Abstract
Many studies report the level of total viral DNA in HIV-infected patients, but few studies report the level of integrated DNA. It is important to measure integrated DNA in HIV-infected patients because the information could shed light on the effectiveness of antiretroviral therapy, especially intensified therapy, when viral loads may remain undetectable. In order to develop an integration assay for patient samples we enhanced the sensitivity of our prior integration assay. To do this, we exploited a technique that we developed, called repetitive sampling, and optimized reaction conditions for rare event detection, rather than large dynamic range. We also redesigned our primers to match more conserved regions of HIV. The result is a new, sensitive, quantitative assay that allows us to measure integrated DNA in HIV-infected patients. When we applied our integration assay to patient PBMCs, we found that use of HAART is associated with reduced levels of integrated DNA.
Keywords: HIV, integration, latency, persistence, reservoirs, HAART, antiretroviral therapy, eradication, ongoing replication, intensification
Introduction
Integration is a central step in the HIV life cycle: it is required for efficient transcription of the genome, for replication, and for establishment of reservoirs. Given that integration is essential for viral survival in the host, it is important to monitor the level of integrated DNA. In addition, unintegrated DNA is only transcribed at low levels (Brussel and Sonigo, 2004; Nakajima, Lu, and Engelman, 2001; Wu and Marsh, 2001; Wu and Marsh, 2003a; Wu and Marsh, 2003b), and is likely to contribute little to productive HIV infection (Nakajima, Lu, and Engelman, 2001). Thus, it may be important to distinguish between integrated an unintegrated HIV DNA. However, measuring integration accurately in patient samples is difficult for two reasons. First, in HIV-infected individuals, unintegrated DNA is present in excess over integrated DNA and so routine PCR does not distinguish unintegrated from integrated DNA. Second, the level of integration in HIV-infected individuals is low and detection is a further challenge since only a fraction of integration events are detectable because of the widely distributed integration sites (Agosto et al., 2007). While many studies have reported the level of total viral DNA in cells from HIV-infected individuals (Brenchley et al., 2004; Burgard et al., 2000; Debiaggi et al., 2000; Douek et al., 2002; Gibellini et al., 2004; Jubault et al., 1998; Kabamba-Mukadi et al., 2005; Kostrikis et al., 2002; Lillo et al., 2004; Ngo-Giang-Huong et al., 2001; Rouzioux et al., 2005; Viard et al., 2004; Vitone et al., 2005), only a few have reported the level of integrated DNA (Carr et al., 2007; Chun et al., 1997a; Chun et al., 1995; Chun et al., 1997b; Ibanez et al., 1999; Izopet et al., 2002; Koelsch et al., 2008; Ostrowski et al., 1999).
Currently there are three PCR assays that measure HIV integration: linker ligation (Vandegraaff et al., 2001), inverse PCR (Chun et al., 1997a), and Alu-PCR (Brussel, Delelis, and Sonigo, 2005; Brussel and Sonigo, 2003; Butler, Hansen, and Bushman, 2001; O'Doherty et al., 2002). Both endpoint and kinetic PCR methods have been applied to these assays, and both methods have their advantages: endpoint PCR is usually more sensitive and kinetic PCR provides superior quantitation. Studies using endpoint PCR integration assays (Chun et al., 1997a; Chun et al., 1998; Chun et al., 1995; Chun et al., 1997b; Ostrowski et al., 1999) revealed the groundbreaking finding that resting CD4+ T cells contain integrated DNA and contribute to viral reservoirs (Chun et al., 1997a; Chun et al., 1995). However, the major limitations of end point PCR are that it is too labor intensive for the high throughput needed for clinical samples, and it provides poor quantitation. For example, two studies using endpoint PCR assays to compare integration levels in total PBMCs from patients on and off highly active anti-retroviral therapy (HAART) reported conflicting results (Ibanez et al., 1999; Izopet et al., 2002). The conflicting results were in part related to the wide uncertainty in early estimates of HIV integration because the variation caused by sampling rare events was not addressed. An additional source of error in the measurements may have come from false positives; unintegrated DNA may have contributed to the signal.
Many kinetic PCR integration assays have been developed, and kinetic PCR can control for false positive signals better than endpoint PCR, as we described by including gag-only controls (Agosto et al., 2007; O'Doherty et al., 2002). However, none of the kinetic PCR assays that have been developed are sensitive enough to measure integration in patient samples. For example, a recent study proposed a kinetic PCR integration assay for clinical use, but the assay did not detect integration in ~50% of patient PBMCs (Carr et al., 2007). Thus, integration data was only obtained for half the patients.
Here, we report a kinetic PCR assay, derived from our original two-step kinetic Alu-PCR integration assay (O'Doherty et al., 2002), that is sensitive enough to measure HIV integration precisely in PBMC samples from patients. To do this, we redesigned our primers and probes to match more conserved regions of the HIV genome and we incorporated a technique that repetitively samples DNA (Agosto et al., 2007). However, even with incorporation of the repetitive sampling technique to patient samples, the assay still lacked the required sensitivity (unpublished observations). Here we show that we can further increase the sensitivity of our integration assay by increasing the number of cycles in the 1st PCR reaction. Previously, we limited the number of cycles to maintain a large dynamic range (i.e. to ensure that substrates are not limiting). However, in patient samples the proviral levels are low and dynamic range is small. Under these conditions, a dramatic increase in sensitivity can be obtained by increasing cycle number without compromising quantitation. We used this new kinetic Alu-PCR integration assay to measure proviral levels in PBMCs from patients on and off HAART. Compared to patients not on HAART, we found that patients on HAART had significantly lower levels of integrated HIV DNA (p=0.007). Furthermore, we found the level of integrated DNA was higher than the original estimates in patients off HAART (Ibanez et al., 1999; Izopet et al., 2002; Koelsch et al., 2008).
Results
Increasing PCR cycle number increases integration assay sensitivity (Table 1)
Table 1. Increasing the number of cycles in the first PCR reaction enhanced the sensitivity of the integration assay.
The integration standard was diluted to 20 proviruses in 15,000 cells and then assayed repetitively with 20, 30 or 40 cycles, while the amount of enzyme, extension time and other variables were kept constant. The number of cycles performed in the first PCR reaction, the average Alu-gag and gag-only signal, as captured by cycle thresholds (Ct), and the difference between the signals are shown. Indicated p-values compare the Alu-gag signals to the gag-only signals.
| Cycles | Alu-gag Ct | gag | Δ | P value |
|---|---|---|---|---|
| 20 | 22.6 | 28.3 | 5.66 | 0.005 |
| 30 | 20.5 | 27.0 | 6.50 | 0.005 |
| 40 | 18.7 | 28.1 | 7.41 | <0.001 |
Previously we showed that the sensitivity of our standard integration assay could be enhanced ~40-fold by repetitive sampling (Agosto et al., 2007), but when applied to patient samples, it was still not sufficiently sensitive to measure integration. Although we had previously optimized our standard integration assay for cycle number, extension time, and enzyme concentration (O'Doherty et al., 2002; Swiggard et al., 2005), now we set out to increase the sensitivity of our repetitive sampling-based assay to detect low integration levels.
To determine the optimal PCR conditions at low proviral numbers we first created a low copy number sample by diluting the integration standard (IS) (Agosto et al., 2007) to 20 proviruses in 15,000 uninfected genomes. We then assayed the diluted standard repetitively while varying cycle number, enzyme concentration, and extension time. For each condition, we measured the average Alu-gag cycle threshold, the average gag-only cycle threshold, and calculated the difference between these values (Table 1). The optimal conditions were defined as those that gave the greatest difference between the Alu-gag and gag-only signals.
We found that, by increasing the number of cycles in the first PCR reaction from 20 to 40, we increased the difference between the Alu-gag and the gag-only signal to a level that was most statistically significant. Under these new conditions, substrates were not limiting because a dose-response was preserved when we performed serial 2-fold dilutions of proviruses from 1,000 to 62.5 proviruses (not shown). Consistent with our prior work, the dose-response was lost at higher concentrations of provirus (between 1,000 and 100,000) when 40 cycles of amplification were used (not shown). We further optimized amplification conditions by doubling the amount of enzyme and by increasing the extension time from 2.5 to 3.5 minutes; however, the effect of these two modifications on increasing the difference between Alu-gag and gag-only signals was small relative to the effect of increasing the cycle number (not shown).
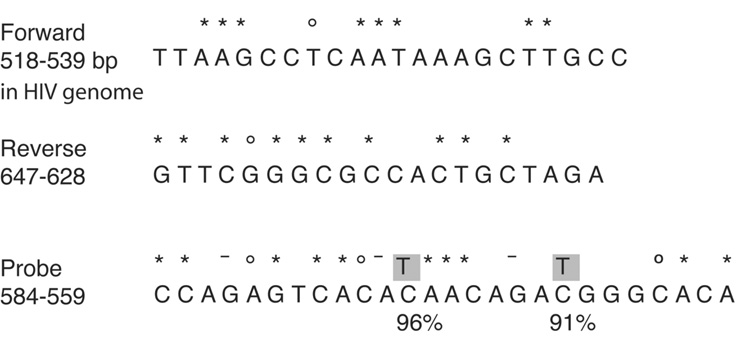
Redesign of the primer-probe pair to a conserved region of the HIV genome to increase the number of patient sequences detected (Figure 1)
Figure 1. Sequence of the primers and probes used in the HIV integration assay.
The nucleotide sequences and their positions relative to the HIV genome of the forward primer, reverse primer and probe are shown. The percent identity to the LANL HIV database consensus is indicated as follows: no symbol denotes 100% identity, an asterisk (*) above a base indicates 99% identity, a circle (○) represents 98% identity, and a dash (-) indicates 97% identity. The forward primer sits at position 518–539 in HIV genome (based on HXB-2 numbering). The reverse primer sits at bases 647–628. The fluorescent probes lie between 584–559. Two degenerate fluorescent probes were synthesized to capture the two most mutated bases, highlighted in grey. These two mutated bases are present in 9% and 4% of sequences. The primer probe sets were used to detect either total DNA (without Alu-gag preamplification) or to detect Alu-gag amplicons as described in the Methods.
In order to better match the sequences in the Los Alamos National Laboratory (LANL) database (Leitner et al., 2005), we redesigned the primers and probes used in our original assay. To do this, we surveyed the LANL database for the most conserved regions of the HIV genome. We slightly modified the original forward and reverse primers by shifting their positions relative to the LANL database by 3 and 6 nucleotides, respectively, compared to our original pair (Agosto et al., 2007). This change resulted in primers that bound to more highly conserved regions in the HIV genome: the last 3 and 4 nucleotides of each primer are perfect matches with the database; the remaining nucleotides are 98–100% conserved (Figure 1).
Although we were unable to identify a region for the probe that was as highly conserved, we prepared a wild-type probe that has been previously described (Yun, Fredriksson, and Sonnerborg, 2002) and two degenerate probes that were perfect matches for the two most frequent mutations which occurr 9% and 4% of the time (Figure 1). We analyzed between 427 and 627 sequences to determine the frequency of mutations in the primers and probes. None of the mutations that are shown are linked. These three probes were mixed in equal concentrations for the kinetic PCR reaction. When we tested the new primer-probe set and compared it to the original primer-probe set, we found that both showed near 100% amplification efficiency, calculated as described (Applied Biosystems, 2004). In other words, both sets of primers amplify efficiently, but the new set is a better match to patient sequences and so will pick up more sequences.
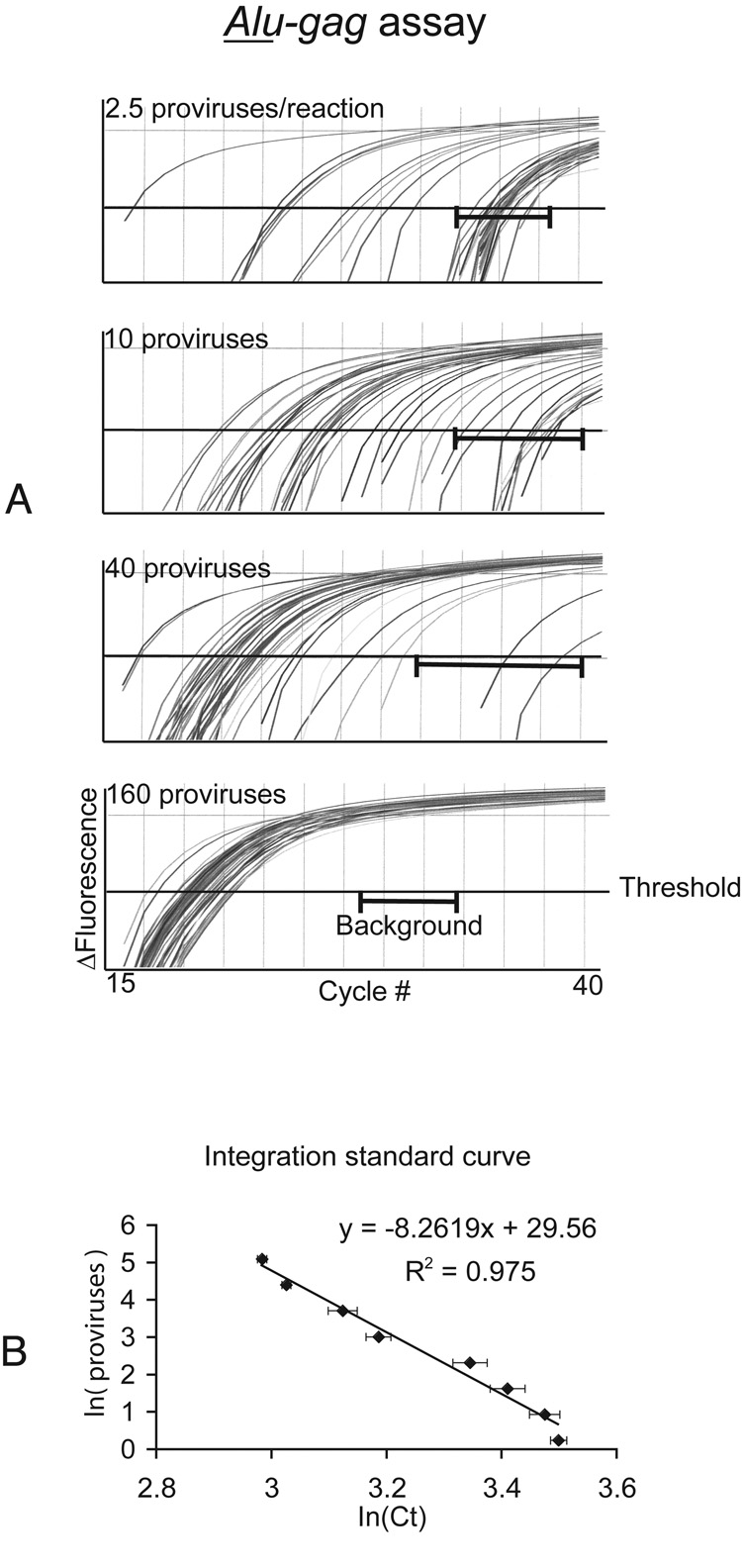
Generation of a standard curve using our integration standard (Figure 2)
Figure 2. IS standard curve and linear regression.
Alu-gag PCR was performed repetitively (N=40) on serial 4-fold dilutions of the integration standard. N represents the number of samples tested.
A. The curves generated at 2.5, 10, 40 and 160 proviruses per reaction are shown for Alu-gag. Background or gag-only signals are not shown, but the brackets ( ) represents the range of gag-only signals. The cycle threshold (Ct) for each sample was calculated by determining where the PCR curve crosses the horizontal line at 0.01 fluorescence units.
) represents the range of gag-only signals. The cycle threshold (Ct) for each sample was calculated by determining where the PCR curve crosses the horizontal line at 0.01 fluorescence units.
B. The natural log of each Ct for each curve was calculated and then averaged. A standard curve was then generated by plotting the x-axis as the average of the natural log of Ct and the y-axis as the natural log of the provirus quantity in the diluted standard. The provirus number in our integration standard is known as previously described (Agosto et al., 2007). The error bars represent standard error of the mean.
To mimic the low integration frequencies in patient samples and to determine the sensitivity of our integration assay, we calculated the proviral number in the IS cells and diluted the IS cells in uninfected PBMCs as described (Agosto et al., 2007). We calculated the proviral number in the IS cell line using routine quantitative DNA PCR because all the HIV DNA in the IS is integrated (Agosto et al., 2007). We performed Alu-gag amplification on IS samples in 40 replicates at multiple proviral concentrations (Figure 2A). We consistently assayed 1.5 × 104 cells per reaction (IS cells + uninfected PBMCs) to maintain a constant number of Alu sites. We kept the number of Alu sites constant because we previously determined that the number of Alu sites per reaction affects the efficiency of PCR amplification.
We previously showed that our IS contains diverse integration sites (Agosto et al., 2007; O'Doherty et al., 2002); and because our IS accurately models integration in vivo, some integration events are detectable and some are not (Agosto et al., 2007; O'Doherty et al., 2002). We found that when 160 proviruses per reaction were assayed repetitively, the Alu-gag signals did not overlap with the background gag-only signals (Figure 2A; gag-only curves are not shown, but brackets denote their location). When 40 proviruses per reaction were assayed repetitively, although a few Alu-gag signals overlapped the gag-only signal, the majority of Alu-gag signals did not (Figure 2A). As the number of proviruses per reaction decreased, the fraction of the samples that gave a positive signal decreased (Figure 2A).
We determined there was a linear relationship between the logarithim of the cycle threshold and the logarithm of the proviral number. For the integration standard, we calculated the natural log of the cycle threshold (lnCt) and plotted the average of the lnCt against the natural log of the proviral number (lnn). We observed a linear relationship between lnn and lnCt Figure 2B This linear relationship is represented by the equation lnn = DlnCt + E (Figure 2B), where n represents proviral number, Ct represents cycle threshold response, D= −8.2619, E= 29.56, and R2= 0.975. Having derived a standard equation, we could apply it to the results of Alu-gag PCR on patient samples, and calculate the proviral number in these samples.
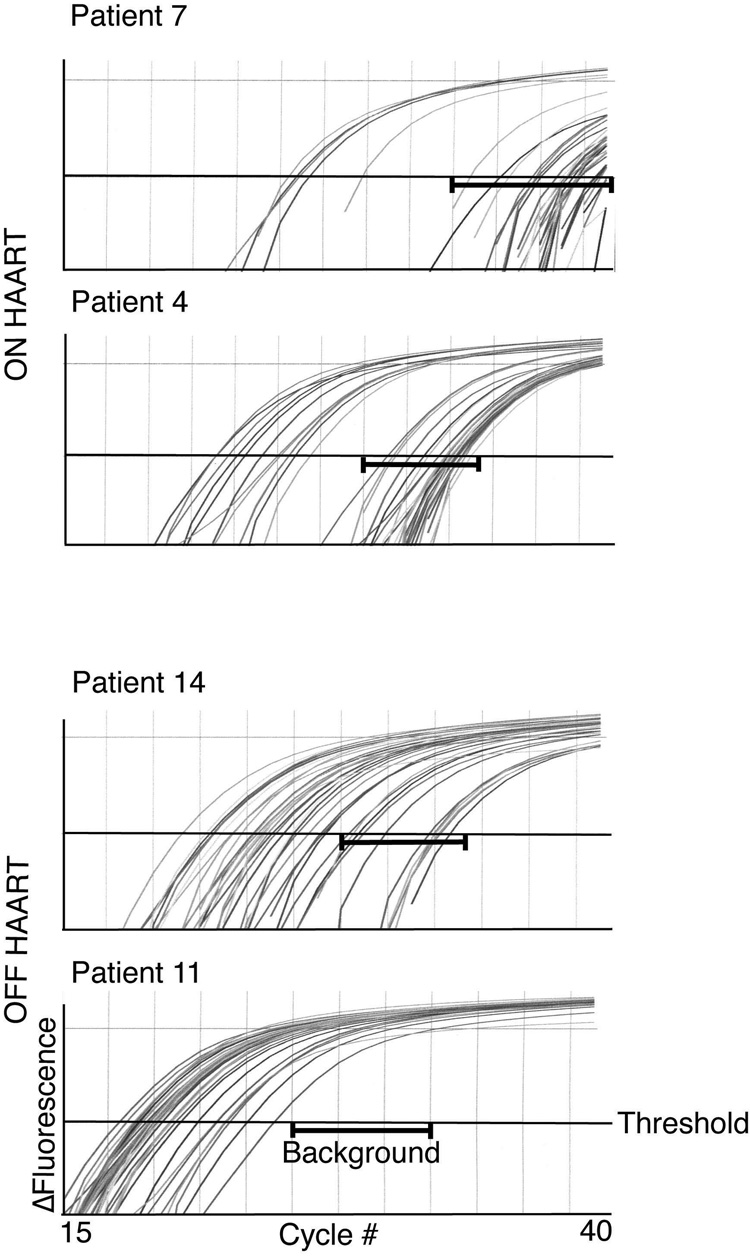
Our new integration assay is sufficiently sensitive to detect integration in patient samples (Figure 3, Table 2)
Figure 3. Representative Alu-gag PCR amplification curves.
PBMC DNA was prepared from HIV infected individuals (Patients 4 and 7 were on HAART and Patients 11 and 14 were not on HAART). Alu-gag PCR was performed in 40 replicates and then HIV-1 specific kinetic PCR was performed on each replicate.
Table 2. Proviral level in patients on and off HAART.
The calculated number of proviruses per 10,000 cells is listed for each patient. In addition, the upper and lower bounds for 95% confidence intervals are also provided. The errors are asymmetrical because a logarithmic transformation was performed before taking averages of the ln (Ct) of the patient samples (as well as the integration standard). The p-value in the final column represents the probability that the Alu-gag and gag-only signals are not different.
| HAART | Patient | Provirus per 104 cells | Lower 95% CI | Upper 95% CI | p-value |
|---|---|---|---|---|---|
| 1 | 0.66 | 0.63 | 0.70 | 2×10−02 | |
| 2 | 2.9 | 2.1 | 4.2 | 7×10−07 | |
| 3 | 2.2 | 1.5 | 3.4 | 1×10−04 | |
| 4 | 3.0 | 2.1 | 4.4 | 3×10−03 | |
| ON | 5 | 2.5 | 1.7 | 3.9 | 2×10−06 |
| 6 | 0.58 | 0.49 | 0.69 | 2×10−02 | |
| 7 | 0.55 | 0.39 | 0.78 | 3×10−05 | |
| 8 | 0.43 | 0.33 | 0.55 | 2×10−02 | |
| 9 | 3.0 | 1.5 | 6.0 | 3×10−04 | |
| 10 | 1.9 | 1.3 | 2.6 | 6×10−05 | |
| Average | 1.8 | 1.2 | 2.7 | ||
| 11 | 46 | 38 | 54 | 2×10−40 | |
| 12 | 19 | 15 | 25 | 2×10−15 | |
| OFF | 13 | 9.1 | 6.8 | 12 | 2×10−25 |
| 14 | 6.4 | 4.5 | 8.9 | 2×10−11 | |
| 15 | 27 | 23 | 32 | 2×10−13 | |
| Average | 18 | 15 | 22 |
We wanted to test whether our improved assay was sensitive enough to measure integration in patient PBMCs. To do this, we purified DNA from PBMCs of 10 patients on HAART and 6 patients off HAART, then performed our new two-step PCR integration assay on 40 samples for each patient repetitively. We obtained a distribution of Alu-gag amplification curves for each patient (Figure 3, representative curves). To measure integration levels in patients, we first determined if integration was detectable by performing a one-tailed t-test to make sure that our Alu-gag was significantly lower than gag-only. Then, we compared their Alu-gag signals to the Alu-gag signals generated by our diluted IS samples. We determined the proviral number for each patient by obtaining the cycle threshold (Ct) for each Alu-gag amplification curve, taking the natural log of the cycle threshold (ln(Ct)). Next, we calculated the average ln(Ct) for each patient and inserted the average ln(Ct) into the equation lnn = D*average(lnCt) + E, where D= −8.26, E= 29.56. Lastly, we calculated the proviral number by taking the exponential of ln(n) and calculated the 95% confidence intervals, which are asymmetric because of the logarithmic transformation. We found that our improved assay was sensitive enough to detect proviral loads as low as 0.5 integration events in 10,000 PBMCs (Table 2, patients 1, 6, 7, and 8). The average Alu-gag and gag-only signals are provided (Supplemental Table) and demonstrate that the Alu-gag signal was always significantly lower than the gag-only signal by a one-tailed Student’s t-test.
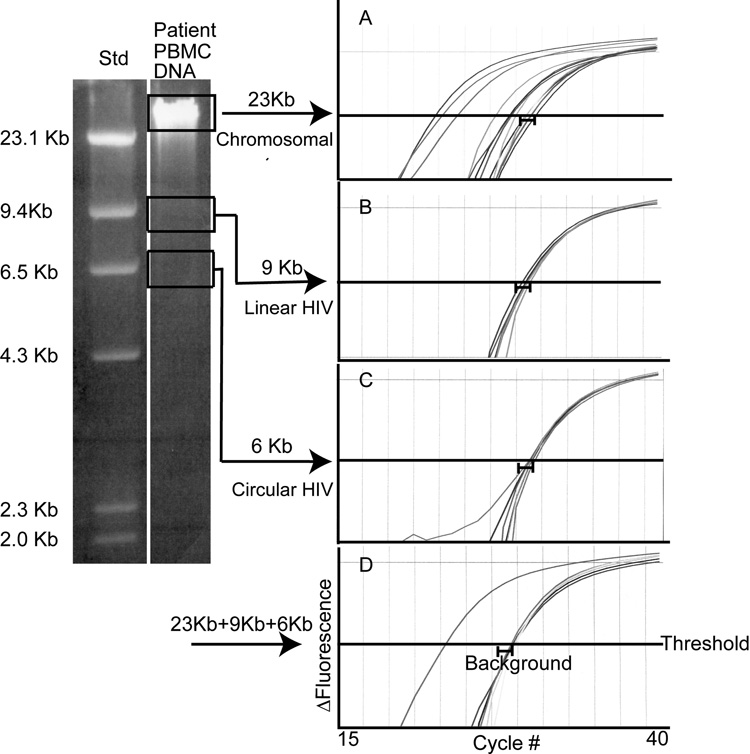
Unintegrated patient DNA does not contribute significantly to the integration assay signal (Figure 4, Table 3)
Figure 4. In a patient sample, unintegrated HIV DNA does not contribute significantly to the Alu-gag integration assay signal.
DNA from patient PBMCs was purified and size-separated into three fractions (chromosomal, linear, and circular) by 0.7% agarose gel electrophoresis. Chromosomal DNA was ~23kb (due to fragmentation during processing), linear HIV DNA ~9kb, and circular HIV DNA ~6kb. The DNA from each fraction was extracted and resuspended in 100ul of 10 mM Tris. Alu-gag and gag-only PCR were performed on equal volumes (8.3 ul) of each fraction separately, and then on a mixture (8.3 ul of each) of the 3 fractions. The Alu-gag and gag-only curves generated from PCR are shown in plots on the right: panel (A) chromosomal, (B) linear HIV DNA, (C) circular HIV DNA, and (D) mixture of chromosomal, linear, and circular. Alu-gag and gag-only PCR were performed 6 times each on sample A and 3 times each on samples B-D (due to limited sample). Alu-gag and gag-only signals (Ct) are defined as the cycle number at which a curve crosses the threshold (0.01). A bracket ( ) indicates the range of signal obtained with gag-only priming. An Alu-gag signal is defined as positive when it is significantly different from the gag-only signal by Student’s t-test.
) indicates the range of signal obtained with gag-only priming. An Alu-gag signal is defined as positive when it is significantly different from the gag-only signal by Student’s t-test.
Table 3. Positive Alu-gag signals are only detected in the chromosomal fraction of patient PBMC DNA.
The average Alu–gag and gag-only signals were determined in Figure 4.
| Cycle number at which signal above threshold |
||
|---|---|---|
| Alu-gag (Ct) | gag only (Ct) | |
| Chromosomal | 26.1 +/− 1.22 | 30.0 +/− 0.15 |
| Linear HIV | 29.6 +/− 0.11 | 29.9 +/− 0.07 |
| Circular HIV | 30.1 +/− 0.04 | 30.2 +/− 0.10 |
| Mixture | 26.8 +/− 1.76 | 28.6 +/− 0.11 |
DNA extracted from patient PBMCs contains a mixture of HIV DNA that is integrated into the host chromosome, and HIV DNA that is unintegrated and present as linear or circular forms. We considered the possibility that unintegrated HIV DNA may contribute to the Alu-gag integration assay signal. If this were the case, then our measurements of integrated HIV DNA would be higher than the actual number of proviruses.
To determine whether unintegrated DNA contributes to the Alu-gag signal attributed to integrated DNA, we prepared DNA from patient PBMCs, separated the chromosomal, linear, and circular fractions by agarose gel electrophoresis, extracted DNA from the gel, and performed Alu-gag PCR on individual fractions as well as on a mixture. When we assayed chromosomal DNA samples, the integration signal was stronger than the background signal: on average the Alu-gag curves crossed the cycle threshold (Ct) at lower cycle numbers than the background gag-only curves (Fig 4, panel A; Table 4, row 1). By comparison, when we assayed linear and circular DNA samples, the integration signal equaled the background signal: Alu-gag and gag-only curves crossed the Ct at the same cycle number (Figure 4, panels B and C; Table , rows 2 and 3). Therefore, HIV integration was detected only in chromosomal DNA samples. Finally, we found that the Alu-gag signal from a mixture of the three fractions, was comparable to that from the chromosomal fraction alone (Figure 4, compare panel D to A; Table 3, compare bolded values). Therefore, we conclude that, in a patient sample containing a mixture of integrated and unintegrated HIV DNA, unintegrated DNA does not contribute significantly to the Alu-gag signal.
Table 4. Unintegrated HIV plasmid DNA contributes minimally to the Alu-gag signal from an integration standard.
In this experiment, unintegrated HIV plasmid DNA was diluted in PBMC DNA, integration standard DNA was diluted in PBMC DNA, and unintegrated and integrated DNA were mixed in a 100:1 ratio. From these, three samples of DNA were prepared. The first sample contained 800 copies/reaction of unintegrated DNA (800 U). The second sample contained 8 copies/reaction of integration standard DNA (8 IS). The third sample was a mixture of 800 copies of unintegrated DNA and 8 integrated proviruses (800 U + 8 IS). Each sample was assayed 42 times using Alu-gag and gag-only PCR and the cycle numbers at which the Alu-gag and gag-only signals were detected above the cycle threshold are shown. Also shown are the number of proviruses calculated from the Alu-gag signal for each sample. The standard curve used to calculate proviruses in this figure is slightly different than the one generated in Figure 2 and used to calculate the number of proviruses for the patient samples because a different master mix was used for the experiment in Table 4. All of the other experiments described in this paper used the same master mix as was used to generate the standard curve in Figure 2.
| HIV DNA copies per well | Provirus per well | Cycle number at which signal detected |
p-value | |
|---|---|---|---|---|
| Alu-gag | gag only | |||
| 800 U | 0 | 27.3 | 27.3 | NS |
| 8 IS | 7 | 29.7 | 36.4 | ≪0.01+ |
| 800 U + 8 IS | 15 | 25.3 | 27.3 | ≪0.01* |
Unintegrated DNA contributes minimally to the integration signal (Table 4)
In a second approach to determine whether unintegrated HIV DNA contributes to the Alu-gag signal from integrated DNA, we modeled the ratio of unintegrated-to-integrated DNA in patient samples and measured integration. We added 100-fold excess of unintegrated HIV plasmid DNA (U), NG38, to our integration standard (IS) DNA, because a 100:1 ratio of unintegrated-to-integrated DNA was close to the highest ratio that we detected in patient samples. We found that when we performed Alu-gag and gag-only PCR on 800 copies of unintegrated DNA (800 U), the Alu-gag and gag-only signals were equivalent indicating that the sample was negative for integration (Table 4). When 8 copies of integrated DNA (8 IS) were assayed, the Alu-gag signal was stronger (detected at a lower cycle number) than the gag-only signal, indicating that integration was detected (Table 4). When we added 100-fold excess of unintegrated DNA to integrated DNA (800 U + 8 IS), the Alu-gag signal remained stronger than the gag-only signal (Table 4). The Alu-gag signal from 800 U + 8 IS was slightly stronger than that from 8 IS (Table 4). This slightly stronger signal translated into only a 2-fold difference when we calculated the number of proviruses per well: 7 proviruses/well in the 8 IS sample, 15 proviruses/well in the 800 U+ 8 IS sample (Table 2). Therefore, we concluded that unintegrated DNA contributes minimally to the integration assay signal.
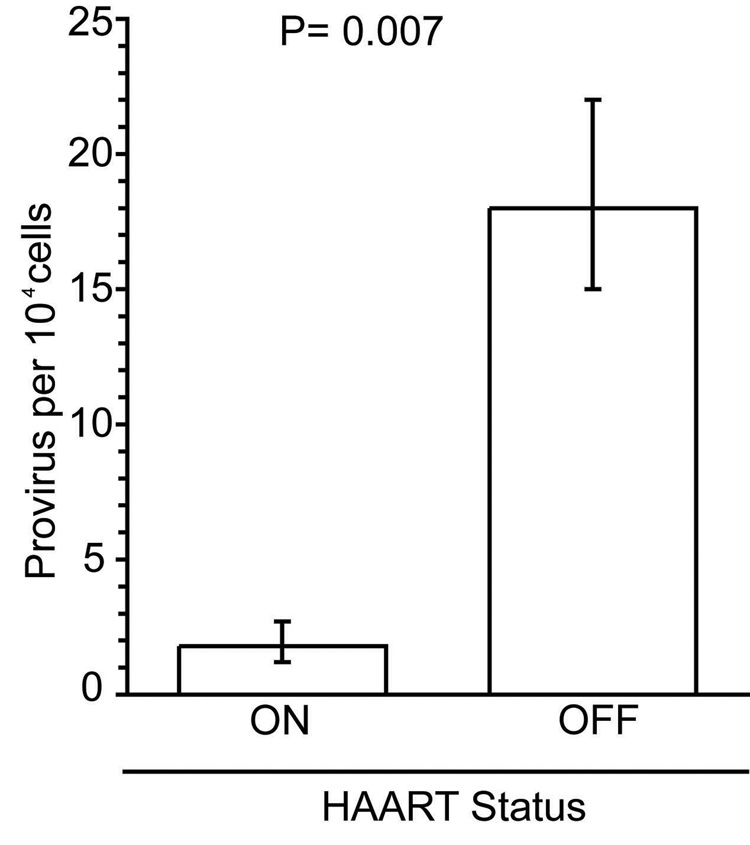
Patients on HAART have lower levels of integrated HIV DNA in PBMCs (Figure 5, Table 2)
Figure 5. Patients on HAART have lower proviral loads in PBMCs than patients off HAART.
The average level of provirus in patients on and off effective HAART are shown with their 95% confidence intervals and are significantly different by the Wilcoxon signed-rank test (p=0.007).
We hypothesized that patients on HAART have lower levels of integrated DNA than patients not on HAART. To test this hypothesis, we measured integration levels in patients on HAART and off HAART, averaged the integration level in each group, and performed a Wilcoxon signed-rank test (Figure 5, p= 0.007). We found that there was a difference between the proviral levels in the two groups: patients on HAART had lower levels of integration than patients off HAART (Figure 5).
All the patients off HAART had a viral load >1000, except for patient 11 with an unknown viral load (Table 5). All the patients on HAART had an undetectable viral load the day of donation (Table 5) except patients 4 and 5 who had barely detectable viral loads. Both populations had relatively preserved CD4 counts (≥290 cells/µl) (Table 5). Prior studies suggest that HAART treatment results in modestly lower concentrations of viral DNA (Ngo-Giang-Huong et al., 2001; Viard et al., 2004). We wanted to measure the amount of total DNA in our patient samples. Using the primer probe set shown in Figure 1, but omitting Alu-gag pre-amplification, we measured the level of total HIV DNA. Consistent with prior literature, there is a significant difference in the level of total DNA between the patients on and off HAART (Table 5) as assessed by the Wilcoxon signed-rank test, p= 0.02. We also calculated the ratio of total DNA to integrated DNA in the two patient groups. We found that HAART affected the level of integrated DNA to a greater extent than the level of unintegrated DNA (Table 2 and Table 5); however, larger numbers of samples need to be examined before additional conclusions can be made about the ratio of integrated to unintegrated DNA.
Table 5. Patient Characteristics.
The patient’s HAART status, CD4 count, viral loads, total HIV DNA (RU5/10,000 cells +/− standard error), and the ratio of total DNA to integrated DNA are shown for each patient. Total DNA was measured using the primers and probe described in Figure 1 without the Alu-gag amplification as described in the Methods.
| Patient | HAART Status | CD4 Count | Viral Load | RU5 per 104 cells | Provirus 104 cells | RU5 per Provirus |
|---|---|---|---|---|---|---|
| 1 | 370 | <400 | 12 ± 1.8 | 0.66 | 18 | |
| 2 | 809 | <50 | 440 ± 31 | 2.9 | 150 | |
| 3 | 596 | <50 | 25 ± 3.7 | 2.2 | 11 | |
| 4 | 745 | 400 | 32 ± 3.8 | 3.0 | 11 | |
| 5 | ON | 515 | 57 | 61 ± 12 | 2.5 | 24 |
| 6 | 461 | <50 | 84 ± 4.0 | 0.58 | 140 | |
| 7 | 903 | <50 | 200 ± 6.1 | 0.55 | 360 | |
| 8 | 749 | <50 | 1.4 ± 0.13 | 0.43 | 3.3 | |
| 9 | 506 | <75 | 14 ± 0.32 | 3.0 | 4.7 | |
| 10 | 322 | <75 | 5.5 ± 0.85 | 1.9 | 2.9 | |
| Average | 87 ± 43 | 1.8 | 72 ± 37 | |||
| 11 | 290 | unknown | 310 ± 13 | 46 | 6.7 | |
| 12 | 400 | 2.5×104 | 320 ± 21 | 19 | 17 | |
| 13 | OFF | 529 | 6.8×104 | 87 ± 17 | 9.1 | 9.6 |
| 14 | 383 | 1.3×104 | 85 ± 14 | 6.4 | 13 | |
| 15 | 630 | 8.0×104 | 610 ± 30 | 27 | 23 | |
| 16 | 587 | 5.5×104 | 79 ± 15 | 1.5 | 53 | |
| Average | 250 ± 86 | 18 | 20 ± 6.9 | |||
Discussion
Here we describe a new integration assay that is a significant advance over prior technology. Our improved kinetic PCR assay is sufficiently sensitive to measure HIV integration and provide error estimates in PBMC samples from infected individuals (Table 2). Furthermore, results obtained with the assay suggest that the level of integrated DNA is lower in patients on effective HAART than in patients not on HAART. Although this result may seem obvious, two prior reports failed to detect this difference (Carr et al., 2007; Ibanez et al., 1999). Our ability to detect this difference suggests our assay may be more precise than prior assays. Nonetheless, longitudinal studies following the level of integration over time after HAART is initiated will be required to determine the effect of HAART.
Our new integration assay overcomes the disadvantages of endpoint PCR, as well as the low sensitivity of prior kinetic PCR techniques, by incorporating repetitive sampling and optimizing conditions for rare event detection. Repetitive sampling increased the sensitivity of the assay, permitted calculation of proviral numbers and permitted calculation of confidence intervals. Notably, prior assays did not provide calculations of confidence intervals. We further optimized the assay by recognizing that we could sacrifice the range of detection of integration to enhance sensitivity by increasing the number of PCR cycles.
Prior to the development of our new assay, studies using endpoint PCR assays to measure HIV integration in patient samples reported several important findings (Chun et al., 1997a; Chun et al., 1998; Chun et al., 1995; Chun et al., 1997b; Ostrowski et al., 1999) such as the level of integration, the concentration of proviruses in memory cells, and the failure of HAART to lower the level of integrated HIV DNA in resting CD4+ T cells. However, endpoint PCR has several disadvantages: it is semi-quantitative, labor-intensive, and does not permit calculation of confidence intervals.
Our new assay can be used to expand upon the findings obtained with endpoint PCR. Specifically, it can be used to refine the initial estimates of the level of integrated DNA in several CD4+ subsets. Since our assay is more sensitive, it should also provide proviral numbers in cell subsets where integration was previously undetectable. In addition, because our assay has tighter confidence intervals, it can be used to follow small fluctuations in integration and so may be useful in determining if HAART treatment leads to clearance of integrated DNA from subsets of CD4+ cells.
To our knowledge, our integration assay is the most sensitive assay currently available and the only assay that provides calculation of confidence intervals. Our measurements may also be more accurate because we used a polyclonal integration standard. This standard more closely models in vivo integration and was used to generate the standard curve used to calculate proviral number in patient samples. Nonetheless, we may be able to refine our assay to make it more accurate and more sensitive. For example, we are developing mathematical models that will correct for the small signal that unintegrated DNA contributes; although at most, the contribution from unintegrated DNA results in a 2-fold overestimate in the number of proviruses and only in a subset of patient samples with strong gag-only signals. This correction is not likely required in our patients on HAART because these patients show very little gag-only priming (Supplemental Table), i.e. the signal from total DNA and gag-only priming is similar. We speculate that little gag-only priming occurs in some patients with relatively high levels of unintegrated DNA (such as patients 2 and 7 on HAART) because the unintegrated DNA may be discontinuous. This correction may be relevant for 3 of the patients off HAART (such as patients 11, 12, and 14) since there was significant gag-only priming in these 3 patients (Supplemental Table). This is the first report that we are aware of that quantitates the contribution from unintegrated DNA.
Application of our assay to larger numbers of patient samples may address important issues in HIV pathogenesis. Even if the majority of integrated proviruses are defective (Coffin, 1995; Han et al., 2007; Lassen et al., 2004; Lehrman et al., 2005), our finding that patients on HAART have lower integration levels suggests that some populations of mononuclear cells bearing proviruses can be cleared of provirus with therapy and so should be monitored. In addition, with more precise measurements of HIV integration, it is possible to test whether the level of integrated DNA correlates with disease progression as has been shown for viral RNA load (Mellors et al., 1995). It has been shown, using kinetic PCR, that the level of total viral DNA within PBMCs also correlates weakly with disease progression (Kostrikis et al., 2002; Ngo-Giang-Huong et al., 2001; Rouzioux et al., 2005; Viard et al., 2004). With this assay, we could also measure the rate of decline of integrated DNA in total PBMCs and even within cellular subsets after a patient begins HAART to assess the effect of therapy longitudinally.
It remains unclear why only a small fraction of CD4+ resting T cells bearing integrated HIV DNA are latently infected, i.e. capable of producing infectious HIV when stimulated (Coffin, 1995; Han et al., 2007; Lassen et al., 2004). Possible explanations are that many of these cells bear proviruses that are either not transcribed, or do not produce infectious virions when stimulated. Rigorous quantitation of the level of integrated DNA, the frequency of cells capable of transcribing HIV proviruses (Haase, 2005), and the frequency of cells capable of producing infectious virions (Han et al., 2007; Lassen et al., 2004) may give us a better understanding of latency. In addition, with the advent of additional anti-HIV therapies, such as fusion inhibitors and integrase inhibitors, it is now possible to treat patients with intensified therapies. A sensitive assay for monitoring integration levels may be an important tool for monitoring and quantitating the additional benefits of intensified therapy when viral load is already undetectable.
Methods
Cell lines, plasmids, and viruses
The CD4+ T-lymphoblastoid cell line CEM-ss (Foley et al., 1965; Nara and Fischinger, 1988) was maintained at 1–5 × 105 cells per ml. The culture medium was RPMI 1640 supplemented with 10% heat-inactivated fetal bovine serum, 25 mM HEPES, with 100µg/ml penicillin-streptomycin (Invitrogen Life Technologies). Virions pseudotyped with vesicular stomatitis virus protein G (VSV-G) (Blumenthal et al., 1987; Emi, Friedmann, and Yee, 1991) were collected 24 hours after transfection of 293T cells with pVSV-G (pHIT) (Fouchier et al., 1997) and pNL4-3Δenv/GFP/HygR. The reporter virus was derived from pNL4-3, an X4-tropic HIV clone (Adachi et al., 1986). The nef open reading frame (ORF) was replaced with a green fluorescent protein (GFP) and hygromycin resistance cassette which contained an internal ribosome entry site between the two ORFs. The plasmid (pNG38) was prepared by removing the human genomic sequence form HXB-2 (personal communications with Nathan Gaddis).
Preparation of the Integration Standard (IS) cell line
The integration standard cell line (IS) was prepared as described (Agosto et al., 2007). The standard contains diverse integration sites with some integration sites close to an Alu repeat and others far away. We estimated that about 10% of proviruses are detectable by our methods at low proviral copy numbers (Figure 2A).
Patients’ blood samples
Patients on and off HAART therapy (both for at least three months) were recruited using an IRB approved protocol to donate blood. Patients 12 and 13 were never on HAART. Patients 11, 14, 15, and 16 were on HAART for 3months, 9, 4 and 7 years, respectively. Patients on HAART had ≤400 copies of viral RNA/ml. PBMCs were prepared from anticoagulated blood by Ficoll-Hypaque separation (Swiggard et al., 2004). Total DNA was prepared (Blood and Cell Culture DNA Maxi Kit, QIAGEN).
PCR conditions
Two step PCR amplification was performed as described (Agosto et al., 2007; O'Doherty et al., 2002) with some modifications. Briefly, the first amplification was performed on dilutions of the IS cells as well as patient samples and no template controls. The sequence of the first step amplification primers were: genomic Alu forward 5’ GCC TCC CAA ACT GCT GGG ATT ACA G-3’ and HIV gag reverse 5’ GTT CCT GCT ATG TCA CTT CC-3’. The gag primer is different from the previously described primer (O'Doherty et al., 2002). It provides a better consensus sequence than the original primer and provides more efficient Alu-gag amplification. The IS was diluted in uninfected PBMC DNA to keep the number of Alu sites per reaction constant. The reactions were conducted in 50µl: 10 mM Tris-HCl, pH 8.0; 1.8 mM MgCl2; 1mM mixed dNTPs; 100 nM Alu forward primer; 600 nM gag reverse primer; and 0.05 units of Platinum Taq DNA polymerase/µl (Invitrogen Life Technologies). The thermal cycler (DNA Engine PTC-200, MJResearch) was programmed to perform a two-minute hot start at 95°C, followed by 40 cycles of the following: denaturation at 95°C for 15s, annealing at 50°C for 15s and extension at 72°C for 3 min 30 sec. Linear, one strand amplification (priming) was also monitored by performing the first amplification PCR with the gag primer alone.
The second round real-time quantitative PCR was performed using 10 µl of the material from the first amplification. These were run with an HIV-1 copy number standard (ACH-2) (Folks et al., 1989). The sequences of the primers were: LTR (R) forward, 5'-TTA AGC CTC AAT AAA GCT TGC C-3’; LTR (U5) reverse, 5'-GTT CGG GCG CCA CTG CTA GA-3’. The LTR Taqman probes were labeled at their 5' terminus with the reporter fluorophore 6-carboxyfluorescein (FAM) and at their 3' terminus with the quencher tetramethylrhodamine (TAMRA). The wild type probe had the following sequence: 5’-FAM-CCA GAG TCA CAC AAC AGA GGG GCA CA-TAMRA-3' as described (Yun, Fredriksson, and Sonnerborg, 2002). The two degenerate probes had the sequences listed in Figure 1. Reactions were carried out in a volume of 20µl containing: 10 mM Tris-HCl, pH 8.0; 75 mM KCl; 5.5 mM MgCl2; 500 nM carboxy-X-rhodamine (ROX, Molecular Probes) as a passive reference; 1.2 mM freshly added dNTPs; 250 nM LTR forward and reverse primers; 100 nM each Taqman probe; and 0.05 units of Platinum Taq DNA polymerase/µl. The reactions were performed on an ABI7500 instrument running 7500 Fast System SDS software with the following thermal program: 20 sec hot start at 95°C, followed by 40 cycles of: denaturation at 95°C for 3 sec, annealing and extending at 60°C for 30 sec. One step reactions were performed to measure the total HIV (RU5) DNA by simply eliminating the first Alu-gag amplification step.
We constructed two degenerate probes, because two mutations were relatively common. We found no difference in amplification efficiency of HIV integrated DNA in the presence or absence of the degenerate probes (not shown).
The quantity of DNA was determined by OD260/280. The number of genomes was calculated given that ~3.18 × 109 bp are present per genome. These numbers were used to calculate the level of total and integrated HIV DNA per cell.
Repetitive sampling to measure integration at low proviral copy number
Alu-gag PCR and gag-only PCR were performed repetitively on 10 µl aliquots of DNA at 2 µg/ml from the IS line and from patient samples. Then HIV-1 specific kinetic PCR was performed on 10µl aliquots of the first reaction. After the kinetic PCR reaction, the cycle thresholds (Ct) for each well were determined, i.e. the cycle value where the accumulating fluorescence (from fluorescently labeled amplicons) crossed a specified fluorescent threshold value. In all of the experiments described here, the fluorescent threshold value was set at 0.01 for data analysis. As previously shown (Agosto et al., 2007), the average natural log of the cycle thresholds are linearly related to the natural log of the proviral number. The standard equation was generated by finding the linear regression of the average natural log of the cycle threshold values from each concentration of IS and the natural log of the proviral number. The proviral number for patient samples was then calculated using this standard curve. To do this, the natural logarithm of each cycle threshold was obtained and averaged and standard errors were calculated. These values (average ln(Ct) +/− std error ln(Ct) were entered into the equation described in Figure 2b. Because of the logarithmic transformation to our data, the errors are asymmetrical for proviral numbers.
Supplementary Material
Acknowledgments
This study was supported in part by NIH grants R01 AI058862-01 (U.O.) and R21 AI064031 (U.O.). We would like to thank Liz Colston, Luis Agosto, Drew Weissman, and Rick Bushman for critical reading of this manuscript. We are grateful to Penn CFAR and Farida Shaheen for excellent technical guidance regarding real time kinetic PCR. We are grateful to Joseph Quinn and Debbie Gudonis for recruiting patients for blood donation and to the ACTG program at Penn, directed by Ian Frank.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adachi A, Gendelman HE, Koenig S, Folks T, Willey R, Rabson A, Martin MA. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J Virol. 1986;59(2):284–291. doi: 10.1128/jvi.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agosto LM, Yu JJ, Dai J, Kaletsky R, Monie D, O'Doherty U. HIV-1 integrates into resting CD4+ T cells even at low inoculums as demonstrated with an improved assay for HIV-1 integration. Virology. 2007;368(1):60–72. doi: 10.1016/j.virol.2007.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Applied Biosystems. Guide to Performing Relative Quantitation of Gene Expression Using Real-Time Quantitative PCR. 2004:12. http://www3.appliedbiosystems.com/cms/groups/mcb_marketing/documents/generaldocuments/cms_042380.pdf.
- Blumenthal R, Bali-Puri A, Walter A, Covell D, Eidelman O. pH-dependent fusion of vesicular stomatitis virus with Vero cells. Measurement by dequenching of octadecyl rhodamine fluorescence. J Biol Chem. 1987;262(28):13614–13619. [PubMed] [Google Scholar]
- Brenchley JM, Hill BJ, Ambrozak DR, Price DA, Guenaga FJ, Casazza JP, Kuruppu J, Yazdani J, Migueles SA, Connors M, Roederer M, Douek DC, Koup RA. T-cell subsets that harbor human immunodeficiency virus (HIV) in vivo: implications for HIV pathogenesis. J Virol. 2004;78(3):1160–1168. doi: 10.1128/JVI.78.3.1160-1168.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brussel A, Delelis O, Sonigo P. Alu-LTR real-time nested PCR assay for quantifying integrated HIV-1 DNA. Methods Mol Biol. 2005;304:139–154. doi: 10.1385/1-59259-907-9:139. [DOI] [PubMed] [Google Scholar]
- Brussel A, Sonigo P. Analysis of early human immunodeficiency virus type 1 DNA synthesis by use of a new sensitive assay for quantifying integrated provirus. J Virol. 2003;77(18):10119–10124. doi: 10.1128/JVI.77.18.10119-10124.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brussel A, Sonigo P. Evidence for gene expression by unintegrated human immunodeficiency virus type 1 DNA species. J Virol. 2004;78(20):11263–11271. doi: 10.1128/JVI.78.20.11263-11271.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgard M, Izopet J, Dumon B, Tamalet C, Descamps D, Ruffault A, Vabret A, Bargues G, Mouroux M, Pellegrin I, Ivanoff S, Guisthau O, Calvez V, Seigneurin JM, Rouzioux C. HIV RNA and HIV DNA in peripheral blood mononuclear cells are consistent markers for estimating viral load in patients undergoing long-term potent treatment. AIDS Res Hum Retroviruses. 2000;16(18):1939–1947. doi: 10.1089/088922200750054666. [DOI] [PubMed] [Google Scholar]
- Butler SL, Hansen MST, Bushman FD. A quantitative assay for HIV DNA integration in vivo. Nature Medicine. 2001;7:631–634. doi: 10.1038/87979. [DOI] [PubMed] [Google Scholar]
- Carr JM, Cheney KM, Coolen C, Davis A, Shaw D, Ferguson W, Chang G, Higgins G, Burrell C, Li P. Development of methods for coordinate measurement of total cell-associated and integrated human immunodeficiency virus type 1 (HIV-1) DNA forms in routine clinical samples: levels are not associated with clinical parameters, but low levels of integrated HIV-1 DNA may be prognostic for continued successful therapy. J Clin Microbiol. 2007;45(4):1288–1297. doi: 10.1128/JCM.01926-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun TW, Carruth L, Finzi D, Shen X, DiGiuseppe JA, Taylor H, Hermankova M, Chadwick K, Margolick J, Quinn TC, Kuo YH, Brookmeyer R, Zeiger MA, Barditch-Crovo P, Siliciano RF. Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection. Nature. 1997a;387:183–188. doi: 10.1038/387183a0. [DOI] [PubMed] [Google Scholar]
- Chun TW, Engel D, Berrey MM, Shea T, Corey L, Fauci AS. Early establishment of a pool of latently infected, resting CD4(+) T cells during primary HIV-1 infection. Proceedings of the National Academy of Sciences USA. 1998;95:8869–8873. doi: 10.1073/pnas.95.15.8869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun TW, Finzi D, Margolick J, Chadwick K, Schwartz D, Siliciano RF. In vivo fate of HIV-1-infected T cells: quantitative analysis of the transition to stable latency. Nat Med. 1995;1(12):1284–1290. doi: 10.1038/nm1295-1284. [DOI] [PubMed] [Google Scholar]
- Chun TW, Stuyver L, Mizell SB, Ehler LA, Mican JA, Baseler M, Lloyd AL, Nowak MA, Fauci AS. Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proceedings of the National Academy of Sciences USA. 1997b;94:13193–13197. doi: 10.1073/pnas.94.24.13193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffin JM. HIV population dynamics in vivo: implications for genetic variation, pathogenesis, and therapy. Science. 1995;267:483–489. doi: 10.1126/science.7824947. [DOI] [PubMed] [Google Scholar]
- Debiaggi M, Zara F, Pistorio A, Bruno R, Sacchi P, Patruno SF, Achilli G, Romero E, Filice G. Quantification of HIV-1 proviral DNA in patients with undetectable plasma viremia over long-term highly active antiretroviral therapy. Int J Infect Dis. 2000;4(4):187–193. doi: 10.1016/s1201-9712(00)90107-3. [DOI] [PubMed] [Google Scholar]
- Douek DC, Brenchley JM, Betts MR, Ambrozak DR, Hill BJ, Okamoto Y, Casazza JP, Kuruppu J, Kunstman K, Wolinsky S, Grossman Z, Dybul M, Oxenius A, Price DA, Connors M, Koup RA. HIV preferentially infects HIV-specific CD4+ T Cells. Nature. 2002 May;417:95–98. doi: 10.1038/417095a. [DOI] [PubMed] [Google Scholar]
- Emi N, Friedmann T, Yee JK. Pseudotype formation of murine leukemia virus with the G protein of vesicular stomatitis virus. J Virol. 1991;65(3):1202–1207. doi: 10.1128/jvi.65.3.1202-1207.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley GE, Lazarus H, Farber S, Uszman BG, Boone BA, McCarthy RE. Continuous culture of human lymphoblasts from periphral blood of a child with acute leukemia. Cancer. 1965;18(4):522–529. doi: 10.1002/1097-0142(196504)18:4<522::aid-cncr2820180418>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- Folks TM, Clouse KA, Justement J, Rabson A, Duh E, Kehrl JH, Fauci AS. Tumor necrosis factor alpha induces expression of human immunodeficiency virus in a chronically infected T-cell clone. Proc Natl Acad Sci U S A. 1989;86(7):2365–2368. doi: 10.1073/pnas.86.7.2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouchier RAM, Meyer BE, Simon JHM, Fischer U, Malim MH. HIV-1 infection of non-dividing cells: evidence that the amino-terminal basic region of the viral matrix protein is important for Gag processing but not for post-entry nuclear import. European Molecular Biology Organization Journal. 1997;16:4531–4539. doi: 10.1093/emboj/16.15.4531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibellini D, Vitone F, Schiavone P, Ponti C, La Placa M, Re MC. Quantitative detection of human immunodeficiency virus type 1 (HIV-1) proviral DNA in peripheral blood mononuclear cells by SYBR green real-time PCR technique. J Clin Virol. 2004;29(4):282–289. doi: 10.1016/S1386-6532(03)00169-0. [DOI] [PubMed] [Google Scholar]
- Haase AT. Perils at mucosal front lines for HIV and SIV and their hosts. Nat Rev Immunol. 2005;5(10):783–792. doi: 10.1038/nri1706. [DOI] [PubMed] [Google Scholar]
- Han Y, Wind-Rotolo M, Yang HC, Siliciano JD, Siliciano RF. Experimental approaches to the study of HIV-1 latency. Nat Rev Microbiol. 2007;5(2):95–106. doi: 10.1038/nrmicro1580. [DOI] [PubMed] [Google Scholar]
- Ibanez A, Puig T, Elias J, Clotet B, Ruiz L, Martinez MA. Quantification of integrated and total HIV-1 DNA after long-term highly active antiretroviral therapy in HIV-1-infected patients. Aids. 1999;13(9):1045–1049. doi: 10.1097/00002030-199906180-00007. [DOI] [PubMed] [Google Scholar]
- Izopet J, Cazabat M, Pasquier C, Sandres-Saune K, Bonnet E, Marchou B, Massip P, Puel J. Evolution of total and integrated HIV-1 DNA and change in DNA sequences in patients with sustained plasma virus suppression. Virology. 2002;302(2):393–404. doi: 10.1006/viro.2002.1621. [DOI] [PubMed] [Google Scholar]
- Jubault V, Burgard M, Le Corfec E, Costagliola D, Rouzioux C, Viard JP. High rebound of plasma and cellular HIV load after discontinuation of triple combination therapy. Aids. 1998;12(17):2358–2359. [PubMed] [Google Scholar]
- Kabamba-Mukadi B, Henrivaux P, Ruelle J, Delferriere N, Bodeus M, Goubau P. Human immunodeficiency virus type 1 (HIV-1) proviral DNA load in purified CD4+ cells by LightCycler real-time PCR. BMC Infect Dis. 2005;5(1):15. doi: 10.1186/1471-2334-5-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koelsch KK, Liu L, Haubrich R, May S, Havlir D, Gunthard HF, Ignacio CC, Campos-Soto P, Little SJ, Shafer R, Robbins GK, D'Aquila RT, Kawano Y, Young K, Dao P, Spina CA, Richman DD, Wong JK. Dynamics of total, linear nonintegrated, and integrated HIV-1 DNA in vivo and in vitro. J Infect Dis. 2008;197(3):411–419. doi: 10.1086/525283. [DOI] [PubMed] [Google Scholar]
- Kostrikis LG, Touloumi G, Karanicolas R, Pantazis N, Anastassopoulou C, Karafoulidou A, Goedert JJ, Hatzakis A. Quantitation of human immunodeficiency virus type 1 DNA forms with the second template switch in peripheral blood cells predicts disease progression independently of plasma RNA load. J Virol. 2002;76(20):10099–10108. doi: 10.1128/JVI.76.20.10099-10108.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassen K, Han Y, Zhou Y, Siliciano J, Siliciano RF. The multifactorial nature of HIV-1 latency. Trends Mol Med. 2004;10(11):525–531. doi: 10.1016/j.molmed.2004.09.006. [DOI] [PubMed] [Google Scholar]
- Lehrman G, Hogue IB, Palmer S, Jennings C, Spina CA, Wiegand A, Landay AL, Coombs RW, Richman DD, Mellors JW, Coffin JM, Bosch RJ, Margolis DM. Depletion of latent HIV-1 infection in vivo: a proof-of-concept study. Lancet. 2005;366(9485):549–555. doi: 10.1016/S0140-6736(05)67098-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitner TFB, Hahn B, Marx P, McCutchan F, Mellors J, Wolinsky S, Korber B. Published by Theoretical Biology and Biophysics Group; Los Alamos National Laboratories, HIV Sequence Compendium. 2005 http://www.hiv.lanl.gov/
- Lillo FB, Grasso MA, Lodini S, Bellotti MG, Colucci G. Few modifications of the Cobas Amplicor HIV Monitor 1.5 test allow reliable quantitation of HIV-1 proviral load in peripheral blood mononuclear cells. J Virol Methods. 2004;120(2):201–205. doi: 10.1016/j.jviromet.2004.05.008. [DOI] [PubMed] [Google Scholar]
- Mellors JW, Kinsley LA, Rinaldo CRJ, et al. Quantitation of HIV-1 RNA in plasma predicts outcome after seroconversion. Annals of Internal Medicine. 1995;122:573–579. doi: 10.7326/0003-4819-122-8-199504150-00003. [DOI] [PubMed] [Google Scholar]
- Nakajima N, Lu R, Engelman A. Human immunodeficiency virus type 1 replication in the absence of integrase-mediated dna recombination: definition of permissive and nonpermissive T-cell lines. J Virol. 2001;75(17):7944–7955. doi: 10.1128/JVI.75.17.7944-7955.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nara PL, Fischinger PJ. Quantitative infectivity assay for HIV-1 and -2. Nature. 1988;332:469–470. doi: 10.1038/332469a0. [DOI] [PubMed] [Google Scholar]
- Ngo-Giang-Huong N, Deveau C, Da Silva I, Pellegrin I, Venet A, Harzic M, Sinet M, Delfraissy JF, Meyer L, Goujard C, Rouzioux C. Proviral HIV-1 DNA in subjects followed since primary HIV-1 infection who suppress plasma viral load after one year of highly active antiretroviral therapy. Aids. 2001;15(6):665–673. doi: 10.1097/00002030-200104130-00001. [DOI] [PubMed] [Google Scholar]
- O'Doherty U, Swiggard WJ, Jeyakumar D, McGain D, Malim MH. A sensitive, quantitative, assay for Human Immunodeficiency Virus type 1 integration. Journal of Virology. 2002;76(21):10,942–10,950. doi: 10.1128/JVI.76.21.10942-10950.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrowski MA, Chun T-W, Justement SJ, Motola I, Spinelli MA, Adelsberger J, Ehler LA, Mizell SB, Hallahan CW, Fauci AS. Both memory and CD45RA(+)/CD62L(+) naive CD4(+) T cells are infected in human immunodeficiency type 1-infected individuals. Journal of Virology. 1999;73:6430–6435. doi: 10.1128/jvi.73.8.6430-6435.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouzioux C, Hubert JB, Burgard M, Deveau C, Goujard C, Bary M, Sereni D, Viard JP, Delfraissy JF, Meyer L. Early levels of HIV-1 DNA in peripheral blood mononuclear cells are predictive of disease progression independently of HIV-1 RNA levels and CD4+ T cell counts. J Infect Dis. 2005;192(1):46–55. doi: 10.1086/430610. [DOI] [PubMed] [Google Scholar]
- Swiggard WJ, Baytop C, Yu JJ, Dai J, Li C, Schretzenmair R, Theodosopoulos T, O'Doherty U. Human immunodeficiency virus type 1 can establish latent infection in resting CD4+ T cells in the absence of activating stimuli. J Virol. 2005;79(22):14179–14188. doi: 10.1128/JVI.79.22.14179-14188.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiggard WJ, O'Doherty U, McGain D, Jeyakumar D, Malim MH. Long HIV Type 1 reverse transcripts can accumulate stably within resting CD4+ T cells while short ones are degraded. AIDS Research and Human Retroviruses. 2004;20(3):285–295. doi: 10.1089/088922204322996527. [DOI] [PubMed] [Google Scholar]
- Vandegraaff N, Kumar R, Burrell CJ, Li P. Kinetics of Human Immunodeficiency Virus type 1 DNA integration in acutely infected cells as determined using a novel assay for detection of integrated HIV DNA. Journal of Virology. 2001;75:11253–11260. doi: 10.1128/JVI.75.22.11253-11260.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viard JP, Burgard M, Hubert JB, Aaron L, Rabian C, Pertuiset N, Lourenco M, Rothschild C, Rouzioux C. Impact of 5 years of maximally successful highly active antiretroviral therapy on CD4 cell count and HIV-1 DNA level. Aids. 2004;18(1):45–49. doi: 10.1097/00002030-200401020-00005. [DOI] [PubMed] [Google Scholar]
- Vitone F, Gibellini D, Schiavone P, Re MC. Quantitative DNA proviral detection in HIV-1 patients treated with antiretroviral therapy. J Clin Virol. 2005;33(3):194–200. doi: 10.1016/j.jcv.2004.11.003. [DOI] [PubMed] [Google Scholar]
- Wu Y, Marsh JW. Selective transcription and modulation of resting T cell activity by pre-integrated HIV DNA. Science. 2001;293:1503–1506. doi: 10.1126/science.1061548. [DOI] [PubMed] [Google Scholar]
- Wu Y, Marsh JW. Early transcription from nonintegrated DNA in human immunodeficiency virus infection. J Virol. 2003a;77(19):10376–10382. doi: 10.1128/JVI.77.19.10376-10382.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Marsh JW. Gene transcription in HIV infection. Microbes Infect. 2003b;5(11):1023–1027. doi: 10.1016/s1286-4579(03)00187-4. [DOI] [PubMed] [Google Scholar]
- Yun Z, Fredriksson E, Sonnerborg A. Quantification of human immunodeficiency virus type 1 proviral DNA by the TaqMan real-time PCR assay. J Clin Microbiol. 2002;40(10):3883–3884. doi: 10.1128/JCM.40.10.3883-3884.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.