla, K. (2005) J. Biol. Chem. 280 26729-26734 [DOI] [PubMed] [Google Scholar]
- 62.Deleted in proof
- 63.Kawaguchi, Y., Kovacs, J. J., McLaurin, A., Vance, J. M., Ito, A., and Yao, T. P. (2003) Cell 115 727-738 [DOI] [PubMed] [Google Scholar]
- 64.Hopkins, S. C., Vale, R. D., and Kuntz, I. D. (2000) Biochemistry 39 2805-2814 [DOI] [PubMed] [Google Scholar]
- 65.Hu, L., and Kopecko, D. J. (1999) Infect. Immun. 67 4171-4182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dompierre, J. P., Godin, J. D., Charrin, B. C., Cordelieres, F. P., King, S. J., Humbert, S., and Saudou, F. (2007) J. Neurosci. 27 3571-3583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yoshida, S., and Sasakawa, C. (2003) Trends Microbiol. 11 139-143 [DOI] [PubMed] [Google Scholar]
- 68.Kawano, Y., Yoshimura, T., Tsuboi, D., Kawabata, S., Kaneko-Kawano, T., Shirataki, H., Takenawa, T., and Kaibuchi, K. (2005) Mol. Cell. Biol. 25 9920-9935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Takenawa, T., and Suetsugu, S. (2007) Nat. Rev. Mol. Cell. Biol. 8 37-48 [DOI] [PubMed] [Google Scholar]
- 70.Wirtz-Peitz, F., Nishimura, T., and Knoblich, J. A. (2008) Cell 135 161-173 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.