Abstract
The low-density lipoprotein receptor-related protein LRP1 is a cell surface receptor with functions in diverse physiological pathways, including lipid metabolism. Here we show that LRP1-deficient fibroblasts accumulate high levels of intracellular cholesterol and cholesteryl-ester when stimulated for adipocyte differentiation. We demonstrate that LRP1 stimulates a canonical Wnt5a signaling pathway that prevents cholesterol accumulation. Moreover, we show that LRP1 is required for lipolysis and stimulates fatty acid synthesis independently of the noradrenergic pathway, through inhibition of GSK3β and its previously unknown target acetyl-CoA carboxylase (ACC). As a result of ACC inhibition, mature LRP1-deficient adipocytes of adult mice are hypotrophic, and lower uptake of fatty acids into adipose tissue leads to their redistribution to the liver. These results establish LRP1 as a novel integrator of adipogenic differentiation and fat storage signals.
The number of adipocytes in an organism is determined by a tightly regulated differentiation process of fibroblast-like preadipocytes (1, 2). Fat cell differentiation (adipogenesis) is controlled by hormonal-induced coordinated expression and activation of two main groups of transcription factors, the CCAAT/enhancer-binding protein (C/EBP) family and peroxisome proliferator-activated receptor γ (PPARγ)4 (2). PPARγ, a member of the nuclear hormone receptors superfamily, is a crucial component of this cascade, as adipogenesis is impaired in PPARγ-deficient mesenchymal stem cells (3). Activation of PPARγ induces the expression of lipogenic genes, such as adipocyte fatty acid-binding protein (422/aP2) (4), CD36 and lipoprotein lipase (LPL) (5). Accumulation of intracellular triglyceride (TG) droplets ultimately gives rise to the morphologically distinct fat cell (2). During periods of caloric restriction, TGs stored in adipocytes are catabolized into glycerol and fatty acids, to provide energy. Mobilization of lipids involves the sequential activation of hormone-sensitive lipase (HSL) and adipose triglyceride lipase (ATGL) (6), two lipolytic enzymes responsible for more than 95% of the TG hydrolase activity in the adipose tissues of mammals (7).
The low-density lipoprotein receptor-related protein 1 (LRP1) is a multifunctional cell surface receptor. Two NPXY motifs in the intracellular domain (ICD) serve as docking sites for several cytoplasmic adaptor proteins including Shc, Disabled-1, JIP1, PSD-95, CED-6/GULP, ARH, and Fe65, which control intracellular trafficking, as well as signaling events (8). LRP1 interacts with and mediates endocytosis of more than 40 unrelated ligands ranging from viruses to protease/protease inhibitor complexes, cytokines, and growth factors (9). In the liver, LRP1 and the LDL receptor (LDLr) share the endocytosis and subsequent degradation of TG-rich very-low-density lipoproteins and chylomicron remnants. However, endocytosis and clearance of macromolecules is only one function of LRP1. There is now substantial evidence that LRP1 also serves as a regulator of several fundamental signal transduction pathways that are essential for cell migration, cell proliferation, and vascular remodeling (8, 10). For instance, LRP1 post-translationally modulates and integrates TGFβ1 and PDGF signals in vascular smooth muscle cells (VSMC), which is essential for protecting the vessel wall from atherosclerosis (11, 12). A prominent feature of atherosclerotic lesions is the accumulation of cholesterol in the vascular wall, raising the possibility that LRP1 might also physiologically modulate lipid trafficking and storage in adipocytes. Here, we show that LRP1 controls signaling pathways involved in cholesterol storage and fatty acid synthesis during adipocyte differentiation.
EXPERIMENTAL PROCEDURES
Animal Studies—The generation of aP2-CreERT2 (13), LRP1flox/flox (14), and LDL receptor knock-out animals (15) have been reported earlier. Mice in which LRP1 was inducibly ablated in mature adipocytes by tamoxifen injection (13) are referred to as adLRP1(-/-)Tam (aP2Cre+ERT2;LRP1flox/flox; LDLr(-/-)), and their littermate controls are referred to as adLRP1+/+Tam (LRP1flox/flox/LDLr(-/-)). Animals were maintained on a 12-h light/12-h dark cycle and fed a standard laboratory chow (UAR, Villemoison sur Orge, France) or a high fat diet for 5 weeks (11). Sex-matched and age-matched animals were used for all experiments. Genotype was confirmed by PCR using previously described primers and PCR conditions (11, 13). Constitutive adipose tissue LRP1 knock-out mice (adLRP1(-/-)) were reported earlier (16). Purification of adipocytes by collagenase II treatment of adipose tissue was performed as described (13). For histology, livers were perfused in situ with paraformaldehyde (4% in PBS), post-fixed in formaldehyde (20% in PBS), and frozen in tissue-Tek OCT compound (SAKURA, Zoeterwoude, The Netherlands). 10-μm cryosections were stained with hematoxylin and eosin or Oil Red O and examined by light or confocal fluorescence microscopy.
Cell Culture—Cells were seeded in 100-mm dishes and grown to 80% confluence in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% (v/v) new born calf serum (NCS). Adipocyte differentiation was induced using the adipogenic mixture containing insulin, dexamethasone, IBMX, and the PPARγ agonist rosiglitazone, and Oil Red O staining was performed as described (17). For post-differentiation starvation, cells were cultured for 4 additional days in Dulbecco's modified Eagle's medium containing 0.5% fetal bovine serum. For experiments with conditioned media from L-M(TK-) (L cells stably overexpressing mouse Wnt5a (CRL-2814; American Type Culture Collection, Manassas, VA) (18) and MEFs, one set of cells was subjected to the adipocyte differentiation protocol. The differentiation media was then removed and cleared by centrifugation. The supernatant was then supplemented with fresh adipogenic mixture and used to monitor lipid accumulation in a second set of cells. GSK-3α (Aloisine A, 7-n-butyl-6-(4-hydroxyphenyl[5H]pyrrolo [2, 3]pyrazine, 100 nm) and GSK-3β (inhibitor VIII, N-(4-methoxybenzyl)-N′-(5-nitro-1,3-thiazol-2-yl) urea, 300 nm) inhibitors were purchased from Calbiochem (San Diego, CA).
Plasmids and Probes—Wnt5a cDNA (NM_009524) was subcloned into the EcoRV and HindIII site of pcDNA 3.1 Zeo (Invitrogen). Full-length and a truncated form of human LRP1 cDNA (NM_02332) containing nts 13049–14101, which encode the transmembrane segment and the cytoplasmic tail, as well as nts 467–523, which encode the native LRP1 leader peptide, were subcloned into XhoI and NotI of pcDNA 3.1 Zeo (Invitrogen). LRP1(-/-) MEFs were stably transfected with Wnt5a expression vector using FuGENE 6 (Roche Applied Sciences) according to the manufacturer's instructions. Positive clones were selected using zeocin.
Adenovirus Protocol Infection—Mouse skin fibroblasts were isolated from LRP1lox/lox mice (14). Cells were cultured in 10% NCS Dulbecco's modified Eagle's medium and infected with adenovirus expressing Cre recombinase (14) for 5 days at 6 × 108 pfu/ml. Cells were subsequently subjected to the adipocyte differentiation protocol as described above.
Recombinant Murine Wnt5a—Murine Wnt5a cloned into the pcDNA3.1zeo expression vector was subcloned into the pAcGP67B baculovirus transfer vector (BD Biosciences, Le Pont de Claix, France) with BamHI and BglII restriction sites, in-frame with the viral secretion signal gp67. The Sf 21 cell line, derived from Spodoptera frugiperda, was grown in Sf 902 medium containing 10% fetal bovine serum. Cells were infected with Wnt5a recombinant baculovirus. Infected cell supernatant was cleared by centrifugation at 4 °C, aliquoted, and stored at -20 °C until use.
Antibodies and Immunoblotting—SDS-polyacrylamide gel electrophoresis and immunoblot analysis were performed according to standard procedures. Proteins were transferred onto nitrocellulose membranes and immunoblot analyses were carried out using antibodies directed against β-catenin (Santa Cruz Biotechnology), LRP1, Wnt5a (RD systems, Minneapolis, MN), p-AMPK, p-ACC (Upstate Biotechnology Inc., Lake Placid, NY), or β-actin (Sigma), AP2 (Abcam, Cambridge, UK), C/EBP-α (Santa Cruz Biotechnology). The CD36 antibody was generated against a synthetic peptide (NH2-FMISYCACKSKNGK-COOH).
Real Time PCR—RNA was isolated using TRIzol reagent (Sigma) according to the manufacturer's instructions. 50 ng of RNA were converted to cDNA using the high capacity cDNA Archive kit (Applied Biosystems, Foster City, CA). PCR amplification was performed using SYBRGreen PCR master mix (Applied Biosystems) according to the manufacturer's instructions.
Lipid Contents and Lipase Activity—Cellular triglyceride, cholesterol, and cholesteryl ester contents were measured using the triglyceride Enzymatic PAP1000 kit (Biomerieux, Craponne, Fr), and the cholesterol/cholesteryl-ester quantification kit (Calbiochem, EMD Biosciences, San Diego, CA), respectively, according to the manufacturer's instructions. Cellular lipase activities (lipoprotein lipase and hepatic lipase activities) were measured using the Roar LPL activity assay kit (Roar Biomedical, New York, NY), according to the manufacturer's instructions. For acetyl-CoA carboxylase activity, cells were harvested in a nondenaturing buffer (150 mm KCl, 10 mm Tris, pH 7.4, 1 mm EDTA, 1 mm EGTA 1 μm phenylmethylsulfonyl, 1 μm pepstatin). Sample were extracted for 30 min on ice and centrifuged at 10,000 × g at 4 °C for 15 min. ACC activity was determined, using the [14C]bicarbonate fixation assay as described (20).
RESULTS
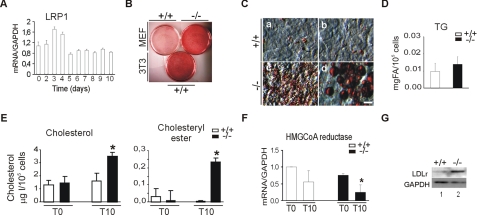
Increased Intracellular Cholesterol and Cholesteryl-ester Storage in LRP1-deficient Cells during Adipocyte Differentiation—To determine whether LRP1 controls differentiation of preadipocytes in vitro, we used a protocol well established in several cell types such as 3T3-L1 preadipocytes or murine embryonic fibroblasts (MEFs), based on a standard adipogenic mixture containing insulin, dexamethasone, IBMX, and the PPARγ agonist rosiglitazone (21). We first determined LRP1 expression in 3T3-L1 cells during the course of adipogenesis. Interestingly, LRP1 mRNA levels increased during the first 2 days of clonal expansion, and returned to basal levels at the end of the differentiation period (Fig. 1A), indicating that LRP1 might play a role during early stages of differentiation.
FIGURE 1.
LRP1 governs adipocyte differentiation. Cells were treated with the adipogenic mixture containing the PPARγ agonist rosiglitazone for 10 days to stimulate differentiation. A, quantitative RT-PCR analysis of LRP1 in 3T3 preadipocytes, B, plates of LRP1(+/+), LRP1(-/-) MEFs, and 3T3-L1 preadipocytes. The extent of cellular lipid accumulation was determined by Oil Red O staining. C, micrographs of LRP1(+/+) and LRP1(-/-) MEFs. c and d show two different morphologies of lipid droplets that accumulate in LRP1(-/-) MEFs. D, triglyceride (at day 10) and (E) cholesterol and cholesteryl-ester quantifications during differentiation of LRP1(+/+) and LRP1(-/-) MEFs. F, quantitative RT-PCR analysis of HMG CoA reductase in MEFs during differentiation. G, Western blot analysis of LDL receptor in MEFs at day 10. T0, day 0; T10, day 10 of treatment. Scale bar, 50 μm. Results are means ± S.D. *, p < 0.05.
We then analyzed the effects of the absence of LRP1 on neutral lipid accumulation during in vitro MEFs differentiation. Oil Red O staining revealed that MEFs lacking LRP1 accumulate more neutral lipids than wild-type MEFs after 10 days of growth in differentiating medium (Fig. 2B and supplemental Fig. S1A). The extent of lipid accumulation was similar to that seen in 3T3-L1 adipocytes after differentiation (Fig. 1B). As lipid content in LRP1(-/-) MEFs stably retransfected with full-length LRP1 expression vector was similar to that observed in wild-type cells (supplemental Fig. S1, B and C), lipid accumulation in LRP1(-/-) MEFs is indeed caused by lack of LRP1 expression. That this striking difference in lipid storage was not caused by artifactual clonal aberrations was confirmed in LRP1flox/flox primary cultured skin fibroblasts in which LRP1 gene disruption was induced with a Cre-expressing recombinant adenovirus (14) (supplemental Fig. S1D). Microscopic analysis revealed that LRP1(-/-) cells accumulate more lipid droplets than wild-type MEFs (Fig. 1C). However, these droplets are morphologically different from those seen in differentiated 3T3-L1 cells. They are small, numerous (Fig. 1C), whereas in 3T3-L1 cells fat accumulates essentially within a limited number of large droplets.
FIGURE 2.
LRP1 controls fat accumulation through a Wnt5a and β-catenin-dependent signaling pathway. MEFs were stimulated with the adipogenic mixture containing the PPARγ agonist rosiglitazone (A), Oil Red O staining of LRP1(+/+) and LRP1(-/-) in the absence (-CM) or presence of conditioned medium from LRP1(-/-) MEFs ((-/-) CM) and LRP1(+/+)(+/+ CM), respectively. B, quantification (top) and immunoblotting (bottom) for nuclear β-catenin accumulation over the course of the differentiation regimen. C, Oil Red O staining of LRP1(+/+) and LRP1(-/-) MEFs differentiated in the absence (-LiCl) or presence (+LiCl) of lithium chloride. D, Wnt5a mRNA and protein levels over the course of the differentiation regimen. E, MEFs were stimulated for differentiation in the absence (-CM) or presence of conditioned medium from L-M(TK-) cells overexpressing (Wnt5a + CM) or not (Wnt5a- CM) Wnt5a. F, lipid accumulation in differentiated LRP1(-/-) MEFs transfected with Wnt5a (-/-, Wnt5a), LRP1(+/+) MEFs (+/+) and mock-transfected LRP1(-/-) MEFs. G, cholesterol and cholesteryl-ester quantifications during the course of differentiation of LRP1(-/-)-transfected with Wnt5a (-/-, Wnt5a) and mock-transfected LRP1(-/-) MEFs. Oil Red O staining of (H) plates and (I) representative micrographs of LRP1(+/+) and LRP1(-/-) MEFs differentiated in the presence of recombinant murine Wnt5a (rWnt5a+) or control medium (rWnt5a-). J, top panel, intracellular cholesteryl-ester quantification and (bottom panel) Western blot analysis with anti-Wnt5a antibodies of supernatant from differentiated control cells (+/+) or LRP1(-/-) MEFs untreated (-/-) or treated with recombinant mouse Wnt5a (rWnt5a+) or control medium (rWnt5a-). K, immunoblot analysis of nuclear β-catenin during the course of differentiation of LRP1(-/-) cells transfected with Wnt5a (-/-, Wnt5a) and mock-transfected LRP1(-/-) MEFs. Scale bar, 50 μm. Results are means ± S.D. *, p < 0.05.
Surprisingly, analysis of neutral lipid showed no significant increase in intracellular triacylglycerol accumulation in LRP1(-/-) MEFs (Fig. 1D) but revealed a strong increase of cholesterol and cholesteryl-ester levels in LRP1-deficient cells (Fig. 1E). Analysis of the mRNA expression of genes involved in cholesterol export such as apolipoprotein A1 (ApoA1), phospholipid transfer protein (Pltp), scavenger receptor class B member 1 (SCARB1), ATP-binding cassette subfamily G member 1 (ABCG1) and ATP-binding cassette subfamily G member 2 (ABCG2), did not show significant differences between LRP1(-/-) and control cells (supplemental Fig. S1E). However, the transcript levels of HMG-CoA reductase (Fig. 1F), a rate-limiting enzyme for cholesterol synthesis were reduced in LRP1(-/-) cells, whereas LDLr protein levels were increased at day 10 (Fig. 1G), indicating that increased cholesterol storage in the absence of LRP1 is due to cholesterol uptake.
LRP1 Controls Intracellular Cholesterol Content via Canonical Wnt5a Signaling Pathway—To investigate whether the LRP1-mediated control of cellular cholesterol content may involve an autocrine mechanism, we induced adipocyte differentiation in LRP1(+/+) MEFs, collected the conditioned medium and added it to LRP1(-/-) MEFs during the process of differentiation. Strikingly, under these conditions, cholesterol accumulation in LRP1(-/-) cells was similar to that seen in wild-type cells, whereas conditioned medium from LRP1(-/-) cells did not affect neutral lipid content in LRP1(+/+) cell (Fig. 2A and supplemental Fig. S2A).
Several laboratories have shown that Wnt/β-catenin signaling is involved in controlling cholesterol homeostasis (22, 23). Cholesterol depletion increase β-catenin expression, its nuclear translocation and activation of the Wnt pathway (23). Similarly, a reduction in Wnt signaling increases plasma cholesterol in mice fed a high fat diet (22). To test whether Wnt signaling might be altered in LRP1(-/-)-deficient cells, we compared the activation of the canonical Wnt pathway in LRP1(+/+) and LRP1(-/-) MEFs over the course of the adipocyte differentiation program. Before differentiation, nuclear β-catenin levels were reduced in the absence of LRP1, and its induction was severely blunted throughout the time course compared with wild type (Fig. 2B). This finding suggests that increased cholesterol accumulation in LRP1(-/-) cells in response to the adipogenic mixture was caused, at least in part, by impaired activation of canonical Wnt/β-catenin signaling. This is further supported by our finding that downstream activation of the Wnt pathway, through inhibition of the glycogen synthase kinase-3 (GSK3) with LiCl, reduced neutral lipid accumulation in the LRP1-deficient MEFs to levels seen in wild-type cells (Fig. 2C).
To identify the Wnt proteins involved in the LRP1(-/-)-dependent cholesterol accumulation, we first compared their transcript levels in LRP1(+/+) and LRP1(-/-) MEFs. Interestingly, whereas Wnt5a transcript levels were induced during the course of adipocyte differentiation in wild-type MEFs, they were almost undetectable in LRP1(-/-) MEFs (Fig. 2D). In contrast, none of the other tested Wnt transcripts were significantly reduced in the knock-out cells (supplemental Fig. S2C). To determine whether LRP1 controls cholesterol accumulation through a Wnt5a signaling pathway, we treated LRP1(+/+) and LRP1(-/-) MEFs with the differentiation mixture in presence of the conditioned medium from L cells (L-MTK) overexpressing (Wnt5a+ CM) or not (Wnt5a- CM) mouse Wnt5a (18). As shown in Fig. 2E, conditioned medium containing Wnt5a+ (Wnt5a+ CM) inhibits neutral lipid accumulation in LRP1(-/-) MEFs, whereas Wnt5a- CM has no effect (Fig. 2E). These data were confirmed by stably transfecting a Wnt5a expression vector into LRP1(-/-) MEFs. Indeed, although Wnt5a was expressed at lower levels in transfected LRP1(-/-) cells than in wild-type MEFs (supplemental Fig. S2B), this was nevertheless sufficient to reduce cholesterol and cholesterylester content to that seen in wild-type cells (Fig. 2, F and G). Similarly, treatment over a period of 7 days with recombinant murine Wnt5a protein was sufficient to efficiently decrease neutral lipid accumulation in LRP1(-/-) MEFs (Fig. 2, H–J). Furthermore in agreement with impaired activation of a canonical Wnt/β-catenin signaling pathway, reexpression of Wnt5a in LRP1(-/-) MEFs increases nuclear β-catenin expression (Fig. 2K). Taken together, these data suggest that LRP1 positively controls a Wnt5a canonical signaling pathway that regulates cholesterol homeostasis during differentiation.
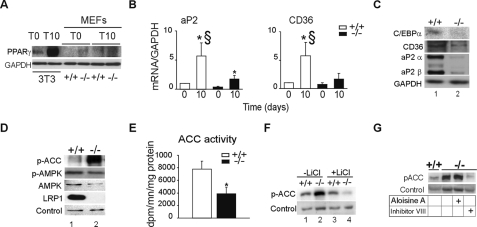
Fatty Acid Synthesis Is Impaired in the Absence of LRP1—Even though cholesterol and cholesterylester accumulate in the absence of LRP1 during differentiation, no significant TG accumulation was observed during differentiation. To test whether the expression of the nuclear receptor PPARγ, which is required for TG storage is affected in the absence of LRP1, we examined its expression during differentiation in these cells. Western blot analysis shows that PPARγ protein levels are very low compared with levels in 3T3L1 preadipocytes and barely induced during the course of differentiation in LRP1(-/-) MEFs, (Fig. 3A). As expected, mRNA and protein expressions of adipocyte marker genes, such as 422/aP2, CD36, and C/EBPα were not higher at day 0 in the absence of LRP1, and were barely increased during differentiation (Fig. 3, B and C and supplemental Fig. S3A). Furthermore, analysis of the transcript levels of acylCoA:diacylglycerol acetyltransferase 1 (DGAT1) and DGAT2, two enzymes that mediate esterification of monoacylglycerol in adipocytes (supplemental Fig. S3B), as well as the regulators of fatty acid synthesis AGPAT1, AGPAT2, GPAT, and SREBP1c did not reveal any major difference between LRP1(+/+) and LRP1(-/-) MEFs (supplemental Fig. S3, C and D).
FIGURE 3.
Impaired adipogenic program and inhibition of fatty acid synthesis in LRP1(-/-) fibroblasts. A, Western blot analysis of PPARγ in 3T3-L1 preadipocytes and MEFs LRP1(+/+) and LRP1(-/-). B, quantitative RT-PCR analysis of the indicated genes in MEFs LRP1(+/+) and LRP1(-/-) during the course of adipogenesis. C, Western blot analysis of the indicated genes in MEFs LRP1(+/+) and LRP1(-/-) during the course of adipogenesis. D, Western blot analysis of p-ACC, p-AMPK, AMPK, LRP1, and loading control in LRP1(+/+) and LRP1(-/-) MEFs. E, ACC activity in MEFs upon differentiation. F, Western blot analysis of p-ACC in LRP1(+/+) and LRP1(-/-) MEFs after 10 days of lithium chloride treatment. G, Western blot analysis of p-ACC in LRP1(+/+) and LRP1(-/-) MEFs after 10 days of treatment with GSK3α (Aloisine A) or GSK3β (inhibitor VIII) inhibitor. Results are means ± S.D. *, p < 0.05 day 0 versus day 10. §, p < 0.05 LRP1(+/+) versus LRP1(-/-).
The rate-limiting enzyme for fatty acid synthesis is acetyl-CoA carboxylase (ACC) (25). ACC inhibition through phosphorylation blocks fatty acid synthesis, and acutely increases fatty acid oxidation. Whereas no change in ACC mRNA levels was observed (supplemental Fig. S3E), immunoblotting of cellular extracts revealed that the phosphorylated form of ACC (p-ACC) was greatly enhanced in LRP1(-/-) MEFs compared with wild type (Fig. 3D), suggesting inhibition of fatty acid synthesis. In agreement with this, LRP1(-/-) MEFs also exhibit reduced ACC activity compared with wild type (Fig. 3E).
AMP-activated protein kinase (AMPK) and its phosphorylated form, p-AMPK is a kinase known to phosphorylate ACC in response to noradrenergic stimulation (25). No difference in AMPK and p-AMPK levels was observed between LRP1(+/+) and LRP1(-/-) MEFs (Fig. 3D). This suggested that, in the absence of noradrenergic stimulation another kinase might phosphorylate ACC. Because GSK3 activity is required for adipocyte development (26), and its inhibition by LiCl or Wnt5a signaling resulted in decreased LRP1(-/-) MEFs cholesterol content, GSK3 might also participate in ACC phosphorylation in these cells. Immunoblot analysis showed a severe decreased in p-ACC levels in differentiated LRP1(-/-) MEFs treated with LiCl (Fig. 3F) or with the GSK3 inhibitor SB216763 (supplemental Fig. S3F) compared with similarly treated LRP1(+/+) MEFs. To determine which isoform of GSK3 is involved in ACC phosphorylation, immunoblot analysis was performed. Treatment with Aloisine A, a GSK-3α inhibitor did not changed p-ACC protein expression, whereas treatment with inhibitor VIII, an inhibitor of GSK-3β severely blunted p-ACC protein expression in LRP1(-/-) MEFs (Fig. 3G). Thus, in the absence of LRP1, canonical Wnt5a signaling pathway is blocked, leading to intracellular cholesterol accumulation and activation of GSK-3β and its target ACC. This previously unrecognized alternative pathway regulates TG synthesis independently of the noradrenergic pathway.
Lipolysis Is Impaired in the Absence of LRP1—As TG synthesis is impaired, but TG storage not decreased in LRP1(-/-) cells (Fig. 1D), we tested whether lipase activity might be affected in these cells. We found reduced lipase activity in differentiated LRP1(-/-) MEFs (Fig. 4A). Analysis of the mRNA of hormone-sensitive lipase (HSL), which encodes a major lipolytic enzyme in white adipose tissue, revealed that whereas its levels were 2-fold increased in LRP1(+/+) MEFs after differentiation, they remained unchanged in LRP1(-/-) cells (Fig. 4B). This absence of induction in LRP1(-/-) cells was independent of Wnt5a signaling, as HSL levels were not increased in LRP1(-/-) MEFs reexpressing Wnt5a (supplemental Fig. S4A). The transcript levels of the PPARγ target gene lipoprotein lipase (LPL), which coordinately with HSL degrade most of the adipose triglycerides (6), were also much lower in LRP1(-/-) cells than in LRP1(+/+) cells after differentiation (Fig. 4B). Interestingly, reexpression of Wnt5a in LRP1(-/-) MEFs restored the expression of LPL (Fig. 4C), but not of other PPARγ target genes (data not shown).
FIGURE 4.
Decreased lipolysis in LRP1(-/-) fibroblasts. A, lipase activity in MEFs upon differentiation. B, relative mRNA levels of HSL and LPL determined by quantitative RT-PCR analysis during adipocyte differentiation. C, LPL mRNA levels analyzed by RT-PCR during the course of differentiation of LRP1(-/-) transfected with Wnt5a (-/-, Wnt5a) and mock-transfected LRP1(-/-) MEFs. D, mobilization of stored lipids; MEFs were exposed to adipogenic differentiation mixture (Diff) (Day 10), followed by starvation in medium containing only 0.5% serum for 4 days (Day 14). Quantitative RT-PCR analysis of HSL and LPL at 0, 10, and 14 days and (E), plates of cells. Results are means ± S.D. **, p = 0.05 day 0 versus day 10. *, p < 0.05 day 0 versus day 10 or day 14. §, p < 0.05 LRP1(+/+) versus LRP1(-/-).
To test whether mobilization of stored fat is impaired in LRP1-deficient cells, we subjected wild type and LRP1(-/-) MEFs to the adipocyte differentiation protocol, removed the adipogenic mixture after 10 days, and starved the cells for 4 days by reducing serum concentrations (0.5%). Over this time period, the transcript levels of HSL, LPL, and ATGL encoding enzymes, which degrade adipose triglycerides (6), sharply increased in LRP1(+/+) cells (Fig. 4D and supplemental Fig. S4B), whereas the amount of stored lipids decreased (Fig. 4E and supplemental Fig. S4C). In contrast, HSL, ATGL, and LPL transcript levels did not increase in LRP1(-/-) cells (Fig. 4D and supplemental Fig. S4B) and neutral lipid stores remained unchanged in these cells (Fig. 4E and supplemental Fig. S4C). Thus, decreased levels of lipolytic enzymes might counterbalance the inhibition of TG synthesis, resulting in similar TG levels in LRP1(-/-) and LRP1(+/+) MEFs.
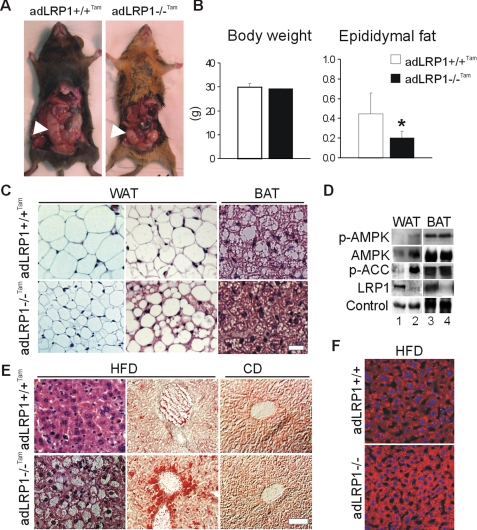
Absence of LRP1 in Mature Adipocytes Results in Lipodystrophy and Hepatosteatosis—To determine the physiological consequences of LRP1 deficiency in mature adipocytes of mice, we crossed LRP1flox/flox mice with aP2-CreERT2 mice that express the tamoxifen-dependent CreERT2 recombinase under the control of the aP2 promoter (27). This system allows experimental temporal control of the Cre recombinase activity, and deletion of LRP1 in adipocytes of adult mice. To increase circulating lipid levels, animals were maintained on a LDL receptor-deficient background (designated adLRP1(-/-)Tam) and fed a high fat diet for 5 weeks.
adLRP1(-/-)Tam mice exhibited decreased epididymal fat mass without difference in total body weight compared with control littermates (Fig. 5, A and B). LRP1 deficiency in adipocytes resulted in a hypercellular white adipose tissue (WAT), consisting of heterogeneous populations of smaller adipocytes, and greatly reduced amounts of stored fat in brown adipose tissue (BAT) (Fig. 5C).
FIGURE 5.
Adipocyte-selective LRP1 knock-out mice exhibit decreased epididymal fat and hepatosteatosis. A, aspect (arrows) and (B), quantification of the epididymal fat and body weight in 20-week-old tamoxifen-treated aP2-CreERT2; LRP1flox/flox;LDLr(-/-) mice (adLRP1(-/-)Tam). C, H&E staining of epididymal WAT and BAT sections from 12-week-old tamoxifen-treated aP2-CreERT2;LRP1flox/flox;LDLr(-/-) mice (adLRP1(-/-)Tam) and tamoxifen-treated controls (aP2CreERT2;LRP1flox/flox;LDLr(-/-)) mice (adLRP1+/+Tam) fed 5 weeks with a high fat diet. D, immunoblot analysis of p-AMPK, AMPK, p-ACC, and loading control in epididymal WAT and BAT, and of LRP1 in BAT and purified epididymal WAT adipocytes from adLRP1+/+Tam (lanes 1, 3) and adLRP1(-/-)Tam (lanes 2, 4) mice. E, H&E (left) and Oil Red O (middle and right) staining of liver sections from adLRP1(+/+)Tam and adLRP1(-/-)Tam mice fed 5 weeks with a high fat diet (HFD) or a regular chow diet (CD). F, Oil Red O staining of liver sections from aP2Cre; LRP1flox/flox;LDLr(+/+) mice. Panels show adLRP1+/+ (aP2Cre-; LRP1flox/flox; LDLr(+/+)) and adLRP1(-/-) (aP2Cre+; LRP1flox/flox;LDLr(+/+)) mice that had been fed a high fat diet (HFD) for 4 weeks. Results are means ± S.D. *, p < 0.05. Scale bar, 50 μm.
Furthermore, in agreement with our in vitro findings, the levels of the essential regulator of fatty acid synthesis, p-ACC were strikingly increased in WAT of adLRP1(-/-)Tam mice (Fig. 5D) without any significant difference in ACC mRNA levels between adLRP1(-/-)Tam mice and control littermates (supplemental Fig. S5C). This increased level of p-ACC was accompanied by an increase in AMPK and its phosphorylated form (Fig. 5D), most likely in response to noradrenergic stimulation. However, no differences in circulating lipid (cholesterol, triglyceride, and free fatty acid) and glucose concentrations levels were found between adLRP1(-/-)Tam mice and control littermates (supplemental Fig. S5A). No difference in the adipogenic marker genes or Wnt5a expression and only a moderate decrease in HSL mRNA expression are observed in LRP1-deficient WAT (supplemental Fig. S5B and data not shown). Transcripts of genes involved in fatty acid oxidation were either increased (PGC1α) or showed an upward trend (LPL, HSL, adiponectin, and UCP1) in BAT from adLRP1(-/-)Tam mice (supplemental Fig. S5B). Thus, absence of LRP1 in mature adipocytes of adult mice results in inhibition of ACC, the rate-limiting enzyme for fatty acid synthesis and lower lipid content in adipocytes.
As lipodystrophic syndrome is often accompanied by a fatty liver (28, 29), we examined livers from adLRP1(-/-)Tam mice fed a high fat diet for 5 weeks. Even though no difference in gross morphology between mutant and control liver was observed, adLRP1(-/-)Tam mice showed hepatocellular lipid accumulation with many more lipid-ladden vacuoles in hepatocytes than control littermates (Fig. 5E). We also examined fatty liver formation in mice where LRP1 is deleted earlier during adipocyte differentiation by using another mouse model, where LRP1 was inactivated by Cre-recombinase expressed under the control of the aP2 promoter (16). Even in the absence of an LDL receptor defect (adLRP1(-/-)), we found the hepatic steatosis phenotype when mice were fed a high fat diet for 5 weeks. Indeed hepatocellular lipid accumulation in adLRP1(-/-) mice was higher than in wild-type controls, but lower than in high fat fed adLRP1(-/-)Tam mice (Fig. 5F and supplemental Fig. S5D). These results indicate that lack of LRP1 in adipocytes impairs lipid uptake in adipocytes, resulting in their redistributed to the liver.
DISCUSSION
In the present study we show that in the absence of LRP1 during the adipogenic program, cells fail to induce the typical spectrum of adipogenic marker genes, and contain larger amounts of cholesterol and cholesteryl ester after in vitro differentiation (Fig. 2). Increased cholesterol storage is due at least in part to impaired canonical Wnt5a signaling in cells lacking LRP1. Wnt proteins are important signaling molecules, which have been shown to regulate cell proliferation and differentiation during embryonic development and in adult. However, Wnt signaling is also an important regulator of cholesterol metabolism (22, 23). LRP5, a co-receptor of Frizzled receptors (30) is essential for normal cholesterol metabolism, and mice lacking LRP5 exhibit decreased hepatic clearance of chylomicron remnants when fed a high fat diet (22). Moreover, Wnt/β-catenin pathway is induced by cholesterol depletion and membrane cholesterol is involved in Wnt/β-catenin signaling in the early steps of myogenic differentiation. Here we show that induction of canonical Wnt signaling with LiCl, and more specifically through restoration of Wnt5a, corrects the abnormal cholesterol and cholesteryl-ester storage in LRP1(-/-) MEFs (Fig. 3) via an autocrine mechanism. Thus, through a canonical Wnt5a signaling pathway, LRP1 plays an important role in controlling cholesterol homeostasis during differentiation.
In the present study, we also show that in vitro, inactivation of ACC by phosphorylation in LRP1-deficient cells is mediated by GSK3-β (Fig. 3, F and G and supplemental Fig. S3), a serine/threonine kinase implicated in Wnt signaling, but also described as a negative regulator of insulin signaling in adipocytes (34), a process that favors fatty acid oxidation. In vivo, inhibition of ACC in WAT of adLRP1(-/-)Tam mice is mediated by AMPK (Fig. 5D), a kinase known to phosphorylate ACC in response to noradrenergic stimulation and exercise (25). These findings reveal that two enzymes, AMPK and GSK3β control TG synthesis through ACC phosphorylation. In vitro, AMPK is not significantly altered in LRP1-deficient cells. In vivo, AMPK is activated (Fig. 5D), probably because of increased noradrenergic stimulation. Thus, our data highlight a new essential physiological role for LRP1 in controlling a novel alternative pathway for TG synthesis, independently of the noradrenergic pathway.
In contrast in WAT from adLRP1(-/-)Tam mice, since LRP1 is ablated only in mature adipocytes, no difference in adipogenic genes and Wnt5a, and only a moderate decrease in HSL mRNA expression, were observed (supplemental Fig. S5B and data not shown). Therefore, mutant mice exhibit reduced levels of fatty acid synthesis than controls, reduced neutral fat storage in adipose tissue, and lipodystrophy, a phenotype comparable to that of ACC knock-out mice (35).
Deficiency in adipose tissue function can elicits dramatic secondary liver phenotype such as hepatosteatosis or increased gluconeogenesis (28, 29). Hofmann et al. (16) have recently shown, using a mouse model where LRP1 was inactivated by Cre-recombinase expressed under the control of the aP2 promoter (adLRP1(-/-)), that such mice show features of lipodystrophy, with reduced white and brown fat mass, as well as post-prandial hypertriglyceridemia, which suggests that fatty acid uptake into adipose tissue is impaired. In the present study, we found that imbalance in the systemic handling of fatty acids in adLRP1(-/-)Tam mice, as well as in adLRP1(-/-) mice, causes pathological lipid storage in the liver (Fig. 5). As in both models LRP1 deficiency is adipocyte-selective, the mechanism for this increased hepatocellular lipid accumulation is indirect. In the absence of LRP1, lipids might be inefficiently taken up by mature white adipocytes, and redistributed to the liver.
In summary, our findings demonstrate that LRP1, through a canonical Wnt5a signaling pathway is an endogenous regulator of cholesterol homeostasis during adipocyte differentiation. Moreover, LRP1 is required for lipolysis and regulates TG synthesis in vitro and in vivo through a previously unknown pathway that requires inhibition of GSK3β and its target ACC (supplemental Fig. S6). Pharmacological modulation of LRP1 activity might emerge as a novel rational approach for treating diabetes and obesity, as well as atherosclerosis.
Supplementary Material
Acknowledgments
We thank Pierre Chambon (IGBMC, University of Strasbourg) for insightful discussions and critical reading of the manuscript.
This work was supported, in whole or in part, by Grants HL20948, HL63762, NS43408, and DK067320 from the National Institutes of Health. This work was also supported by grants from the center national pour la recherche scientifique (CNRS), Fondation pour la Recherche Médicale (FRM), Fondation de France, Université Louis Pasteur de Strasbourg, agence nationale de la recherche (ANR-05-PCOD-020). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S6.
Footnotes
The abbreviations used are: PPAR, peroxisome proliferator-activated receptor; PBS, phosphate-buffered saline; ACC, acetyl-CoA carboxylase; TG, triglyceride; HSL, hormone-sensitive lipase; LPL, lipoprotein lipase; LRP, low-density lipoprotein receptor-related protein; GSK, glycogen synthase kinase; AMPK, AMP-activated protein kinase.
References
- 1.Rosen, E. D., and MacDougald, O. A. (2006) Nat. Rev. Mol. Cell. Biol. 7 885-896 [DOI] [PubMed] [Google Scholar]
- 2.Rosen, E. D., Walkey, C. J., Puigserver, P., and Spiegelman, B. M. (2000) Genes Dev. 14 1293-1307 [PubMed] [Google Scholar]
- 3.Rosen, E. D., Sarraf, P., Troy, A. E., Bradwin, G., Moore, K., Milstone, D. S., Spiegelman, B. M., and Mortensen, R. M. (1999) Mol. Cell 4 611-617 [DOI] [PubMed] [Google Scholar]
- 4.Tontonoz, P., Hu, E., Graves, R. A., Budavari, A. I., and Spiegelman, B. M. (1994) Genes Dev. 8 1224-1234 [DOI] [PubMed] [Google Scholar]
- 5.Schoonjans, K., Peinado-Onsurbe, J., Lefebvre, A. M., Heyman, R. A., Briggs, M., Deeb, S., Staels, B., and Auwerx, J. (1996) EMBO J. 15 5336-5348 [PMC free article] [PubMed] [Google Scholar]
- 6.Zimmermann, R., Strauss, J. G., Haemmerle, G., Schoiswohl, G., Birner-Gruenberger, R., Riederer, M., Lass, A., Neuberger, G., Eisenhaber, F., Hermetter, A., and Zechner, R. (2004) Science 306 1383-1386 [DOI] [PubMed] [Google Scholar]
- 7.Schweiger, M., Schreiber, R., Haemmerle, G., Lass, A., Fledelius, C., Jacobsen, P., Tornqvist, H., Zechner, R., and Zimmermann, R. (2006) J. Biol. Chem. 281 40236-40241 [DOI] [PubMed] [Google Scholar]
- 8.Herz, J., and Hui, D. Y. (2004) Curr. Opin. Lipidol. 15 175-181 [DOI] [PubMed] [Google Scholar]
- 9.Herz, J., and Strickland, D. K. (2001) J. Clin. Invest. 108 779-784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lillis, A. P., Mikhailenko, I., and Strickland, D. K. (2005) J Thromb Haemost 3 1884-1893 [DOI] [PubMed] [Google Scholar]
- 11.Boucher, P., Gotthardt, M., Li, W. P., Anderson, R. G., and Herz, J. (2003) Science 300 329-332 [DOI] [PubMed] [Google Scholar]
- 12.Boucher, P., Liu, P., Gotthardt, M., Hiesberger, T., Anderson, R. G., and Herz, J. (2002) J. Biol. Chem. 277 15507-15513 [DOI] [PubMed] [Google Scholar]
- 13.Imai, T., Takakuwa, R., Marchand, S., Dentz, E., Bornert, J. M., Messaddeq, N., Wendling, O., Mark, M., Desvergne, B., Wahli, W., Chambon, P., and Metzger, D. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 4543-4547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rohlmann, A., Gotthardt, M., Willnow, T. E., Hammer, R. E., and Herz, J. (1996) Nat Biotechnol 14 1562-1565 [DOI] [PubMed] [Google Scholar]
- 15.Ishibashi, S., Brown, M. S., Goldstein, J. L., Gerard, R. D., Hammer, R. E., and Herz, J. (1993) J. Clin. Investig. 92 883-893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hofmann, S. M., Zhou, L., Perez-Tilve, D., Greer, T., Grant, E., Wancata, L., Thomas, A., Pfluger, P. T., Basford, J. E., Gilham, D., Herz, J., Tschop, M. H., and Hui, D. Y. (2007) J. Clin. Investig. 117 3271-3282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ross, S. E., Hemati, N., Longo, K. A., Bennett, C. N., Lucas, P. C., Erickson, R. L., and MacDougald, O. A. (2000) Science 289 950-953 [DOI] [PubMed] [Google Scholar]
- 18.Willert, K., Brown, J. D., Danenberg, E., Duncan, A. W., Weissman, I. L., Reya, T., Yates, J. R., 3rd, and Nusse, R. (2003) Nature 423 448-452 [DOI] [PubMed] [Google Scholar]
- 19.Deleted in proof
- 20.Kudo, N., Barr, A. J., Barr, R. L., Desai, S., and Lopaschuk, G. D. (1995) J. Biol. Chem. 270 17513-17520 [DOI] [PubMed] [Google Scholar]
- 21.Student, A. K., Hsu, R. Y., and Lane, M. D. (1980) J. Biol. Chem. 255 4745-4750 [PubMed] [Google Scholar]
- 22.Fujino, T., Asaba, H., Kang, M. J., Ikeda, Y., Sone, H., Takada, S., Kim, D. H., Ioka, R. X., Ono, M., Tomoyori, H., Okubo, M., Murase, T., Kamataki, A., Yamamoto, J., Magoori, K., Takahashi, S., Miyamoto, Y., Oishi, H., Nose, M., Okazaki, M., Usui, S., Imaizumi, K., Yanagisawa, M., Sakai, J., and Yamamoto, T. T. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 229-234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mermelstein, C. S., Portilho, D. M., Mendes, F. A., Costa, M. L., and Abreu, J. G. (2007) Differentiation 75 184-192 [DOI] [PubMed] [Google Scholar]
- 24.Deleted in proof
- 25.Kahn, B. B., Alquier, T., Carling, D., and Hardie, D. G. (2005) Cell Metab. 1 15-25 [DOI] [PubMed] [Google Scholar]
- 26.Ross, S. E., Erickson, R. L., Hemati, N., and MacDougald, O. A. (1999) Mol. Cell. Biol. 19 8433-8441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Imai, T., Jiang, M., Chambon, P., and Metzger, D. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 224-228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He, W., Barak, Y., Hevener, A., Olson, P., Liao, D., Le, J., Nelson, M., Ong, E., Olefsky, J. M., and Evans, R. M. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 15712-15717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shimomura, I., Hammer, R. E., Richardson, J. A., Ikemoto, S., Bashmakov, Y., Goldstein, J. L., and Brown, M. S. (1998) Genes Dev. 12 3182-3194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bhanot, P., Brink, M., Samos, C. H., Hsieh, J. C., Wang, Y., Macke, J. P., Andrew, D., Nathans, J., and Nusse, R. (1996) Nature 382 225-230 [DOI] [PubMed] [Google Scholar]
- 31.Deleted in proof
- 32.Deleted in proof
- 33.Deleted in proof
- 34.Orena, S. J., Torchia, A. J., and Garofalo, R. S. (2000) J. Biol. Chem. 275 15765-15772 [DOI] [PubMed] [Google Scholar]
- 35.Abu-Elheiga, L., Matzuk, M. M., Abo-Hashema, K. A., and Wakil, S. J. (2001) Science 291 2613-2616 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.