Abstract
We observed previously that combined small interfering RNAs (siRNAs) targeting CrkII and CrkL, known activators of guanine nucleotide exchange factor DOCK1, strongly inhibit Caco-2 intestinal epithelial cell spreading and migration on collagen IV. DOCK1 siRNA reduced its expression >95% in Caco-2 cells but inhibited spreading much less than combined CrkII/CrkL siRNAs, suggesting that CrkII/CrkL interact with additional DOCK proteins. siRNA targeting DOCK5, a closely related DOCK1 family member, inhibited Caco-2 spreading similarly to DOCK1 siRNA, and the combined siRNAs synergistically inhibited spreading. Similar results were observed in human umbilical vein endothelial cells, and reverse transcriptase PCR demonstrated DOCK5 siRNA reduction of DOCK5 expression in both cell types. Combined DOCK1/DOCK5 siRNAs also inhibited Caco-2 migration and lamellipodial extension. Expression of DOCK5 cDNA, with silent mutations in the siRNA target region allowing expression simultaneously with DOCK5 siRNA, required CrkII/CrkL to restore cell spreading and DOCK5 coimmunoprecipitated with CrkII and CrkL. DOCK5 association with CrkII and CrkL was greatly reduced by mutations in their NH2-terminal SH3 domains. Expression of the DOCK5 COOH-terminal region (Met1738–Gln1870), containing potential Src homology 3 domain-binding proline-rich sites but lacking other functional regions, inhibited Caco-2 spreading and coimmunoprecipitated with CrkL. Coimmunoprecipitation of full-length DOCK5 with CrkL was strongly reduced by deletion of DOCK5 COOH-terminal amino acids 1832–1870. Green fluorescent protein-tagged DOCK5 localized to the membrane of Caco-2 cells spreading on collagen IV. In these studies, we describe human DOCK5 cloning and expression, our results indicating that, along with DOCK1, DOCK5 is an important mediator of CrkII/CrkL regulation of Caco-2 spreading and migration on collagen IV.
The Rac guanine-nucleotide exchange factor (GEF)2 DOCK1 (dedicator of cytokinesis 1) (1) belongs to a family of GEFs that activate either the Rac or CDC42 small G proteins (reviewed in Refs. 2–4). Genome analysis indicates that there are 11 family members in humans (5) and that DOCK1 orthologues are present in other organisms, including Drosophila melanogaster (Myoblast city (6)), Caenorhabditis elegans (Ced-5 (7)), and zebrafish (8). Association with the amino-terminal CrkII SH3 domain via a proline-rich region in the carboxyl-terminal part of DOCK1 contributes to DOCK1 translocation to focal adhesions (9, 10) and activation of RacI (11) and regulates formation of the DOCK1-ELMOI complex (12) that contributes to DOCK1 function in apoptotic cell phagocytosis in C. elegans and mammalian cell migration (9). Within the human DOCK1 family of proteins, DOCK5 has the closest sequence similarity to DOCK1. Although DOCK5 contains several proline-rich sequences in its carboxyl-terminal region, this region diverges considerably from the corresponding region of DOCK1, and human DOCK5 does not contain the consensus CrkII amino-terminal SH3 domain binding sequence (PXLPXK) that is present twice in the carboxyl-terminal region of DOCK1 (1, 13). siRNA knockdown studies indicate a role for DOCK5 in osteoclast differentiation (14), and morpholino-oligo knockdown of zebrafish DOCK5 (8) and knock-out of DOCK5 in mouse (15) indicate that DOCK5 functions in myoblast fusion in these organisms. Additionally, a mutation in DOCK5 is correlated with the rupture of lens cataract in mice (16). The DOCK5 functional mechanism, however, including its potential upstream activators and its role in integrin-mediated cell movement, has not been fully characterized.
Although intestinal epithelial cell adhesion and migration on matrix proteins have been described by several investigators, including our own group (17–23), the signaling mechanisms by which matrix regulates intestinal epithelial cells are poorly understood. Our previous work in human Caco-2 intestinal epithelial cells, a cell line that progressively differentiates as it grows past confluence and is thus a widely used human intestinal epithelial cell in vitro model system, suggests that Src kinase phosphorylation of p130cas and subsequent association of p130cas with the adaptor proteins CrkII and CrkL regulates intestinal epithelial cell spreading, lamellipodial extension, and sheet migration on collagen IV (24–26), which activates the α1β1 and α2β1 integrins (25) and is a major component of the intestinal epithelial basement membrane in vivo. The downstream effectors of CrkII and CrkL in this system, however, have not been identified. In this paper, we examine the role of DOCK1 and DOCK5 in regulation of Caco-2 intestinal epithelial cell spreading and migration on collagen IV using siRNAs that target these proteins and confirm these results in human umbilical vein endothelial cells (HUVECs). Additionally, we report the cloning and expression of the complete human DOCK5 cDNA, examine its association with CrkII and CrkL through coimmunoprecipitation studies using mutant forms of these proteins, and characterize the function of the expressed DOCK5 protein in cell spreading assays. We also characterize the intracellular localization of DOCK5 in Caco-2 cells by expression of green fluorescent protein-tagged DOCK5.
EXPERIMENTAL PROCEDURES
Materials—Dulbecco's modified Eagle's medium, Oligofectamine, Lipofectamine, and Plus Reagent and the β-galactosidase detection kit were obtained from Invitrogen. Collagen IV was obtained from Sigma. CrkII monoclonal and polyclonal antibodies were obtained from Transduction Laboratories (Lexington, KY) and Santa Cruz Biotechnology, Inc. (Santa Cruz, CA), respectively. CrkL polyclonal antibody was obtained from Santa Cruz Biotechnology. Monoclonal antibodies to α-tubulin and HA tag (12CA5) were obtained from Calbiochem and Roche Applied Science, respectively. Glutathione S-transferase (GST) polyclonal antibody was obtained from Cell Signaling Technology (Danvers, MA). Protein A/G-agarose was obtained either from Santa Cruz Biotechnology or EMD Biosciences (Gibbstown, NJ). siRNAs to human DOCK1, DOCK5, CrkII, and CrkL and control nontargeting siRNA 1 (NT1 control siRNA) were purchased from Dharmacon (Lafayette, CO). siRNA sequences were selected using Dharmacon Smartdesign and corresponded to the following sequences: DOCK1, GTACCGAGGTTACACGTTA; DOCK5, AGATTTACCTTCCGACAC (DOCK5-1), ATCCATTGCTATAGAAGAA (DOCK5-2), and TGATCCACGGTGAGTTTGA (DOCK5-3); CrkII, ACACTATTTGGACACTACA; CrkL, GTCACAAGGATGAATATAA. DNA primers were obtained from Integrated DNA Technologies (Coralville, IA). pKH3 expression vector was generously provided by Dr. Jun-Lin Guan (University of Michigan, Ann Arbor, MI). Aequorea coerulescens green fluorescent protein (AcGFP) vector and monoclonal antibody (JL-8) to AcGFP were obtained from Clontech.
Human DOCK5 Cloning and Expression—RNA from Caco-2 cells and HUVECs was obtained using TRIzol reagent (Invitrogen), and cDNA libraries were made using Superscript reverse transcriptase (Invitrogen). The full-length DOCK5 cDNA clone was assembled from two partial Caco-2 cDNA clones and one partial cDNA clone (AK126249) obtained from the NITE Biological Resource Center (Kisarazu-shi, Chiba Prefecture, Japan), extending from 137 to 1700, 1682 to 2977, and 2833 to 6266, respectively, of the most recently compiled DOCK5 sequence (accession number NM024940; open reading frame from 138 to 5750). Silent mutations were introduced into one of the DOCK5 siRNA target regions (DOCK5-2 targeting 1682–1700) by using overlapping primers for the two Caco-2 cDNA library clones containing the silent mutations in the sequences at the start of the 49-base pair primers. Caco-2 cDNA library clones were amplified using Vent polymerase (New England Biolabs, Beverly, MA) for PCR and then verified by sequencing. Aside from the silent mutations introduced into the siRNA target region, the Caco-2 cDNA clones differed from the DOCK5 sequence given in NM024940 at three positions, none of which affected the protein sequence: A for G at 1028, which changes a valine codon from GTG to GTA; C for T at 1946, which changes a serine codon from CTT to CTC; and G for A at 2786, which changes a threonine codon from ACA to ACG.
DOCK5 carboxyl-terminal deletion mutants were made by PCR using Vent polymerase or by using internal restriction enzyme sites to create in-frame stop codons. Eukaryotic expression expression vectors containing CrkII and CrkII W169L were obtained from Dr. Michiyuki Matsuda (Kyoto University, Kyoto, Japan), and PCR was used for cloning of cDNAs into pGEX4T-1 GST fusion protein expression vector (GE Healthcare). GST-CrkL expression vector was obtained from Dr. Ayako Arai and Dr. Osamu Miura (Tokyo Medical and Dental University), and PCR was used to introduce W160L mutation into the amino-terminal SH3 domain of this protein. All expression vectors derived by PCR were confirmed by sequencing.
Cell Culture—The Caco-2 cell line used for this work was a subclone (Caco-2BBE) of the original Caco-2 cell line that was selected for its ability to differentiate in culture, as indicated by the formation of an apical brush border and brush border enzyme expression, and has been previously described (27, 28). Passage 45–67 Caco-2 cells were cultured as described previously (26). HUVECs were obtained from the American Type Culture Collection (Manassas, VA) and cultured according to their instructions.
Coating of Cell Culture Dishes—Cell culture dishes were coated with a saturating concentration (29) of collagen IV (12.5 μg/ml) in precoating buffer (15 mm Na2CO3, 35 mm NaHCO3, pH 9.4). Collagen IV-coated tissue culture dishes were overlaid with 1% heat-inactivated (80 °C, 30 min) bovine serum albumin in phosphate-buffered saline for 45 min at 37 °C prior to spreading studies.
Cell Lysis—Cells were lysed on ice in modified radioimmunoprecipitation buffer (50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 1% Triton X-100, 10% glycerol, 1% deoxycholic acid, 0.1% SDS, 1 mm EDTA, 1 mm ethylene glycol-bis[β-aminoethyl ether]-N,N,N′,N′-tetraacetic acid, 50 mm phenylmethylsulfonyl fluoride, 1 mm Na3VO4, 1 mm NaF, 10 mm sodium pyrophosphate, 2 μg/ml aprotinin, 2 μg/ml leupeptin). For coimmunoprecipitation experiments, deoxycholic acid and SDS were omitted from the lysis buffer. Lysates were centrifuged at 15,000 × g for 10 min at 4 °C, and supernatants were stored at –80 °C or, for coimmunoprecipitation experiments, used directly. Protein concentrations were determined by the BCA® method (Pierce). Blots were detected by the ECL method (GE Healthcare) after transfer of gels to Immobilon P membranes (Millipore, Bedford, MA). Densitometry on autoradiographs of immunoblots was performed using an Eastman Kodak Co. 440CF Image Station within the linear range of exposure.
Expression of Glutathione S-Transferase Fusion Proteins—BL21(DE3)pLysS Escherichia coli (Invitrogen) were transformed with control pGEX vector or pGEX vectors expressing CrkII, CrkL, or the corresponding mutant proteins (CrkII W169L and CrkL W160L) with the invariant tryptophan in the amino-terminal SH3 domain changed to leucine. 1 mm isopropyl 1-thio-β-d-galactopyranoside was added to 10-ml log phase cultures of BL21(DE3)pLysS for 2 h at 33 °C to induce expression of GST fusion proteins. Cultures were then lysed in modified radioimmunoprecipitation buffer, and lysates were incubated with glutathione-Sepharose (GE Healthcare) for 2–3 h at 4 °C. After beads were rinsed with modified radioimmunoprecipitation buffer, lysates of Caco-2 cells expressing HA-DOCK5 were added, and immunoprecipitations were continued overnight at 4 °C.
Transfections—Caco-2 cells and HUVECs were transfected with siRNA using Oligofectamine and Plus reagent as described previously (26). For plasmid transfections, p100 dishes of Caco-2 cells were transfected with 5 μg of vector control or DOCK5 plasmid DNA, as described previously (26). For cell spreading experiments, cells were cotransfected with 1 μg of β-galactosidase expression vector to indicate cells transfected with plasmid DNA.
Cell Spreading and Migration—Cell spreading experiments were performed as described previously (26). Briefly, trypsinized cells were allowed to initially adhere for 15 min at 37 °C in serum-free medium to collagen IV-coated dishes blocked with bovine serum albumin. Nonadherent cells were then rinsed off with serum-free medium, and cells were allowed to continue spreading in serum-free medium at 37 °C for 90 min (Caco-2) or 30 min (HUVECs). Cells were plated at low density (∼5000 cells/well of a 6-well dish) to minimize cell-cell contacts that might interfere with cell spreading. Cells were stained with Harris modified hematoxylin after fixation with 10% formalin. Measurements of cell size were based on at least 150 cells for each condition in each experiment. For spreading experiments in which cells were transfected with plasmid DNA, cells were fixed and stained for β-galactosidase expression to identify transfected cells. In these experiments, measurements of cell size were based on at least 150 lacZ-positive cells for each condition in each experiment.
For migration experiments, cells transfected with NT1 control or DOCK1 and DOCK5 siRNAs were replated 2 days after transfection on collagen IV-coated p60 Petri dishes in 0.3% serum medium (to minimize proliferation after replating) at 3 × 106 cells/dish. A uniform migrating front was then created the next day in the confluent cell monolayer, as described previously (25), and migrations were allowed to continue for 16–20 h. Measurements of migration area were based on at least five random measurements for each condition in each of five or more independent experiments.
Statistical Analysis—Where indicated, results were compared using Student's t test and considered statistically significant when p was <0.05. When indicating significance, an asterisk indicates p < 0.05, a double asterisk indicates p < 0.01, and a triple asterisk indicates p < 0.001. All experiments were done independently at least three times unless otherwise indicated.
RESULTS
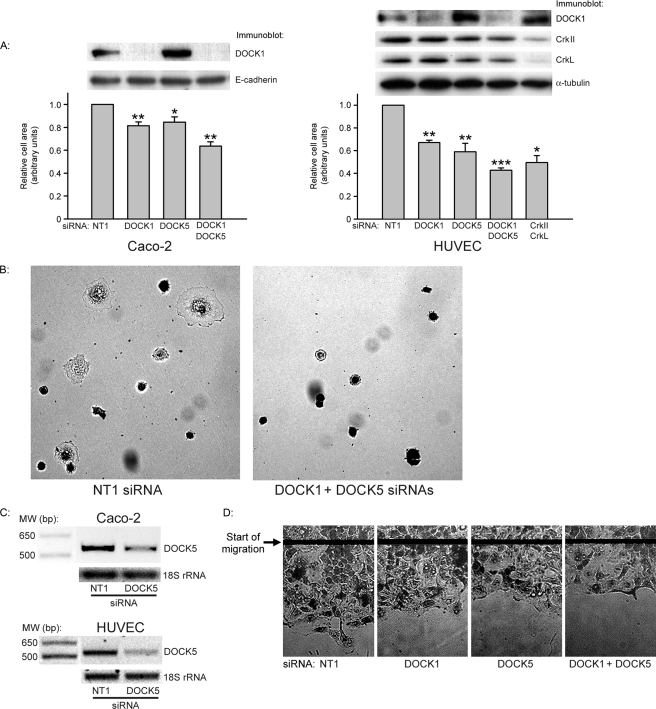
DOCK1 siRNA Only Partially Reduces Caco-2 Cell Spreading—We have previously observed that combined siRNA inhibition of the adaptor proteins CrkII and CrkL strongly inhibits Caco-2 cell spreading and migration on collagen IV by more than 40% and strongly inhibits lamellipodial formation at the leading edge of the migrating front (26). siRNA targeting the GEF DOCK1, which is a downstream effector of CrkII and CrkL in numerous cell systems (1, 30, 31), only reduced cell spreading by 15% despite reducing DOCK1 protein levels by more than 95% (Fig. 1A). Among the DOCK1 family members most closely related to DOCK1 is DOCK5. Two siRNAs specifically targeting different regions of the DOCK5 mRNA each reduced cell spreading similarly to DOCK1 siRNA, whereas the combined DOCK1 and DOCK5 siRNAs inhibited cell spreading synergistically (36 ± 4% inhibition, n = 8, p < 0.001 compared with NT1 control siRNA; Fig. 1A; images in Fig. 1B). In order to confirm our results in a nonepithelial cell line, we examined HUVEC spreading on collagen IV (Fig. 1A). The DOCK1 and DOCK5 siRNAs had an even greater effect in this cell type, and, as in Caco-2 cells, the combined siRNAs acted synergistically (33 ± 2, 41 ± 7, and 57 ± 2% inhibition, respectively, for DOCK1, DOCK5, and combined siRNAs; n ≥ 3, p < 0.01 for each condition). Additionally, the combined DOCK1 and DOCK5 siRNAs inhibited HUVEC spreading similarly to the combined Crk and CrkL siRNAs (50 ± 6% inhibition, n = 3, p < 0.05 compared with NT1 control). As in Caco-2 cells, DOCK5 results were confirmed in HUVECs with two different DOCK5 siRNAs. Reverse transcriptase PCR using primers to a carboxyl-terminal portion of the DOCK5 open reading frame confirmed expression of DOCK5 in Caco-2 cells and HUVECs and reduction of DOCK5 mRNA by DOCK5 siRNA (Fig. 1C). TA vector cloning of the Caco-2 PCR product additionally confirmed its identity as DOCK5. Although the individual siRNAs to DOCK1 and DOCK5 slightly, but not significantly, inhibited Caco-2 cell migration (11 ± 5 and 9 ± 4% inhibition, respectively, for individual DOCK1 and DOCK5 siRNAs, n = 7 for each), as for cell spreading, the combination of DOCK1 and DOCK5 siRNAs synergistically inhibited Caco-2 sheet migration on collagen IV by 20 ± 3% (Fig. 1D, n = 5, p < 0.01 compared with NT1 control siRNA-transfected cells). Similarly to migrating Caco-2 cells in which CrkII and CrkL are reduced by siRNA (26), Caco-2 cells transfected with the combination of DOCK1 and DOCK5 siRNAs had fewer lamellipodial extensions at the migrating front compared with NT1 control siRNA-transfected cells (Fig. 1D).
FIGURE 1.
A, effect of DOCK1 and DOCK5 siRNAs on Caco-2 cell and HUVEC spreading. Caco-2 or HUVECs were transfected with 200 nm NT1 control siRNA, 100 nm NT1 siRNA, and either 100 nm DOCK1 or DOCK5 siRNA or combined 100 nm DOCK1 and 100 nm DOCK5 siRNA and, after trypsinization, were replated on collagen IV and allowed to spread as described under “Experimental Procedures.” For comparison, HUVECs were also transfected with combined 100 nm CrkII and 100 nm CrkL siRNAs. Results are based on measurements of at least 150 cells for each condition in each of three or more experiments. *, p < 0.05 compared with NT1 siRNA; **, p < 0.01 compared with NT1 siRNA; ***, p < 0.001 compared with NT1 siRNA. B, representative images of NT1 control and combined DOCK1 and DOCK5 siRNA-transfected Caco-2 cells measured in A. C, PCR using primers to the carboxyl-terminal region of DOCK5 on cDNA libraries of Caco-2 cells or HUVECs transfected with either 100 nm NT1 control or DOCK5-2 siRNA. One of two (Caco-2) or three (HUVECs) similar experiments is shown. Expression of 18 S ribosomal RNA was used as a control. D, representative images of migrating Caco-2 cells on collagen IV transfected with either NT1 control, individual DOCK1 or DOCK5 siRNAs, or combined DOCK1 and DOCK5 siRNAs. The migration area was quantified from at least five random measurements for each condition in each of four independent experiments.
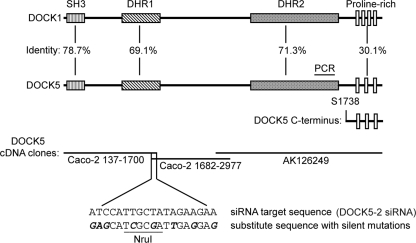
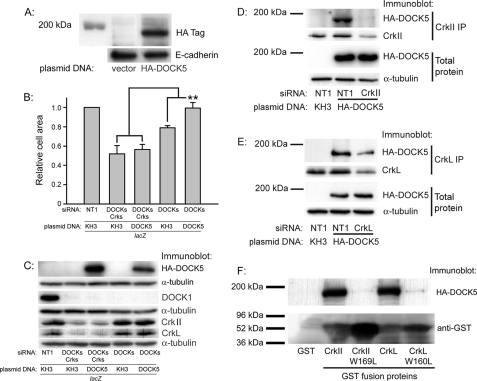
Dock5 Requires CrkII/CrkL in Order to Restore Cell Spreading—Full-length DOCK5 was assembled from Caco-2 cDNA library clones and from a cDNA containing the carboxyl-terminal portion of DOCK5 obtained from the NITE Biological Resource Center (Fig. 2). During the process of cloning, eight silent mutations were introduced into one of the DOCK5 siRNA target regions (DOCK5-2) to allow expression of the HA-tagged DOCK5 in the presence of the DOCK5 siRNA used in Fig. 1A and 1C. The HA-tagged DOCK5 protein was expressed at the predicted size of ∼190 kDa (Fig. 3A). HA-DOCK5 reexpression in Caco-2 cells transfected with both DOCK1 and DOCK5-2 siRNAs restored cell spreading to control levels, verifying the function of DOCK5 in cell spreading in Caco-2 cells. HA-DOCK5 reexpression, however, did not affect cell spreading after siRNA reduction of CrkII and CrkL (Fig. 3B; immunoblots in Fig. 3C), indicating that DOCK5 function in cell spreading requires CrkII/CrkL. Additionally, HA-DOCK5 was present in both CrkII (Fig. 3D) and CrkL (Fig. 3E) immunoprecipitates from lysates of subconfluent spreading Caco-2 cells on either collagen I (Fig. 3D) or collagen IV (not shown). Treatment of cells with CrkII or CrkL siRNA greatly reduced the amount of HA-DOCK5 in the CrkII or CrkL immunoprecipitates, confirming the specificity of the association. Together, these results suggest that both CrkII and CrkL promote cell movement in part via DOCK5 and that DOCK5 function in cell spreading requires CrkII and CrkL.
FIGURE 2.
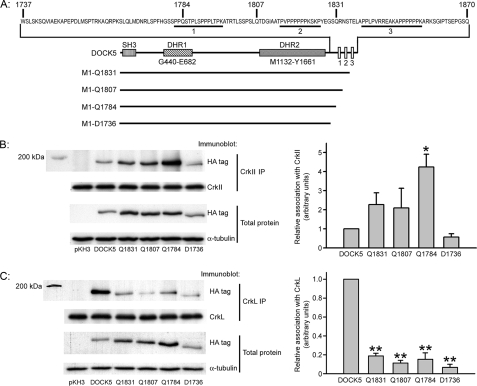
Cloning of Human Docks. Full-length human DOCK5 cDNA was assembled from Caco-2 cDNA clones and from clone AK126249 obtained from the NITE Biological Resource Center as described under “Experimental Procedures” and cloned into the pKH3 expression vector (amino-terminal HA tag) or into the AcGFP expression vector (amino-terminal AcGFP tag). Introduction of silent mutations to the DOCK5-2 siRNA target region, PCR target region for Fig. 1C, and carboxyl-terminal proline-rich region construct (Ser1738–Gln1870) and percentage amino acid identity between functional regions of DOCK1 and DOCK5 are also indicated. DHR1 and DHR2, Docker homology regions 1 and 2; these regions of DOCK1 are as defined by Cote and Vuori (5).
FIGURE 3.
DOCK5 expression and function in Caco-2 cell spreading. A, Caco-2 cells were transfected with pKH3 control vector or HA-tagged DOCK5 cDNA assembled as described under “Experimental Procedures.” B, HA-DOCK5 expression compensates for DOCK1 function only in the presence of CrkII/CrkL. Cells were transfected with 250 nm NT1 siRNA, 50 nm CrkII, CrkL, and DOCK1, and 100 nm DOCK5-2 siRNAs or with 100 nm NT1, 50 nm DOCK1, and 100 nm DOCK5-2 siRNAs and then transfected with pKH3 control vector or HA-DOCK5 cDNA as indicated, along with β-galactosidase expression plasmid to indicate transfected cells, as described under “Experimental Procedures.” For cell spreading studies, at least 150 lacZ-positive cells for each condition in each of three or more independent experiments were measured. **, p < 0.01 compared with pKH3 vector control C. The remaining cells not used for the spreading study in Fig. 3B were lysed and immunoblotted for the indicated proteins. D and E, DOCK5 associates with CrkII and CrkL. Lysates of subconfluent Caco-2 cells on collagen I transfected with the indicated siRNAs and with either pKH3 control vector or HA-DOCK5 were immunoprecipitated with CrkII or CrkL antibody and immunoblotted for HA-tagged DOCK5. One of three or more independent experiments is shown for both CrkII and CrkL. For both CrkII and CrkL coimmunoprecipitations, 100 nm DOCK5-2 siRNA was included in each transfection condition to reduce endogenous DOCK5 expression. F, CrkII and CrkL association with DOCK5 requires the amino-terminal SH3 domain of each protein. Lysates of Caco-2 cells transfected with HA-DOCK5 were immunoprecipitated with GST-CrkII, GST-CrkII W169L, GST-CrkL, or GST-CrkL W160L fusion proteins complexed to glutathione-Sepharose, as described under “Experimental Procedures,” and then immunoblotted for HA tag or GST. One of four similar experiments is shown.
Amino-terminal SH3 Domains of CrkII and CrkL Mediate Their Association with DOCK5—The CrkII amino-terminal SH3 domain mediates its association with the adaptor protein C3G (32) and with DOCK1 (33). The CrkII amino-terminal SH3 domain mutant W169L expressed at 10-fold lower levels than the wild-type protein in transfected Caco-2 cells (not shown), so in order to compare binding of the mutant and wild-type protein to DOCK5, they were expressed in E. coli as GST fusion proteins. Additionally, we compared the binding of DOCK5 to CrkL and CrkL with the corresponding mutation in its amino-terminal SH3 domain (W160L). Both CrkII W169L and CrkL W160L exhibited dramatically reduced association with DOCK5 compared with the respective unmutated protein (Fig. 3F), indicating that for both CrkII and CrkL, the amino-terminal SH3 domain is essential for mediating association with DOCK5.
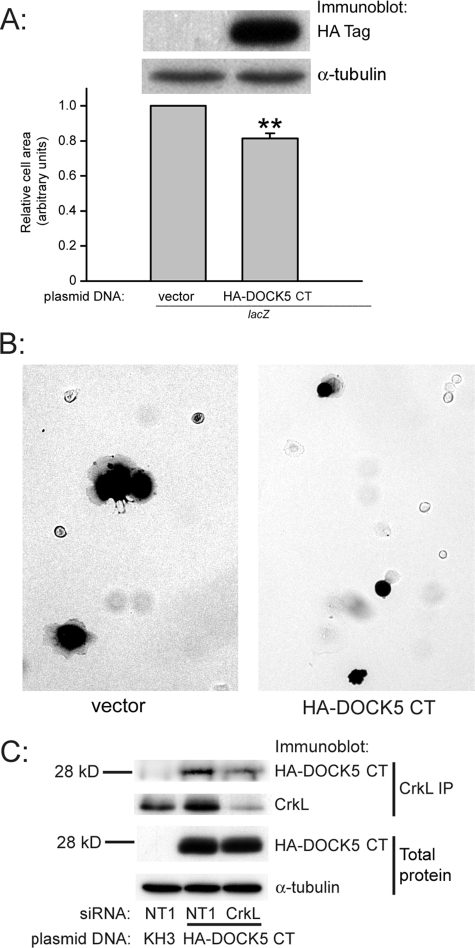
Expression of the DOCK5 Carboxyl-terminal Region Inhibits Caco-2 Cell Spreading—We expressed the carboxyl terminal proline-rich region of DOCK5 to determine its effect on cell spreading. Expression of HA-DOCK5 Ser1738–Gln1870 significantly inhibited Caco-2 cell spreading by 19 ± 3% (n = 6, p < 0.01; Fig. 4A; representative images in Fig. 4B), suggesting a function for this region of DOCK5 in mediating cell spreading. HA-DOCK5 Ser1738–Gln1870 was present in CrkL immunoprecipitates from subconfluent spreading Caco-2 cells on both collagen I (Fig. 4C) and collagen IV (not shown). We were not, however, able to detect HA-DOCK5 Ser1738–Gln1870 in CrkII immunoprecipitates using a CrkII antibody that, like the CrkL antibody, was raised to the carboxyl-terminal end of the protein, or detect CrkII in HA immunoprecipitates from these experiments (not shown).
FIGURE 4.
A, expression of the DOCK5 carboxyl-terminal proline-rich region inhibits cell spreading. Cells were transfected with either pKH3 control vector or HA-DOCK5 Ser1738–Gln1870 (HA-DOCK5 CT; Fig. 2) along with the β-galactosidase expression vector to indicate transfected cells, as described under “Experimental Procedures.” The remaining cells were lysed and immunoblotted for the HA-tagged DOCK5 Ser1738–Gln1870. Results are based on measurements of at least 150 lacZ-positive cells for each condition in each of six independent experiments. **, p < 0.01 compared with pKH3 vector control. B, representative images of cells stained for β-galactosidase expression from A. C, CrkL associates with DOCK5 Ser1738–Gln1870. Subconfluent Caco-2 cells on collagen I transfected with the indicated siRNAs and with either pKH3 control vector or HA-DOCK5 Ser1738–Gln1870 were lysed, immunoprecipitated with CrkL antibody, and immunoblotted for HA-tagged DOCK5 Ser1738-Gln1870. One of three similar experiments is shown.
Identification of DOCK5 Carboxyl-terminal Regions Involved in CrkII and CrkL Binding—Although DOCK1 and DOCK5 are closely related throughout the SH3, Docker homology region 1, and Docker homology region 2 domains, the proteins diverge in the carboxyl-terminal proline-rich region that in DOCK1 mediates interaction with the SH3 domain of CrkII (Fig. 2), and the human DOCK5 C-terminal region does not contain a precise match for the consensus CrkII binding region (PXLPXK) that is present in DOCK1 (Fig. 5A). In the DOCK5 carboxyl-terminal region, there are three proline-rich regions, so we made a series of carboxyl-terminal deletion mutants in which these regions are successively removed and examined their association with CrkII and CrkL (Fig. 5A). Interestingly, CrkII and CrkL exhibited different patterns of association with the DOCK5 deletion mutants. CrkII association with HA-DOCK5 Met1–Gln1831 and Met1–Gln1807 was increased compared with CrkII association with full-length DOCK5. This increase, however, was not significant and appeared to correlate with the increased expression of these deletion mutants compared with full-length DOCK5. CrkII association with HA-DOCK5 Met1–Gln1784 was significantly increased by more than 4-fold compared with association with full-length DOCK5, and this increased association appeared much greater than the slightly increased expression of this deletion mutant compared with full-length DOCK5. Further deletion of amino acids 1737–1784 eliminated this increased association (Fig. 5B). Unlike CrkII, however, CrkL association with DOCK5 was reduced by 81 ± 3% (n = 3, p < 0.01) when amino acids 1832–1870 were deleted, despite the higher expression level of this deletion mutant compared with full-length DOCK5 (Fig. 5C).
FIGURE 5.
Association of DOCK5 carboxyl-terminal deletion mutants with CrkII and CrkL. A, diagram of DOCK5 deletion mutants and proline-rich regions in the DOCK5 carboxyl-terminal region. B, association of DOCK5 deletion mutants with CrkII. Lysates of Caco-2 cells expressing the indicated DOCK5 deletion mutant were immunoprecipitated with CrkII antibody and then immunoblotted for HA tag or CrkII. *, p < 0.05 compared with full-length HA-DOCK5. C, association of DOCK5 deletion mutants with CrkL. Lysates of Caco-2 cells expressing the indicated DOCK5 deletion mutant were immunoprecipitated with CrkL antibody and then immunoblotted for HA tag or CrkL. In both B and C, total cell lysates were also immunoblotted for HA tag to measure relative expression of HA-DOCK5 mutants. **, p < 0.01 compared with full-length HA-DOCK5. Densitometric analysis of DOCK5 mutant association with CrkII or CrkL is based on three or more independent experiments.
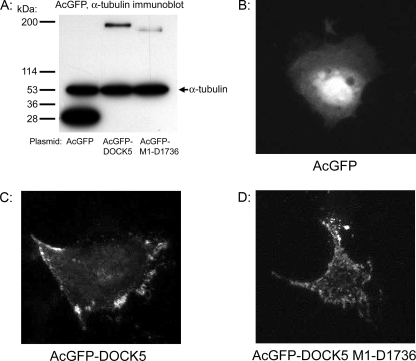
Intracellular Localization of DOCK5—To examine the intracellular localization of DOCK5, we expressed it as an AcGFP-tagged fusion protein. In anti-AcGFP immunoblots of lysates from transfected cells, AcGFP-DOCK5 was expressed at the expected size of ∼200 kDa (Fig. 6A). Although AcGFP exhibited strong nuclear localization with diffuse cytoplasmic expression (Fig. 6B), AcGFP-DOCK5 exhibited punctate membranous localization (Fig. 6C) and was not present in the nucleus of spreading Caco-2 cells on collagen IV, consistent with a role for this protein in spreading and migration of Caco-2 cells. AcGFP-DOCK5 Met1–Gln1736 also exhibited punctate membranous localization similar to full-length DOCK5, suggesting that association with CrkII or CrkL may not be essential for DOCK5 membrane association (Fig. 6D).
FIGURE 6.
Localization of AcGFP-DOCK5 in Caco-2 cells spreading on collagen IV. A, AcGFP immunoblots of lysates of cells transfected with AcGFP control vector, AcGFP-DOCK5 fusion protein, or AcGFP-DOCK5 Met1–Asp1736. B–D, representative images of spreading cells transfected, respectively, with AcGFP, AcGFP-DOCK5, or AcGFP-DOCK5 Met1–Asp1736, as described under “Experimental Procedures.”
DISCUSSION
The differential expression of integrins in intestinal epithelial cells and basement membrane extracellular matrix proteins along the intestinal epithelial crypt-villus axis in vivo (34, 35) (reviewed in Ref. 36) suggests a potential role for cell-matrix interactions in regulating intestinal epithelial cell differentiation and migration. Our previous work with chemical inhibitors of Src, dominant negative forms of Src and p130cas, and siRNAs specifically targeting p130cas and the adaptor proteins CrkII and CrkL indicates that these proteins are important regulators of Caco-2 intestinal epithelial cell spreading, migration, and lamellipodial extension on the intestinal epithelial basement membrane matrix protein collagen IV, which activates the α1β1 and α2β1 integrins in Caco-2 cells (25, 26). In the current work, we determined that the CrkII-binding GEF DOCK1 functions in Caco-2 spreading and migration on collagen IV, as has previously been observed for A549 cells on the α3β1 integrin-activating matrix proteins laminin 10/11 (37). In addition, however, we also determined through siRNA and cDNA expression studies that the closely related DOCK1 family member DOCK5 functions in these processes as well.
DOCK5 Regions Involved in CrkII and CrkL SH3 Binding; Comparison with Other DOCK1 Family Members—Our evidence suggests that CrkII and CrkL are necessary for DOCK5 function in Caco-2 cell spreading (Fig. 3B) and that DOCK5 associates with both CrkII and CrkL (Fig. 3, D–F). This is consistent with recent work in zebrafish in which morpholino oligonucleotide reduction of either CrkII or DOCK5 results in similar defects in myoblast fusion (8). DOCK5 association with CrkII and CrkL is dramatically reduced by mutation of the invariant tryptophan in the amino-terminal SH3 domain of each protein (Fig. 3F). The consensus binding site for the amino-terminal SH3 domain of CrkII and CrkL, PXLPXK, is present at two sites within the carboxyl-terminal proline-rich region of DOCK1. DOCK5 does not contain an exact match for this consensus sequence but has three near matches in its proline-rich region: PPLPVR, at amino acids 1839–1844, and PPPPPK at both 1818–1823 and 1851–1856 (Fig. 6A). We detected the carboxyl-terminal proline-rich fragment of DOCK5, which inhibits cell spreading (Fig. 4A), in CrkL immunoprecipitates (Fig. 4C), indicating that DOCK5 association with CrkL is mediated by sequences in the Ser1738–Gln1870 region. DOCK5 Met1–Gln1831 association with CrkL was dramatically reduced compared with association of the full-length DOCK5 protein with CrkL (Fig. 5C), consistent with a potential role for the sequence PPLPVR at amino acids 1839–1844 or PPPPPK at amino acids 1851–1856 in mediating DOCK5 association with CrkL. More detailed site-directed mutagenesis of the amino acid 1832–1870 region of DOCK5 will be required to precisely determine the sequences within this region of DOCK5 that mediate its association with CrkL.
The pattern of CrkII association with the DOCK5 deletion mutants (Fig. 5B) clearly differed from that of CrkL (Fig. 5C). The significantly increased CrkII association with DOCK5 Met1–Gln1784 compared with full-length DOCK5 might be partially explained by the increased expression of the deletion mutant compared with the full-length protein, but this would not explain the difference in the pattern of association of the DOCK5 deletion mutants with CrkII and CrkL. This is a somewhat surprising result, since CrkII and CrkL have been found to have similar binding properties in most studies (e.g. see Refs. 38–40; reviewed in Ref. 41). One possible explanation for this result might be that deletion of amino acids 1785–1870 prevents DOCK5 interaction with other proteins that may bind to this region and frees up DOCK5 Met1–Gln1784 to interact with CrkII. Another closely related DOCK1 family member, DOCK2, has been found to associate with CrkL in human leukemia cell lines via one region in the amino-terminal part of the protein and another region in the Docker homology region 2 domain, neither of which contains proline-rich sequences (42). Additionally, DOCK2 did not coimmunoprecipitate with CrkII in this study, indicating that at least one other DOCK family member interacts differently with CrkII and CrkL. Association of DOCK5 Met1–Asp1736 with CrkII is strongly reduced compared with DOCK5 Met1–Gln1784, suggesting that the region between amino acids 1736 and 1784 may function in CrkII-DOCK5 association. Although there are no proline-rich sequences in this region conforming to the CrkII consensus binding site, the sequence from amino acid 1757 to 1765 (PTRKAQRPK) is enriched for proline and the basic amino acids arginine and lysine, and numerous recent studies (reviewed in Refs. 43 and 44) have suggested that RK-rich sequences can bind to SH3 domains. If this region mediates the association of DOCK5 with CrkII, this might explain the failure to detect DOCK5 Ser1738-Gln1870 (Fig. 4) in CrkII immunoprecipitates, since the close proximity of the HA tag to this region might affect CrkII-DOCK5 Ser1738–Gln1870 association. Additionally, there may be other regions outside of Ser1738–Gln1870 that contribute to DOCK5-CrkII association. As for CrkL, additional DOCK5 mutagenesis and deletion mutants will need to be examined for their association with CrkII to more precisely map the regions of DOCK5 necessary for interaction with CrkII.
DOCK5 Intracellular Localization—Our results indicate that DOCK5 is associated with the edge of the plasma membrane in Caco-2 cells spreading on collagen IV (Fig. 6C). Although it is possible that the weaker association of DOCK5 Met1–Asp1736 with CrkII and CrkL (Fig. 5) is sufficient to target it to the plasma membrane, the membrane localization of DOCK5 Met1–Asp1736 (Fig. 6D) suggests that association with CrkII and CrkL may not be essential for DOCK5 membrane localization (Fig. 6D) and that other protein interactions may contribute to DOCK5 membrane localization. The sequence homology of DOCK5 to DOCK1 suggests that DOCK5 may associate with ELMO family members. In LR73 Chinese Hamster Ovary cells, DOCK1 localizes to the cytoplasm when expressed alone, but some DOCK1 localizes to membrane-proximal regions when coexpressed with ELMO1 in the absence of CrkII (9). It has been recently determined that amino acids 1–187 of DOCK1 are important for mediating its association with ELMO1 (45). This region is 74% identical between DOCK1 and DOCK5, suggesting DOCK5 may also associate with ELMO1 or the related proteins ELMO2 and ELMO3.
Functional Overlap of DOCK1 and DOCK5—Our data indicate that both DOCK1 and DOCK5 regulate both Caco-2 and HUVEC spreading on collagen IV (Fig. 1A) and that reexpression of HA-DOCK5 is able to completely restore cell spreading in Caco-2 cells transfected with siRNAs targeting DOCK1 and DOCK5 (Fig. 3B). Although this result may be affected by possible overexpression of recombinant HA-DOCK5 relative to the combined endogenous levels of DOCK1 and DOCK5, this result suggests that DOCK5 can compensate for loss of DOCK1 function in Caco-2 cells spreading on collagen IV. Additionally, both DOCK5 and DOCK1 regulate myoblast fusion during zebrafish development (8), although the comparable studies in zebrafish to Fig. 3B were not performed in this study. In DOCK1 knock-out mice, embryogenic myoblast fusion is only partially inhibited, and knock-out of DOCK5 in addition to DOCK1 results in further inhibition of this process (15). Along with the sequence similarity among the two proteins, this suggests that DOCK5 may be able to functionally compensate for loss of DOCK1 function in systems in which the two proteins are coexpressed. It will be of great interest to determine whether DOCK5 can fully or partially compensate for DOCK1 or its orthologues in other cell systems, such as ced-5 mutant Caenorhabditis elegans, in which expression of human DOCK1 is able to restore distal tip cell migration (7). Additionally, it will be of interest to determine whether DOCK5 shares other functional properties of DOCK1, such as the contribution of ELMOI association to activation of Rac found in some studies (46) (see Ref. 5 for differing results), the functional requirement for association with phosphatidylinositol 3,4,5-trisphosphate (47), and the intramolecular association of the amino-terminal SH3 domain with the GEF catalytic domain when the protein is in an inactive conformation (48).
In conclusion, we have observed an important role for DOCK1 and DOCK5 in regulation of human intestinal epithelial Caco-2 cell spreading and migration on collagen IV. Additionally, our data indicate that interaction of DOCK5 with CrkII and CrkL participates in regulation of Caco-2 cell spreading on collagen IV and that DOCK5 localizes to the plasma membrane in spreading Caco-2 cells. We observed that expression of DOCK5 is able to compensate for siRNA inhibition of DOCK1 expression in Caco-2 cell spreading. Taken together with our previous observations (23, 25, 26), the results described in this work suggest that CrkII and CrkL association with Src-phosphorylated p130cas initiated by intestinal epithelial basement membrane matrix proteins, such as type IV collagen, may regulate intestinal epithelial cell migration through multiple DOCK1 GEF family members, including DOCK1 and DOCK5.
Acknowledgments
We thank the following for technical assistance and generous gifts of expression vectors: Dr. Xuebao Zhang (Department of Neurology, Wayne State University Medical School) and Dr. Cheri Owen (Department of Surgery, Wayne State University Medical School) and the laboratory of Dr. Paul Skoff (Department of Anatomy, Wayne State University Medical School) for assistance with microscopy; Dr. Jun-Lin Guan (University of Michigan, Ann Arbor, MI) for providing pKH3; Dr. Michiyuki Matsuda (Graduate School of Medicine, Kyoto University, Kyoto, Japan) for providing CrkII and CrkII W169L expression vectors; Dr. Ayako Arai and Dr. Osamu Miura (Department of Hematology, Tokyo Medical and Dental University) for providing the GST-CrkL expression vector; Dr. Arun Rishi (Department of Internal Medicine, Wayne State University) for helpful discussions about DOCK5 cloning; and the Applied Genomics Technology Center at Wayne State University for DNA sequencing.
This work was supported, in whole or in part, by National Institutes of Health Grant RO1DK067257. This work was also supported by a Veterans Affairs Merit Award (to M. D. B.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: GEF, guanine-nucleotide exchange factor; AcGFP, A. coerulescens green fluorescent protein; GST, glutathione S-transferase; HUVEC, human umbilical vein endothelial cell; siRNA, small interfering RNA; NT1 siRNA, nontargeting control siRNA 1; SH3, Src homology 3; HA, hemagglutinin.
References
- 1.Hasegawa, H., Kiyokawa, E., Tanaka, S., Nagashima, K., Gotoh, N., Shibuya, M., Kurata, T., and Matsuda, M. (1996) Mol. Cell. Biol. 16 1770–1776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cote, J. F., and Vuori, K. (2006) Methods Enzymol. 406 41–57 [DOI] [PubMed] [Google Scholar]
- 3.Cote, J. F., and Vuori, K. (2007) Trends Cell Biol. 17 383–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meller, N., Merlot, S., and Guda, C. (2005) J. Cell Sci. 118 4937–4946 [DOI] [PubMed] [Google Scholar]
- 5.Cote, J. F., and Vuori, K. (2002) J. Cell Sci. 115 4901–4913 [DOI] [PubMed] [Google Scholar]
- 6.Nolan, K. M., Barrett, K., Lu, Y., Hu, K. Q., Vincent, S., and Settleman, J. (1998) Genes Dev. 12 3337–3342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu, Y. C., and Horvitz, H. R. (1998) Nature 392 501–504 [DOI] [PubMed] [Google Scholar]
- 8.Moore, C. A., Parkin, C. A., Bidet, Y., and Ingham, P. W. (2007) Development 134 3145–3153 [DOI] [PubMed] [Google Scholar]
- 9.Gumienny, T. L., Brugnera, E., Tosello-Trampont, A. C., Kinchen, J. M., Haney, L. B., Nishiwaki, K., Walk, S. F., Nemergut, M. E., Macara, I. G., Francis, R., Schedl, T., Qin, Y., Van Aelst, L., Hengartner, M. O., and Ravichandran, K. S. (2001) Cell 107 27–41 [DOI] [PubMed] [Google Scholar]
- 10.Kiyokawa, E., Hashimoto, Y., Kurata, T., Sugimura, H., and Matsuda, M. (1998) J. Biol. Chem. 273 24479–24484 [DOI] [PubMed] [Google Scholar]
- 11.Kiyokawa, E., Hashimoto, Y., Kobayashi, S., Sugimura, H., Kurata, T., and Matsuda, M. (1998) Genes Dev. 12 3331–3336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Akakura, S., Kar, B., Singh, S., Cho, L., Tibrewal, N., Sanokawa-Akakura, R., Reichman, C., Ravichandran, K. S., and Birge, R. B. (2005) J. Cell. Physiol. 204 344–351 [DOI] [PubMed] [Google Scholar]
- 13.Matsuda, M., Ota, S., Tanimura, R., Nakamura, H., Matuoka, K., Takenawa, T., Nagashima, K., and Kurata, T. (1996) J. Biol. Chem. 271 14468–14472 [DOI] [PubMed] [Google Scholar]
- 14.Brazier, H., Stephens, S., Ory, S., Fort, P., Morrison, N., and Blangy, A. (2006) J. Bone Miner. Res. 21 1387–1398 [DOI] [PubMed] [Google Scholar]
- 15.Laurin, M., Fradet, N., Blangy, A., Hall, A., Vuori, K., and Cote, J. F. (2008) Proc. Natl. Acad. Sci. U. S. A. 105 15446–15451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Omi, N., Kiyokawa, E., Matsuda, M., Kinoshita, K., Yamada, S., Yamada, K., Matsushima, Y., Wang, Y., Kawai, J., Suzuki, M., Hayashizaki, Y., and Hiai, H. (2008) Exp Eye Res. 86 828–834 [DOI] [PubMed] [Google Scholar]
- 17.Basson, M. D., Modlin, I. M., and Madri, J. A. (1992) J. Clin. Invest. 90 15–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.De Arcangelis, A., Neuville, P., Boukamel, R., Lefebvre, O., Kedinger, M., and Simon-Assmann, P. (1996) J. Cell Biol. 133 417–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lotz, M. M., Nusrat, A., Madara, J. L., Ezzell, R., Wewer, U. M., and Mercurio, A. M. (1997) Am. J. Pathol. 150 747–760 [PMC free article] [PubMed] [Google Scholar]
- 20.Lotz, M. M., Rabinovitz, I., Mercurio, A. M., Nusrat, A., Madara, J. L., Ezzell, R., and Wewer, U. M. (2000) Am. J. Pathol. 156 985–996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vachon, P. H., and Beaulieu, J. F. (1995) Am. J. Physiol. 268 G857–G867 [DOI] [PubMed] [Google Scholar]
- 22.Wolpert, S., Wong, M. L., and Bass, B. L. (1996) Am. J. Surg. 171 109–112 [DOI] [PubMed] [Google Scholar]
- 23.Yu, C. F., Sanders, M. A., and Basson, M. D. (2000) Am. J. Physiol. 278 G952–G966 [DOI] [PubMed] [Google Scholar]
- 24.Sanders, M. A., and Basson, M. D. (2000) J. Biol. Chem. 275 38040–38047 [DOI] [PubMed] [Google Scholar]
- 25.Sanders, M. A., and Basson, M. D. (2004) Am. J. Physiol. 286 G547–G557 [DOI] [PubMed] [Google Scholar]
- 26.Sanders, M. A., and Basson, M. D. (2005) J. Biol. Chem. 280 23516–23522 [DOI] [PubMed] [Google Scholar]
- 27.Peterson, M. D., Bement, W. M., and Mooseker, M. S. (1993) J. Cell Sci. 105 461–472 [DOI] [PubMed] [Google Scholar]
- 28.Peterson, M. D., and Mooseker, M. S. (1993) J. Cell Sci. 105 445–460 [DOI] [PubMed] [Google Scholar]
- 29.Madri, J. A., Pratt, B. M., and Yannariello-Brown, J. (1988) Am. J. Pathol. 132 18–27 [PMC free article] [PubMed] [Google Scholar]
- 30.Albert, M. L., Kim, J. I., and Birge, R. B. (2000) Nat Cell Biol. 2 899–905 [DOI] [PubMed] [Google Scholar]
- 31.Reddien, P. W., and Horvitz, H. R. (2000) Nat Cell Biol. 2 131–136 [DOI] [PubMed] [Google Scholar]
- 32.Knudsen, B. S., Feller, S. M., and Hanafusa, H. (1994) J. Biol. Chem. 269 32781–32787 [PubMed] [Google Scholar]
- 33.Tanaka, S., Hattori, S., Kurata, T., Nagashima, K., Fukui, Y., Nakamura, S., and Matsuda, M. (1993) Mol. Cell. Biol. 13 4409–4415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Beaulieu, J. F. (1992) J. Cell Sci. 102 427–436 [DOI] [PubMed] [Google Scholar]
- 35.Beaulieu, J. F., and Vachon, P. H. (1994) Gastroenterology 106 829–839 [DOI] [PubMed] [Google Scholar]
- 36.Beaulieu, J. F. (1997) Prog. Histochem. Cytochem. 31 1–78 [DOI] [PubMed] [Google Scholar]
- 37.Gu, J., Sumida, Y., Sanzen, N., and Sekiguchi, K. (2001) J. Biol. Chem. 276 27090–27097 [DOI] [PubMed] [Google Scholar]
- 38.Feller, S. M., Knudsen, B., and Hanafusa, H. (1995) Oncogene 10 1465–1473 [PubMed] [Google Scholar]
- 39.Posern, G., Zheng, J., Knudsen, B. S., Kardinal, C., Muller, K. B., Voss, J., Shishido, T., Cowburn, D., Cheng, G., Wang, B., Kruh, G. D., Burrell, S. K., Jacobson, C. A., Lenz, D. M., Zamborelli, T. J., Adermann, K., Hanafusa, H., and Feller, S. M. (1998) Oncogene 16 1903–1912 [DOI] [PubMed] [Google Scholar]
- 40.Sattler, M., Salgia, R., Okuda, K., Uemura, N., Durstin, M. A., Pisick, E., Xu, G., Li, J. L., Prasad, K. V., and Griffin, J. D. (1996) Oncogene 12 839–846 [PubMed] [Google Scholar]
- 41.Feller, S. M. (2001) Oncogene 20 6348–6371 [DOI] [PubMed] [Google Scholar]
- 42.Nishihara, H., Maeda, M., Oda, A., Tsuda, M., Sawa, H., Nagashima, K., and Tanaka, S. (2002) Blood 100 3968–3974 [DOI] [PubMed] [Google Scholar]
- 43.Kaneko, T., Li, L., and Li, S. S. (2008) Front. Biosci. 13 4938–4952 [DOI] [PubMed] [Google Scholar]
- 44.Li, S. S. (2005) Biochem. J. 390 641–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Komander, D., Patel, M., Laurin, M., Fradet, N., Pelletier, A., Barford, D., and Cote, J. F. (2008) Mol. Biol. Cell 19 4837–4851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brugnera, E., Haney, L., Grimsley, C., Lu, M., Walk, S. F., Tosello-Trampont, A. C., Macara, I. G., Madhani, H., Fink, G. R., and Ravichandran, K. S. (2002) Nat. Cell Biol. 4 574–582 [DOI] [PubMed] [Google Scholar]
- 47.Cote, J. F., Motoyama, A. B., Bush, J. A., and Vuori, K. (2005) Nat. Cell Biol. 7 797–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lu, M., Kinchen, J. M., Rossman, K. L., Grimsley, C., Hall, M., Sondek, J., Hengartner, M. O., Yajnik, V., and Ravichandran, K. S. (2005) Curr. Biol. 15 371–377 [DOI] [PubMed] [Google Scholar]