SUMMARY
Although the hammerhead ribozyme is regarded as a prototype for understanding RNA catalysis, the mechanistic roles of associated metal ions and water molecules in the cleavage reaction remain controversial. We have investigated the catalytic potential of observed divalent metal ions and water molecules bound to a 2 Å structure of the full-length hammerhead ribozyme by using X-ray crystallography in combination with molecular dynamics simulations. A single Mn2+ is observed to bind directly to the A9 phosphate in the active site, accompanying a hydrogen-bond network involving a well-ordered water molecule spanning N1 of G12 (the general base) and 2′-O of G8 (previously implicated in general acid catalysis) that we propose, based on molecular dynamics calculations, facilitates proton transfer in the cleavage reaction. Phosphate-bridging metal interactions and other mechanistic hypotheses are also tested with this approach.
INTRODUCTION
The hammerhead ribozyme is derived from a small, self-cleaving genomic RNA discovered in satellites of various plant RNA virus genomes (Forster and Symons, 1987; Haseloff and Gerlach, 1989; Prody et al., 1986; Uhlenbeck, 1987) and other species (Bourdeau et al., 1999; Ferbeyre et al., 1998; Forster et al., 1988). It consists of a conserved core of ~15 mostly invariant residues (Ruffner et al., 1990) and, for optimal activity, requires the presence of sequences in stems I and II that interact to form tertiary contacts (De la Pena et al., 2003; Khvorova et al., 2003). The hammerhead ribozyme catalyzes an RNA self-cleavage phosphodiester isomerization reaction that involves nucleophilic attack of the C17 2′O upon the adjacent scissile phosphate, producing two RNA product strands. The 5′ product possesses a 2′,3′-cyclic phosphate terminus, and the 3′ product possesses a 5′-OH terminus. The reaction is therefore, in principle, reversible, as the scissile phosphate remains a phosphodiester and may thus act as a substrate for hammerhead RNA-mediated ligation.
There are several catalytic strategies ribozymes such as the hammerhead may invoke to facilitate self-cleavage. These include base-catalyzed abstraction of a proton from the 2′O nucleophile, acid-catalyzed donation of a proton to the 5′O leaving group, and direct electrostatic stabilization of the pentacoordinate oxyphosphorane transition state (Wedekind and McKay, 1998). The detailed molecular mechanism by which hammerhead RNA catalysis occurs has been the subject of much debate (Blount and Uhlenbeck, 2005), and a recent crystal structure of the full-length hammerhead (Martick and Scott, 2006; Nelson and Uhlenbeck, 2007; Przybilski and Hammann, 2007) has now reframed that discussion by revealing how two invariant residues, G12 and G8, are positioned in the active site in a manner consistent with their previously proposed roles (Han and Burke, 2005) in general base and acid catalysis, respectively. G12 is within hydrogen-bonding distance to the nucleophile 2′O of C17, and the ribose of G8 hydrogen bonds to the leaving group 5′O (Figure 1A). The original interpretation of pH activity relations (Han and Burke, 2005) had implicated functional groups on the G8 nucleobase; however, examination of the crystallographic data prompted testing of a correlated G8C/C3G mutant that exhibited a rescue effect (Martick and Scott, 2006), consistent with the role of the G8 nucleobase providing a structural scaffold through base pairing with C3 to position the essential 2′-OH of G8 to act as a general acid. It remains to be established what specific role (Dahm et al., 1993; Dahm and Uhlenbeck, 1991; Pyle, 1993; Zhou et al., 2002), if any (Murray et al., 1998), the divalent metal ions and other solvent components play in the catalytic mechanism.
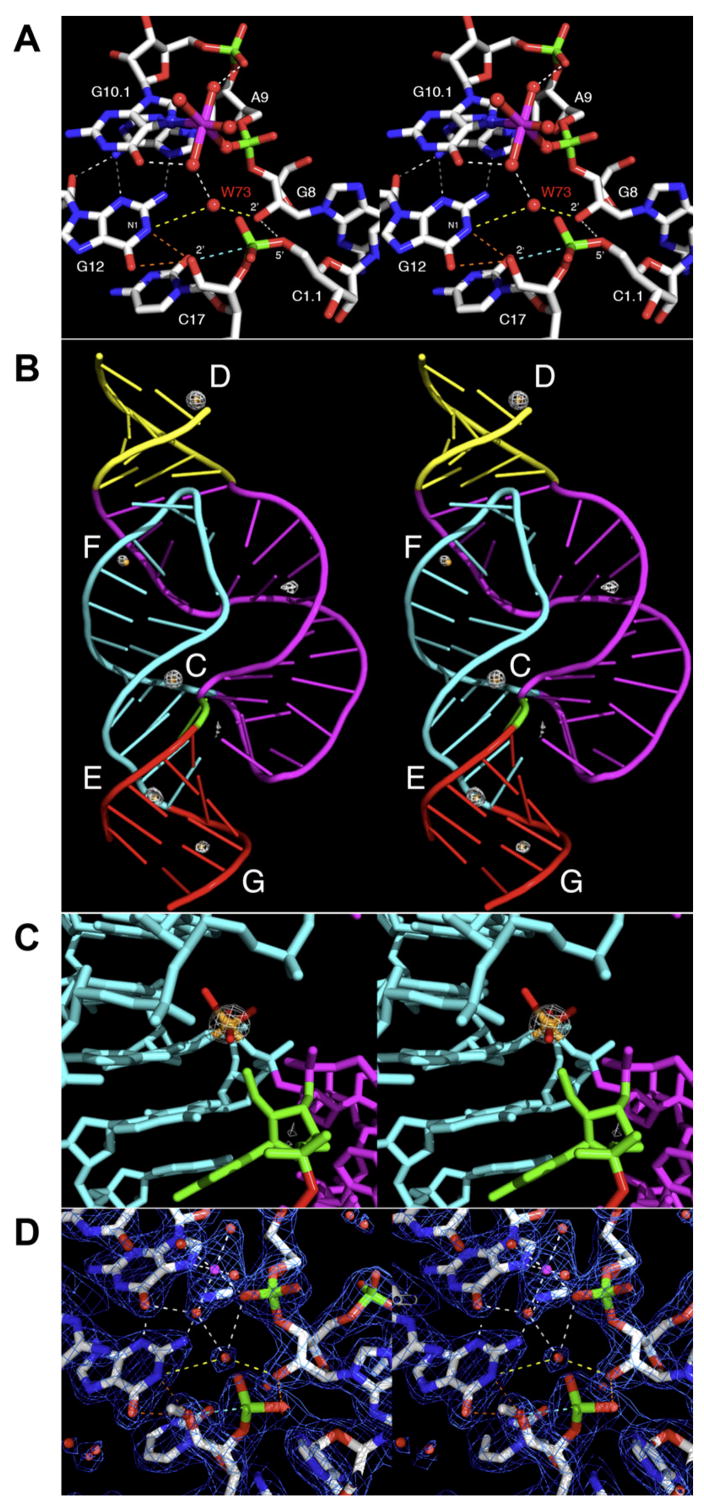
Figure 1. Wall-Eyed Stereo Representation of the Hammerhead Ribozyme.
(A) Stereo view of the active site with mechanistically relevant contacts indicated. G12 adopts a position in the hammerhead ribozyme active site consistent with the role of a general base catalyst in the cleavage reaction (assuming the N1 of G12 is abstracted), and the 2′-OH of G8 is positioned in such a way that it may participate in general acid catalysis by donating a proton to the 5′ oxygen leaving group of C1.1 as negative charge accumulates during bond scission. A Mn2+ ion (magenta) is coordinated by the pro-R oxygen of the A9 phosphate and the N7 of G10.1, as well as four water molecules (red spheres). These two purines also serve to poise G12 for general base catalysis. An ordered water (W, in red) or other solvent molecule is also bound at the cleavage site. It is within hydrogen-bonding distance to the N1 of G12 and the 2′-OH of G8 (yellow, dotted lines), the two functional groups thought to be involved in the proton transfers required for initiating and terminating catalytic cleavage, respectively. The orange, dotted lines indicate possible routes of proton abstraction by G12. Other hydrogen bonds are shown as white, dotted lines, and the covalent bond that will form upon activation of the nucleophile is shown as a light-blue, dotted line. The 2′ -CH3 that prevents cleavage in the crystal has been omitted from the figure for clarity.
(B) Overview of water and Mn2+-ion-binding sites in the hammerhead ribozyme crystal structure. Water (small, orange spheres) and Mn2+ (large, white spheres) sites on the full-length hammerhead ribozyme are indicated. The Mn2+ sites were identified by using an F(+) − F(−) anomalous difference Fourier as described in the text. The color scheme corresponds to that used to describe the full-length hammerhead ribozyme structure (Martick and Scott, 2006).
(C) A close-up of Mn2+ site C near the cleavage site of the hammerhead ribozyme. The F(+) − F(−) anomalous difference Fourier permits unambiguous identification of high-occupancy Mn2+-ion-binding sites. Shown as a magenta mesh corresponding to an anomalous difference Fourier peak contoured at 10σ, and displayed by using COOT (Emsley and Cowtan, 2004), the C site Mn2+ ion coordinates the N7 of G10.1, the pro-R oxygen of the A9 phosphate, and four water molecules. Oxygen atoms, including those of the water molecules, are shown in red, carbon atoms are shown in white, and nitrogen atoms are shown in blue.
(D) Stereo view of the 2Fo −Fc electron density map in the vicinity of the C site Mn2+ ion. The final refined σA-weighted 2Fo − Fc electron density map contoured at 1.25 rmsd is shown as a blue mesh superimposed on the refined structure at 2.0 Å resolution. The atomic coloring scheme, as well as that of the contacts, corresponds to (A).
The original crystal structure of the full-length hammerhead ribozyme reveals an active-site architecture wherein the scissile phosphate resides within 4.3 Å of the A9 phosphate (Figure 1A), suggesting that there is likely an electrostatic requirement for cation occupation that screens the repulsion between these phosphates even prior to the formation of the transition state. Biochemical evidence (Wang et al., 1999), consistent with recent molecular dynamics simulation results (Lee et al., 2007, 2008), suggest that a single divalent metal ion such as Mg2+ may bridge the A9 and scissile phosphates in the transition state. However, no bound Mg2+ ions were observed in the original full-length crystal structure under conditions of (nonphysiological) high monovalent salt (~1 M NH4+) and physiological divalent (1 mM) Mg2+ concentration. Under physiological conditions, a divalent metal ion may be required to bind to and possibly to bridge the two closely approaching phosphates (Nelson and Uhlenbeck, 2006) in a manner similar to that suggested (Wang et al., 1999) based on phosphorothioate substitutions. On the other hand, the potential bridging metal-binding site, when comprised of two phosphorothioates, may simply recruit thiophilic divalent metal ions opportunistically.
To test these and related hypotheses, we have solved the structure of a full-length hammerhead ribozyme to 2.0 Å resolution in the presence of 10 mM Mn2+ (in the presence of ~1 M NH4+). We chose Mn2+ due to the facts that (a) Mn2+ stimulates catalysis in the hammerhead ribozyme to an even greater extent (Dahm et al., 1993; Dahm and Uhlenbeck, 1991; Kim et al., 2005; Kisseleva et al., 2005; Osborne et al., 2005; Zhou et al., 2002) than Mg2+, (b) Mn2+ binds more tightly (Schiemann et al., 2003) to the hammerhead than does Mg2+, and (c) Mn2+ is more electron-rich than Mg2+ and possesses a distinct and unambiguous X-ray absorption K edge at 1.896 Å, permitting positive crystallographic identification of Mn2+-ion-binding sites. The 2.0 Å resolution data allowed for the identification of five Mn2+-ion-binding sites, including one Mn2+ ion at the active site, in addition to ~200 bound water molecules. The crystal structure, together with molecular dynamics simulations, provides detailed structural insight into the possible role of divalent metal ions and other solvent components in hammerhead ribozyme catalysis and establishes a structural basis from which further experimental and theoretical investigations into the mechanism can be initiated.
RESULTS
Crystallography
An Fo(+) −Fo(−) or anomalous difference Fourier map was calculated by using σA-weighted phases generated in Refmac (CCP4, 1994; Murshudov et al., 1997) from the previously determined positions of the RNA atoms in the full-length hammerhead ribozyme crystal structure. Six prominent peaks above 7σ (1σ the rmsd of the map) were detected. One of these (with a peak height of 9.67σ) corresponded to residual absorption from a single bromine atom in 5Br-U1.5 incorporated in Chain B of stem I as a positive control. The other five peaks, labeled C, D, E, F, and G in Figure 1B, correspond to Mn2+-ion-binding sites. These range in prominence from 21σ to 7.8σ. The single strongest site, D, is a Mn2+ ion bound to the 5′-terminal GTP of the enzyme strand and is thus most likely an artificial Mn2+-ion-binding site. Almost as strong is a peak near the active site of the hammerhead, C, in which a Mn2+ ion is observed to coordinate the N7 of G10.1 and the adjacent A9 phosphate, in a manner very similar to that observed in structures of the minimal hammerhead RNA (Pley et al., 1994; Scott et al., 1996; Figure 1C). No other Mn2+ ions are bound near the cleavage site; E, F, and G are all more distantly located. Over 200 ordered water (or similar solvent) molecules bound to the hammerhead ribozyme (Figure 1B) were also identified, tested, and confirmed, based upon difference Fourier analyses and stereochemical criteria implemented within COOT (Emsley and Cowtan, 2004). These include water molecules that complete the octahedral coordination of the Mn2+ ions, along with several well-ordered solvent molecules, such as W73, that participate in extensive hydrogen-bonding interactions with the functional groups implicated in hammerhead catalysis (Figure 1D). Data collection and refinement statistics are reported in Table 1, and key features of the Mn2+-ion-binding sites are summarized in Table 2.
Table 1.
X-Ray Data Collection and Crystallographic Refinement
| Data Collection: Mn remote (1.378 Å) | Overall | Inner Shell | Outer Shell | |
|---|---|---|---|---|
| Resolution limits (Å) | 38.52–2.00 | 38.52–6.32 | 2.11–2.00 | |
| Rmerge | 0.037 | 0.031 | 0.113 | |
| Rmeas (within I+/I−) | 0.049 | 0.042 | 0.147 | |
| Rmeas (all I+ and I−) | 0.054 | 0.052 | 0.149 | |
| Total number of observations | 51995 | 1360 | 7601 | |
| Total number unique reflections | 12527 | 362 | 1814 | |
| <(I)/σ(I)> | 23.2 | 36.4 | 10.4 | |
| Completeness | 97.0 | 85.3 | 96.3 | |
| Multiplicity | 4.2 | 3.8 | 4.2 | |
| Anomalous completeness | 94.3 | 93.3 | 94.3 | |
| Anomalous multiplicity | 2.1 | 2.1 | 2.1 | |
| ΔAnom correlation between half-sets | 0.092 | |||
| Mid-slope of Anom normal probability | 1.295 | |||
|
| ||||
| Space Group C2; Unit Cell Parameters: | a = 50.312 Å, b = 68.981 Å, c = 60.443 Å, α = 90°, β = 112.62°, γ = 90° | |||
|
| ||||
| Crystallographic Refinement | Mn remote (1.378 Å) | |||
|
| ||||
| Resolution range (Å) | 55.8032.00 | |||
| Data cutoff for refinement (σ(F)) | None | |||
| Completeness for range (%) | 96.55 | |||
| Number of reflections used in refinement | 11,277 | |||
| Crossvalidation method | throughout | |||
| Rfree value test set selection | random | |||
| R value (working + test set) | 0.17028 | |||
| R value (working set) | 0.16435 | |||
| Rfree value | 0.22293 | |||
| Rfree value test set size (%) | 9.9 | |||
| Rfree value test set count | 1,234 | |||
| Non-hydrogen atoms used in refinement | 1,556 | |||
| Mean B value (overall, Å2) | 46.609 | |||
| Coordinate error based on R Value (Å) | 0.191 | |||
| Coordinate error based on Rfree (Å) | 0.173 | |||
| Esu For B values based on maximum likelihood (Å2) | 8.629 | |||
| Correlation coefficient Fo − Fc | 0.971 | |||
| Correlation coefficient Fo − Fc Free | 0.950 | |||
|
| ||||
| Rmsd from Ideal Values | Count | Rmsd | Weight | |
|
| ||||
| Bond lengths (Å) | 1,507 | 0.010 | 0.021 | |
| Bond angles (°) | 2,332 | 1.708 | 3.000 | |
| Chiral-center restraints (Å3) | 312 | 0.077 | 0.200 | |
| General planes refined atoms (Å) | 649 | 0.009 | 0.020 | |
| Nonbonded contacts (Å) | 519 | 0.176 | 0.200 | |
| Symmetry Vdw contacts (Å) | 35 | 0.125 | 0.200 | |
| H-bond (X…Y) (Å) | 187 | 0.193 | 0.172 | |
|
| ||||
| X-ray Absorption and Dispersion | Mn2+ Edge Data Set | Mn2+ Remote Data Set | ||
|
| ||||
| f″ | f′ | f″ | f′ | |
| Mn2+ (refined) | 3.25 | −10.3 | 2.09 | 0.31 |
| Br (theoretical) | 1.82 | −0.46 | 1.05 | −0.98 |
| P (theoretical) | 0.63 | 0.34 | 0.35 | 0.25 |
Table 2.
Mn2+-Ion-Binding Sites in the Full-Length Hammerhead Crystal Structure
| Site | AFD | Occ | CN | L1 | Mn2+···L1 | L2 | Mn2+···L2 | <Mn2+···W> |
|---|---|---|---|---|---|---|---|---|
| C | 19.14 | 1.0 | 6 | G10.1:N7 | 2.004 | A9:O2P | 2.008 | 2.32(19) |
| D | 21.46 | 1.0 | 6 | GTP:N7 | 2.508 | GTP:OPβ | 2.013 | 2.28(16) |
| E | 17.42 | 1.0 | 6 | A14:O2P | 2.010 | – | – | 2.32(16) |
| F | 7.86 | 1.0 | 6 | AL2.2:N7 | 1.981 | – | – | 2.55(29) |
| G | 9.38 | 0.5 | 6 | 15.3 | – | 15.4 | – | 2.31(39) |
Listed are the anomalous difference Fourier peak heights (ADF) in units of σ, the refined occupation numbers (Occ), the coordination numbers (CN), RNA ligands involved in inner-sphere coordination (L1 and L2), the corresponding Mn2+-ligand distances (Mn2+···L1/L2), the average Mn2+-water distances (<Mn2+···W>), and standard deviations (in parentheses, in Å). Note: for the G site, the RNA ligands do not form inner-sphere coordination with the metal, i.e., metal binding is at solvent separation. Sites C, E, and F, shown in bold, are most likely relevant binding sites for hammerhead catalysis.
Molecular Dynamics Calculations
Molecular simulations, together with X-ray crystallography, provide a powerful means by which to characterize solvent structure and hydrogen-bond networks and to probe protonation states and ion occupancies. In the present work, molecular dynamics crystal simulations were performed under conditions of the experiment to probe the solvent identity, protonation propensity, and nature of the hydrogen-bonding network in the hammerhead active site (see Experimental Procedures). Three sets of simulations were carried out in which the active-site solvent molecule identified at position 73 was assumed to be an uncharged water molecule (designated W73), a positively charged hydronium ion (designated WH+73), or an ammonium ion (designated NH4+); the latter was considered because ~1 M NH4+ is present in the crystal. As a control, simulations of all three systems were carried out both with and without harmonic B factor restraints that ensure that the RNA simulation structure did not significantly drift from the crystal structure. Hence, a total of four crystal simulations were performed, each containing four copies of the full-length hammerhead RNA, and were carried out to 10 ns (postequilibration). The rms positional deviation for heavy atoms stably fluctuates around values of ~0.35 and 0.70 Å from the starting structure in the restrained and unrestrained simulations, respectively. There is only one monomer per asymmetric unit in the crystal; hence, each of the four crystallographic monomers are identical. In the crystal simulations, each of the hammerhead monomers move independently (i.e., without explicit symmetry constraints). Thus, for finite simulations, each monomer will sample slightly different regions of configurational space, and consequently exhibit correspondingly variable statistics. These statistics are useful in ascertaining the degree of variation of structural properties derived from the simulations. For the unrestrained simulations, the unit cell average structure, created from averaging all four hammerhead monomers over the last 10 ns of simulation, has an overall heavy-atom rmsd of 0.8 Å from the crystal structure. The rmsd values for the individual time-averaged monomers with respect to the crystal structure range from 1.0 to 1.2 Å and, with respect to one another, range from 0.6 to 0.8 Å (see Figure 2; Table S1 in the Supplemental Data available with this article online). A comparison of the simulation results with the crystal structure are shown in Table 3, and a detailed analysis of the hydrogen bonding in the active site are presented in Table 4 (and Table S2). The general features in these tables are quite similar between unrestrained and restrained (control) simulations, but somewhat significant differences arise between simulations with water, hydronium ion, and ammonium ion; the hydronium ion simulations are, in general, most consistent with the crystallographic data. The sugar pucker of the 2′ methylated C17 is C2′-endo in the crystal as in B-form DNA, and remains predominantly in this puckering state in the simulations that contain an unmodified 2′-OH, suggesting that there is no significant structural effect of the 2′ modification.
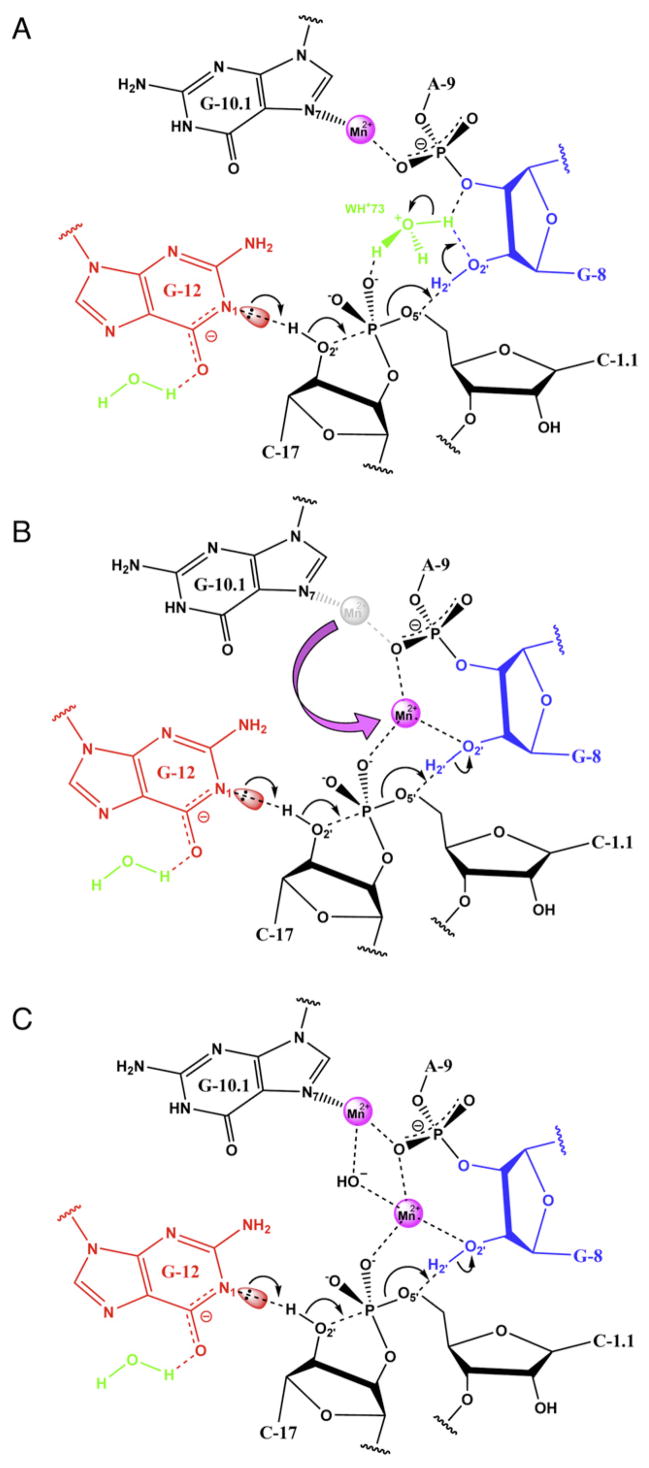
Figure 2. A Schematic Diagram Depicting Three Possible Catalytic Reaction Mechanisms.
(A) Scenario 1, wherein only indirect metal ion participation takes place. This is the most conservative hypothesis in that it is consistent with the present crystal structural analysis as well as the previous observation that the hammerhead ribozyme does not strictly require divalent metal ions for efficient catalysis (Murray et al., 1998; Scott, 1999).
(B) Scenario 2, a possible single-metal mechanism.
(C) Scenario 3, a possible double-metal mechanism.
Each scenario is described in detail in the text.
Table 3.
Non-Hydrogen Atomic Distances
| Restrained
|
Unrestrained
|
||||||
|---|---|---|---|---|---|---|---|
| Heavy Atom A-B | X-Ray | W73 | W73H+ | NH4+ | W73 | W73H+ | NH4+ |
| W(H+)73-A9:O2P | 3.17 | 3.31 (24) | 3.14 (12) | 2.93 (08) | 3.85 (33) | 3.28 (11) | 2.91 (09) |
| W(H+)73-G8:O2′ | 2.69 | 2.94 (21) | 2.79 (07) | 3.00 (08) | 3.40 (34) | 2.77 (06) | 3.02 (09) |
| W(H+)73-G8:O3′ | 3.36 | 4.37 (44) | 2.70 (07) | 2.65 (07) | 4.37 (27) | 2.65 (05) | 2.65 (05) |
| W(H+)73-C1.1:O2P | 2.57 | 2.83 (19) | 2.50 (03) | 2.73 (08) | 3.02 (22) | 2.52 (04) | 2.75 (08) |
| A9:N7-G12:N2 | 2.83 | 2.95 (06) | 2.92 (06) | 2.93 (07) | 2.98 (07) | 2.98 (08) | 3.21 (11) |
| A9:N6-G12:N3 | 2.09 | 3.12 (09) | 3.08 (08) | 3.15 (09) | 3.13 (12) | 3.04 (09) | 3.16 (10) |
| A9:N6-G12:O2′ | 3.04 | 3.08 (10) | 3.11 (10) | 3.20 (09) | 3.02 (10) | 3.02 (10) | 3.43 (15) |
| A9:O2P-C17:O2P | 4.27 | 4.67 (14) | 4.79 (15) | 4.94 (12) | 4.08 (12) | 4.16 (11) | 4.79 (13) |
Heavy atom-heavy atom distances (Å) from the X-ray crystal structure (X-ray) and from restrained and unrestrained crystal simulations of the full-length hammerhead. Standard deviations are shown in parentheses divided by the decimal precision of the average quantity, e.g., 3.31(24) means 3.31 ± 0.24. Statistics were obtained from data taken for all four monomers in the crystal unit cell over 10 ns simulation, with data sampled every 1 ps.
Table 4.
Hydrogen-Bond Distances and Angles from Simulations
| Restrained
|
Unrestrained
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| W73
|
WH+73
|
NH4+ |
W73
|
WH+73
|
NH4+ |
|||||||
| D-H···A | RHD | θDHA | RHD | θDHA | RHD | θDHA | RHD | θDHA | RHD | θDHA | RHD | θDHA |
| W(H+)73···A9:O2P | 4.10 (38) | 77.3 (101) | 2.67 (20) | 110.8 (104) | 2.04 (12) | 149.8 (110) | 3.86 (27) | 74.2 (9.7) | 2.78 (13) | 112.9 (55) | 1.99 (12) | 152.5 (109) |
| W(H+)73···C17:O2′ | 4.41 (39) | 148.3 (104) | 3.10 (12) | 120.2 (57) | 3.12 (16) | 118.9 (74) | 4.67 (26) | 134.4 (10.5) | 3.91 (12) | 134.6 (48) | 3.24 (16) | 125.5 (75) |
| W(H+)73···G8:O2′ | 3.77 (41) | 119.1 (85) | 2.32 (13) | 108.7 (69) | 2.21 (12) | 137.1 (104) | 3.52 (30) | 120.1 (6.4) | 2.32 (11) | 106.9 (55) | 2.19 (11) | 144.5 (99) |
| W(H+)73···C1.1:O2P | 2.16 (26) | 154.8 (124) | 1.53 (04) | 169.5 (48) | 1.80 (13) | 155.4 (65) | 2.37 (18) | 148.0 (8.9) | 1.56 (04) | 166.3 (51) | 1.85 (16) | 154.5 (63) |
| W(H+)73···C1.1:O5′ | 2.69 (30) | 127.0 (97) | 3.29 (09) | 131.0 (59) | 3.35 (20) | 128.1 (95) | 2.45 (17) | 150.2 (9.0) | 3.49 (07) | 143.3 (49) | 3.37 (24) | 123.7 (95) |
| G12:HN2···W(H+)73:O | 2.07 (25) | 115.1 (69) | 3.53 (17) | 139.1 (122) | - | - | 2.78 (34) | 101.3 (119) | 3.68 (37) | 141.3 (114) | - | - |
| G12:H1···C17:O2′ | 2.20 (11) | 155.6 (56) | 2.24 (10) | 155.2 (53) | 2.17 (11) | 154.6 (58) | 2.03 (12) | 156.8 (7.0) | 1.90 (06) | 164.8 (57) | 2.11 (11) | 153.1 (60) |
| C17:H2′···G12:O6 | 4.08 (10) | 26.0 (48) | 3.59 (14) | 57.9 (64) | 3.95 (11) | 29.3 (51) | 4.07 (12) | 26.3 (5.4) | 4.02 (12) | 53.1 (48) | 3.85 (11) | 32.7 (52) |
| G8:H2′···C1.1:O5′ | 2.94 (16) | 105.6 (94) | 1.88 (11) | 153.1 (92) | 2.02 (13) | 145.3 (99) | 1.97 (18) | 152.3 (14.9) | 1.84 (08) | 156.5 (75) | 2.12 (16) | 138.1 (109) |
| G12:HN2···A9:N7 | 2.02 (07) | 152.5 (50) | 2.01 (07) | 149.8 (54) | 2.08 (11) | 143.8 (73) | 2.06 (11) | 142.1 (60) | 2.04 (08) | 153.0 (54) | 2.63 (18) | 125.9 (89) |
| A9:HN6···G12:N3 | 2.15 (09) | 160.6 (49) | 2.11 (08) | 160.0 (49) | 2.18 (10) | 161.3 (50) | 2.16 (11) | 156.6 (48) | 2.07 (09) | 154.6 (48) | 2.27 (11) | 151.6 (65) |
| A9:HN6···G12:O2′ | 2.21 (12) | 142.2 (63) | 2.29 (12) | 142.4 (65) | 2.38 (13) | 138.4 (63) | 2.21 (13) | 134.4 (70) | 2.14 (12) | 139.3 (63) | 2.60 (16) | 139.3 (62) |
Average hydrogen bond (D-H···A) distances (RHD in Å) and angles (θDHA in °) from restrained and unrestrained crystal simulations of the full-length hammerhead. Standard deviations are shown in parentheses divided by the decimal precision of the average quantity, e.g., 4.10(38) means 4.10 ± 0.38. Statistics were obtained from data taken for all four monomers in the crystal unit cell over 10 ns simulation, with data sampled every 1 ps. We designate “strong” and “weak” hydrogen bonds as having average lengths less than 2.2 and 2.5 Å, respectively, and average angles as having greater than 135° and 90°, respectively.
DISCUSSION
To facilitate comparison with other hammerhead RNAs, we have designated the enzyme and substrate strand as Chains A and B, respectively, as in the original full-length structure (Martick and Scott, 2006); the five bound Mn2+ ions and their coordinated waters as “Chains” C–G; and the remaining waters as “Chain” W. A conversion table that identifies each residue according to both the sequential (Martick and Scott, 2006) and canonical (Hertel et al., 1992) hammerhead numbering schemes is provided as Supplemental Data and is also available from the authors’ website (http://xanana.ucsc.edu/hh).
The Five Mn2+-Ion-Binding Sites
Five Mn2+-ion-binding sites in the hammerhead ribozyme were unambiguously detected by using X-ray absorption Fo(+) − Fo(−) anomalous difference Fourier procedures near the K edge of Mn(II). We have labeled these as sites C–G (Figure 1B), and we propose that the site C metal ion is the one most likely to be directly relevant to the ribozyme’s catalytic mechanism. The environments corresponding to each of these binding sites are described below.
C Site
The Mn2+-ion-binding site within the essential active-site region of the full-length hammerhead ribozyme involves coordination with the N7 of G10.1 and a nonbridging phosphate oxygen (O2P) at A9. Contrary to expectations, the mode of binding is identical to what was observed originally for Mn2+ ion binding to minimal hammerhead ribozyme constructs (Pley et al., 1994; Scott et al., 1996), as is shown in Figures 1C and 1D. The C site Mn2+ ion does not bridge the A9 and scissile phosphates, despite the fact that these approach one another within 4.3 Å. The C site, in the context of the minimal hammerhead RNA, binds a variety of divalent metal ions, including Mg2+, Mn2+, Cd2+, and Co2+. The metal complex is a distorted octahedron with a 112° N7-Mn2+-OP2 angle.
D Site
The most prominent anomalous difference Fourier peak corresponds to a Mn2+-ion-binding site formed by the 5′-terminal GTP that is present in the crystal structure as an artifact from in vitro transcription. The Mn2+ ion is octahedrally coordinated by four water molecules as well as by the N7 of the GTP and a nonbridging phosphate oxygen from the β-phosphate of the GTP.
E Site
The O2P of A-14, in which the augmented stem II joins stem III, coordinates a Mn2+ ion. The other five ligands are all waters, two of which, in turn, hydrogen bond to A-15.1 in stem III.
F Site
The N7 of A-L2.2 in the loop of stem II coordinates a Mn2+ ion. The other five ligands are all waters that engage in hydrogen-bonding networks with other structural waters and adjacent nucleotides, presumably to stabilize the interaction between stems I and II. The stem II loop residue A-L2.2 itself is involved in a stacking interaction with A-B1.3 of stem I.
G Site
The weakest (fractionally occupied) site binds a fully hydrated Mn2+ ion between residues 15.3 and 15.4 in a nonconserved region of stem III.
The fully occupied Mn2+ ion binding observed at sites C, E, and possibly F are the most likely to be physiologically relevant divalent metal-ion-binding sites in the full-length hammerhead ribozyme. This is consistent with observations obtained from EPR spectroscopy (Kim et al., 2005) and FRET measurements (Lilley, 2005; Penedo et al., 2004). Of these, only the C site Mn2+ ion is close to the cleavage site (Figure 1C). No anomalous difference density above noise can be detected that might correspond to the divalent metal ion proposed (Wang et al., 1999) to bind simultaneously to the A9 and scissile phosphates, despite their 4.3 Å separation, which could easily accommodate a single bridging Mn2+ ion. Instead, the Mn2+ ion that binds to the A9 phosphate does so in a manner nearly identical to the “McKay Site” binding (Pley et al., 1994) mode originally observed in the minimal hammerhead structures, in which the scissile phosphate lies 18 Å from the A9 phosphate. It is possible that a thio-substituted scissile phosphate helps to recruit a thiophilic divalent cation to the bridging position.
Solvent Structure
Over 200 ordered solvent molecules are readily identifiable in 2Fo − Fc and Fo − Fc Fourier maps and are located with COOT (Emsley and Cowtan, 2004). These have been identified as water molecules (designated W), but the possibility that some may in fact be NH4+ must also be considered, as the crystals form in the presence of ~1 M NH4+. The Mn2+-bound waters form octahedral complexes with coordination distances between 2.0 and 2.4 Å (Table 2). Most of the remaining waters form specific first solvent shell networks on the surface of the RNA. Several highly ordered solvent molecules were observed within the active site (Figures 1A and 1D), including a solvent molecule at position 73 bound to the scissile phosphate. Crystal simulations were used to gain further insight into the specific role of the Mn2+ ion, the identity of the solvent molecule at position 73, and the nature of the hydrogen-bond network in the active site. The solvent structure in the crystal simulations was overall very similar to that of the crystal, particularly in the active site, despite the fact that the initial solvent configuration did not consider the crystallographic water positions (see Figure S6). The active-site waters in the low-temperature crystal simulations maintained an ordered structure for the 10 ns sampling duration. Residence times for waters are sensitive to temperature and environment. In the crystal, with simulations at the experimental temperature (100K), the residence times of waters around the RNA phosphates were typically too long to be reliably determined. Crystal simulations at 300K exhibit water residence times around the phosphates of 1.05 ns. Simulations in a solution environment at 300K exhibited corresponding water residence times of 0.58 ns, considerably less than in the crystal at the same temperature, and in reasonable accord with the typical residence times for ordered waters around RNA reported in other simulation studies (Auffinger and Westhof, 1997, 2000). The water-phosphate radial distribution functions show a persistent first peak at 2.78 Å that increases in height and sharpness that is correlated with increasing water residence time (see Figures S7 and S8).
The Position 73 Solvent Molecule May Favor Protonation
Although W73 is most likely H2O, partial occupancy of the site by H3O+ or NH4+ is also possible and is indistinguishable by X-ray crystallography at 2 Å resolution. Table 3 compares crystallographic data with simulation results with a water molecule (W73), a hydronium ion (WH+73), or an ammonium ion (NH4+) at position 73. In the W73 simulation, the water at position 73 deviates considerably from its crystallographic position and, in the case of the unrestrained simulation, is observed to exchange with bulk solvent and Na+ ions. This observation suggests that, in the crystal, a solvent molecule at W73 possesses an increased propensity to be positively charged.
In the NH4+ and WH+73 simulations, on the other hand, a cation at position 73 will remain well ordered, in closer agreement with the crystallographic data. The WH+73 simulation is overall in the closest agreement with the crystallographic data (in particular the hydronium ion coordination with G8:O2′ [the implicated general acid], and O2P of the scissile phosphate). Despite the presence of an unmodified 2′-OH in the simulations (to simulate an active ribozyme), no significant hydrogen bonding was observed between the protonated nucleophile and the ordered solvent at position 73 (Table 4). In simulations performed modeling the crystallographic conditions, a hydronium ion can occupy position 73 for extended residence times and, along with the Mn2+ bound at site C, can satisfy the electrostatic requirement of the electronegative active-site pocket without forming an explicit bridge between the A9 and scissile phosphates. The extent to which the potential hydronium and Mn2+-binding modes persist under physiological conditions and what changes may occur upon proceeding to the transition state remain open questions. In the crystal, W73 may actually be NH4+, but this does not alter the main conclusion, which is that the solvent molecule observed at W73 possesses an increased propensity to be positively charged.
A Mn2+-Mediated Hydrogen-Bond Network Stabilizes the Catalytic Precursor
The hydrogen-bond network in the active site is examined in more detail in Table 4. In the W73 simulations, the water at position 73 is weakly hydrogen bonded to the O2P and O5′ atoms of C1.1. In the WH+73 simulations, the hydronium ion forms strong hydrogen bonds with the O2P atom of C1.1, and it is weakly hydrogen bonded to O2′ of G8 and, to a lesser extent, O2P of A9. When position W73 is occupied by a cation in the simulations, several key hydrogen bonds in the active site become stabilized relative to water, including interactions between G12:H1···C17:O2′ and G8:H2′ ···C1.1:O5′ and, to a lesser extent, hydrogen bonding between A9 and G12. The hydrogen-bond network in the active site is shown in Figure 1A. It is of interest to note that, in simulations in the absence of Mn2+ at site C, the hydronium ion loses the hydrogen-bonding interaction with G8 and moves into a bridging position between the A9 and scissile phosphates, making strong hydrogen bonds with the nonbridging phosphate oxygens (see the Supplemental Data). These results suggest that Mn2+ binding at site C disperses the negative charge and induces solvent ordering in the active site.
The C site Mn2+, together with W73, establishes a hydrogen-bond network in the active site that stabilizes the catalytically active structure. These stabilizing interactions include orienting G12 with respect to the O2′ nucleophile via hydrogen-bond interactions with A9 and partial stacking interactions with G10.1, as suggested previously by FRET measurements (Lilley, 2005; Penedo et al., 2004), and positioning the 2′-OH of G8 to hydrogen bond to the leaving group via hydrogen bonding with W73.
Testing Three Possible Mechanisms
The Mn2+-binding mode identified in the full-length hammerhead active site is suggestive of a possible mechanistic role. If divalent metal ions do play a role in the chemical steps of hammerhead ribozyme catalysis, it is possible to envision several scenarios that range from indirect participation to direct involvement of one (or possibly two) divalent metal ion(s). We emphasize in the present discussion the scenario that is most consistent with the metal ion and solvent structure observed in the 2.0 Å crystal structure, and we only briefly touch upon other possible mechanisms that rely on predicted, but as yet unobserved, structural and metal-ion-binding modes that may appear during formation of the transition state. It should be emphasized that further functional studies are needed in order to distinguish between these mechanistic scenarios, and the present work presents key structural data from which such studies may begin.
Scenario 1: Indirect or No Metal Ion Participation
Figure 2A shows Scenario 1. The least speculative hypothesis, based on the currently available structural data, is that the C site Mn2+ ion and W73 in the crystal structure simply retain their positions and coordination in proceeding to the transition state of the chemical step of the self-cleavage reaction (Figure 2A). If this binding mode persists in the transition state, the Mn2+ ion likely participates in catalysis, if at all, only indirectly by helping to withdraw some of the accumulating negative charge of the scissile phosphate. In addition, it may introduce order to the solvent molecules near the cleavage site, as indicated in Figures 1A and 1D. Networks of ordered water molecules in the core of ribozymes such as the hepatitis delta virus ribozyme have been proposed to function as proton-relay systems (Rhodes et al., 2006). W73, whether a water or an ammonium ion, can form hydrogen bonds with G12 (the general base) and the 2′-OH of G8 (implicated as the general acid). These potential contacts are highlighted in yellow in Figures 1A and 1D. Since the cleavage reaction must be initiated by deprotonation of the N1 of G12 (to generate a base that is able to abstract the 2′-H of C17), and the cleavage reaction must be terminated by replacing a proton on the 2′-OH of G8 after it has been donated to the 5′-O leaving group, the solvent molecule W73 is ideally positioned for proton relay. Our simulation results indicate that binding of a positively charged ion (hydronium or ammonium) at position W73 is favorable, consistent at least with transient protonation of W73 in a proton-relay mechanism. Scenario 1 is the least speculative hypothesis, as it does not invoke the appearance or repositioning of any ions or atoms in the crystal structure, apart from protons. Although the binding of the divalent metal ion at site C is consistent with previous crystallographic (Pley et al., 1994; Scott et al., 1996) and biochemical data (Wang et al., 1999) that suggest that this is a divalent metal-binding site in the ground state, scenario 1 does not fully explain the thio-effect at the scissile phosphate (Wang et al., 1999), nor does it provide a clear explanation of the observed metal-ion dependence of the apparent pKa of the general acid (Roychowdhury-Saha and Burke, 2006).
Scenario 2: Single-Metal Mechanism
Figure 2B shows the single-metal mechanism. An alternative role for the Mn2+ ion is one in which it migrates from the crystallographically observed position to bridge the A9 and scissile phosphates in response to the negative charge that accumulates as the transition state is formed, in a manner previously suggested (Wang et al., 1999) and refined by recent molecular dynamics simulation results (Lee et al., 2007, 2008). If this (crystallographically unobserved) migration takes place, the Mn2+ ion would then be a direct participant in a transition-state charge stabilization interaction, and it might also shift the pKa of the general acid G8-2′-OH in a metal-dependent fashion (Roychowdhury-Saha and Burke, 2006). The observed equilibrium and kinetic thio-effect at the A9 phosphate and the kinetic thio-effect at the scissile phosphate are consistent with this mechanism (Wang et al., 1999), as are biochemical studies that implicate the importance of the G8 base as a general acid with pH dependence correlated with metal ion pKa (Roychowdhury-Saha and Burke, 2006). This scenario has been presented in considerable detail elsewhere (Lee et al., 2007, 2008).
Scenario 3: Double-Metal Mechanism
Figure 2C shows the double-metal mechanism. A third possibility involves a combination of the first two scenarios in which the C site Mn2+ ion remains bound in the crystallographically observed position, and a second divalent metal ion is recruited during approach to the transition state to displace W73 and bridge the A9 and scissile phosphates. This mechanism is consistent with the simulation results that indicate that the W73 site favors cation binding and is analogous to the arrangement of the observed two Mn2+-ion cluster in arginase (Kanyo et al., 1996). Arginase is thought to exploit an oxo-bridging water between the two Mn2+ ions of its unique binuclear active-site metal cluster to activate specific base catalysis (Kanyo et al., 1996). If an analogous mechanism activated catalysis in the hammerhead, it might do so by helping to abstract the N1 proton from G12, whose removal is required for G12 to function as a general base in the cleavage reaction. Mn2+ ions would then be seen to play two roles in catalysis-transition-state charge stabilization and activation of general base catalysis by G12. A μ-bridging hydoxide ion sandwiched between two Mg2+ ions in the hammerhead active site has been previously proposed based upon electrostatic calculations (Hermann et al., 1997), albeit in a different context, and was shown to be stable when bound to biological phosphates in quantum chemical calculations (Mayaan et al., 2004, 2007) and to be plausible as a mechanistic model (Leclerc and Karplus, 2006).
SIGNIFICANCE
The full-length hammerhead ribozyme is up to 1000-fold more catalytically active than the minimal hammerhead ribozyme (Canny et al., 2004; Khvorova et al., 2003). The structural basis for the observed rate enhancement lies, at least in part, on the dramatic rearrangements of the ground-state conformation of the active site in the full-length hammerhead ribozyme relative to the minimal hammerhead (Martick and Scott, 2006). The addition of Mn2+ to the crystal structure has permitted us to observe not only where the principal divalent metal ion sites are located, but, due to a combination of higher-resolution data and the possible solvent ordering induced by Mn2+ binding near the active site (i.e., site C, between N7 of G10.1 and the A9 phosphate), where apparently catalytically important water molecules reside in the hammerhead active site. One of these well-ordered solvent molecules in particular, W73, is within hydrogen-bonding distance to the putative general base (G12) and general acid (G8) implicated in hammerhead catalysis by both biochemical (Han and Burke, 2005) and crystallographic (Martick and Scott, 2006) analyses. Molecular dynamics calculations indicate that W73 has an increased propensity to be protonated, at least transiently, and is therefore ideally suited for solvent-mediated proton transfer during the cleavage reaction. It is therefore possible that ordered solvent molecules in the active site, such as W73, may participate in a proton-transfer relay or assist in activating the general acid and base through affecting pKa shifts. Three mechanistic scenarios are presented, one of which does not rely on crystallographically unobserved changes in metal ion binding or other structural rearrangements (apart from cleavage of the scissile bond). Our combined 2 Å resolution crystallographic and molecular dynamics investigation of metal ion and solvent binding to the full-length hammerhead ribozyme provides insight that will aid in the elucidation of the detailed chemical mechanism of this prototype ribozyme.
EXPERIMENTAL PROCEDURES
Crystallography
Crystals of the full-length hammerhead ribozyme were prepared as previously described (Martick and Scott, 2006), except only one site, U1.5, was brominated. The crystals were soaked in a solution that included the original crystallization components (including ~1 M NH4+) supplemented with 10 mM MnCl2 for 30 min and were then flash frozen. X-ray data sets were obtained at both the Mn2+ inflection and a “remote” wavelength to 2.0 Å resolution (see Table 1). The space group and unit cell were the same as those previously reported (Martick and Scott, 2006). The structure was initially refined by using the coordinates from the metal-free structure, 2GOZ in the Protein Data Bank, by using Refmac (Murshudov et al., 1997), to an R factor of ~20% (Rfree of 24%). The Mn2+ sites were identified by using an Fo(+) −Fo(−) anomalous differences Fourier obtained from X-ray data collected at the inflection point and remote wavelengths, and all subsequent calculations were carried out by using the remote (γ = 1.378 Å) data set. (Data at the absorption peak were unavailable due to an experimental difficulty, but the inflection data possessed a strong absorption signal.) Five distinct Mn2+ peaks were identified above 7σ in the anomalous differences Fourier calculated with phases derived from the refined, metal-free RNA model based on 2GOZ (Figure 1B). These sites were added to the model by using COOT (Emsley and Cowtan, 2004), refined in Refmac, and, subsequently, ~200 water molecules were identified, refined, and tested by using COOT and Refmac. Mn2+ occupancies were estimated by using the occupancy refinement algorithm in CNS (Brunger et al., 1998). Coordinate, map, and data manipulations and other calculations were carried out by using the CCP4 suite of crystallographic programs (CCP4, 1994; Winn, 2003). Figures were prepared by using COOT (Emsley and Cowtan, 2004), PyMol (DeLano, 2003), and VMD (Humphrey et al., 1996).
Molecular Dynamics Simulations
Simulation Protocol
Crystal simulations were performed to mimic as closely as possible the experimental conditions and environment. The crystallographic unit cell (Martick and Scott, 2006) contains four asymmetric units, of which consists of one full-length hammerhead motif. The unit cell was constructed and hydrogen atom positions were generated by using VMD (Version 1.8.5) (Humphrey et al., 1996). All crystal waters were kept, and an additional 3,795 water molecules, 223 sodium ions, and 11 chloride ions were added, resulting in a molecular weight of 157,699 g/mol and a density of 1.36 g/cm3 for the entire unit cell. All of the crystal Mn2+ ions were replaced with Mg2+ ions due to the availability of reliable parameters (Mayaan et al., 2007), and the C site ion was harmonically restrained to bind to N7 of G10.1 so as to mimic the higher Mn2+-binding affinity and maintain the crystallographically observed coordination. Sodium chloride was chosen as the neutralizing salt based on the established reliability of Na+ in simulations of nucleic acids. All simulations were performed with NAMD (Version 2.6) (Phillips et al., 2005) by using the Cornell et al. (1995) force field prepared with the AMBER9 package (Case et al., 2005). The TIP3P (Jorgensen et al., 1983) water model was used, and hydronium ion parameters were derived from RESP (Bayly et al., 1993) fitting results from the QCRNA (Giese et al., 2006) database with other parameters kept consistent with the TIP3P water model. Crystal simulations were performed in the canonical (NVT) ensemble at 100 K with experimental unit cell parameters (Martick and Scott, 2006). To check the solvent packing density and pressure, validation simulations in the isothermal-isobaric ensemble (NPT) at 1 atm and 100 K were performed for different packing densities to ultimately arrive at the chosen density, which resulted in only a slight (3%) average drift from the experimental unit cell volume when the volume constraints were released. Electrostatic interactions were treated with the smooth particle mesh Ewald (PME) method (Essmann et al., 1995) with a κ value of 0.34 Å −1 and a 50 × 72 × 64 FFT grid. Nonbonded interactions were treated by using an atom-based cutoff of 10 Å with switching of the van der Waals potential between 10 and 12 Å. Numerical integration was performed by using the leap-frog Verlet algorithm with a 1 fs time step (Allen and Tildesley, 1987), and covalent bond lengths involving hydrogen were constrained by using the SHAKE algorithm (Ryckaert et al., 1977).
Equilibration
Water and ions were equilibrated by using an extensive annealing procedure similar to that used in previous work (Lee et al., 2007, 2008) over the course of 10 ns; the solute positions were harmonically restrained to their crystal structure positions with a tight force constant of 50 kcal/mol/Å2 and were then slowly released over the course of 500 ps. A harmonic restraint of 20 kcal/mol/Å2 was used to ensure the experimental coordination distance to G10:N7 was maintained at 2.004 Å to mimic the Mn2+ binding.
Production Simulations
A total of four 10 ns simulations were performed to explore different solvent and metal occupation in the hammerhead active site. A series of two restrained and two unrestrained simulations were performed. The restrained simulations used weak harmonic force constants of kBT·8π2/Bi kcal/mol/Å2, where Bi is the crystal B value for atom i, on heavy-atom solute positions so as to ensure the integrity of the RNA structure while maintaining approximately the estimated fluctuations (the average force constant was ~0.1 kcal/mol/Å2). Restrained and unrestrained simulations in the presence of a divalent ion bound to G10:N7 were performed alternately with a water (W73), a hydronium ion (WH+73), or an ammonium ion (NH4+) at the crystallographically observed W73 position.
Acknowledgments
We thank Harry Noller and others in the Center for the Molecular Biology of RNA at the University of California, Santa Cruz, and Francesca Guerra and George Giambasu in the Department of Chemistry at the University of Minnesota for helpful advice and discussion. The experiments and simulations reported herein were supported by the National Institutes of Health (W.G.S. and D.M.Y), and computational resources were provided in part by the Minnesota Supercomputing Institute, the Molecular Science Computing Facility (MSCF) in the William R. Wiley Environmental Molecular Sciences Laboratory at the Pacific Northwest National Laboratory (operated for the Department of Energy by Battelle) as well as by a generous allocation on the IBM Blue Gene at the On-Demand Center in Rochester, Minnesota. We also thank to Carlos Sosa, Cindy Mestad, Steven Westerbeck, and Geoffrey Costigan for technical assistance.
Footnotes
ACCESSION NUMBERS
The coordinates and Fos for the crystal structure reported herein are currently available in the Protein Data Bank under PDB code 2OEU.
SUPPLEMENTAL DATA
Supplemental data include eight figures, two tables, and two anomalous difference Fourier maps and can be found with this article online at http://www.chembiol.com/cgi/content/full/15/4/332/DC1/.
References
- Allen MP, Tildesley DJ. Computer Simulation of Liquids. Oxford: Oxford University Press; 1987. [Google Scholar]
- Auffinger P, Westhof E. RNA hydration: three nanoseconds of multiple molecular dynamics simulations of the solvated tRNAAsp anticodon hairpin. J Mol Biol. 1997;269:326–341. doi: 10.1006/jmbi.1997.1022. [DOI] [PubMed] [Google Scholar]
- Auffinger P, Westhof E. Water and ion binding around RNA and DNA (C,G) oligomers. J Mol Biol. 2000;300:1113–1131. doi: 10.1006/jmbi.2000.3894. [DOI] [PubMed] [Google Scholar]
- Bayly CI, Cieplak P, Cornell W, Kollman PA. A well-behaved electrostatic potential based method using charge restraints for deriving atomic charges: the RESP model. J Phys Chem. 1993;97:10269–10280. [Google Scholar]
- Blount KF, Uhlenbeck OC. The structure-function dilemma of the hammerhead ribozyme. Annu Rev Biophys Biomol Struct. 2005;34:415–440. doi: 10.1146/annurev.biophys.34.122004.184428. [DOI] [PubMed] [Google Scholar]
- Bourdeau V, Ferbeyre G, Pageau M, Paquin B, Cedergren R. The distribution of RNA motifs in natural sequences. Nucleic Acids Res. 1999;27:4457–4467. doi: 10.1093/nar/27.22.4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Canny MD, Jucker FM, Kellogg E, Khvorova A, Jayasena SD, Pardi A. Fast cleavage kinetics of a natural hammerhead ribozyme. J Am Chem Soc. 2004;126:10848–10849. doi: 10.1021/ja046848v. [DOI] [PubMed] [Google Scholar]
- Case DA, Cheatham TE, III, Darden T, Gohlke H, Luo R, Merz KM, Jr, Onufriev A, Simmerling C, Wang B, Woods RJ. The Amber biomolecular simulation programs. J Comput Chem. 2005;26:1668–1688. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CCP4 (Collaborative Computational Project Number 4) The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Cornell WD, Cieplak P, Bayly CI, Gould IR, Merz KM, Ferguson DM, Spellmeyer DC, Fox T, Caldwell JW, Kollman PA. A second generation force field for the simulation of proteins, nucleic acids, and organic molecules. J Am Chem Soc. 1995;117:5179–5197. [Google Scholar]
- Dahm SC, Uhlenbeck OC. Role of divalent metal ions in the hammerhead RNA cleavage reaction. Biochemistry. 1991;30:9464–9469. doi: 10.1021/bi00103a011. [DOI] [PubMed] [Google Scholar]
- Dahm SC, Derrick WB, Uhlenbeck OC. Evidence for the role of solvated metal hydroxide in the hammerhead cleavage mechanism. Biochemistry. 1993;32:13040–13045. doi: 10.1021/bi00211a013. [DOI] [PubMed] [Google Scholar]
- De la Pena M, Gago S, Flores R. Peripheral regions of natural hammerhead ribozymes greatly increase their self-cleavage activity. EMBO J. 2003;22:5561–5570. doi: 10.1093/emboj/cdg530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano WL. The PyMOL Molecular Graphics System. 2003 http://pymol.sourceforge.net/
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Essmann U, Perera L, Berkowitz ML, Darden T, Hsing L, Pedersen LG. A smooth particle mesh Ewald method. J Chem Phys. 1995;103:8577–8593. [Google Scholar]
- Ferbeyre G, Smith JM, Cedergren R. Schistosome satellite DNA encodes active hammerhead ribozymes. Mol Cell Biol. 1998;18:3880–3888. doi: 10.1128/mcb.18.7.3880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster AC, Symons RH. Self-cleavage of virusoid RNA is performed by the proposed 55-nucleotide active site. Cell. 1987;50:9–16. doi: 10.1016/0092-8674(87)90657-x. [DOI] [PubMed] [Google Scholar]
- Forster AC, Davies C, Sheldon CC, Jeffries AC, Symons RH. Self-cleaving viroid and newt RNAs may only be active as dimers. Nature. 1988;334:265–267. doi: 10.1038/334265a0. [DOI] [PubMed] [Google Scholar]
- Giese TJ, Gregersen BA, Liu Y, Nam K, Mayaan E, Moser A, Range K, Faza ON, Lopez CS, de Lera AR, et al. QCRNA 1.0: a database of quantum calculations of RNA catalysis. J Mol Graph Model. 2006;25:423–433. doi: 10.1016/j.jmgm.2006.02.011. [DOI] [PubMed] [Google Scholar]
- Han J, Burke JM. Model for general acid-base catalysis by the hammerhead ribozyme: pH-activity relationships of G8 and G12 variants at the putative active site. Biochemistry. 2005;44:7864–7870. doi: 10.1021/bi047941z. [DOI] [PubMed] [Google Scholar]
- Haseloff J, Gerlach WL. Sequences required for self-catalysed cleavage of the satellite RNA of tobacco ringspot virus. Gene. 1989;82:43–52. doi: 10.1016/0378-1119(89)90028-0. [DOI] [PubMed] [Google Scholar]
- Hermann T, Auffinger P, Scott WG, Westhof E. Evidence for a hydroxide ion bridging two magnesium ions at the active site of the hammerhead ribozyme. Nucleic Acids Res. 1997;25:3421–3427. doi: 10.1093/nar/25.17.3421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertel KJ, Pardi A, Uhlenbeck OC, Koizumi M, Ohtsuka E, Uesugi S, Cedergren R, Eckstein F, Gerlach WL, Hodgson R, et al. Numbering system for the hammerhead. Nucleic Acids Res. 1992;20:3252. doi: 10.1093/nar/20.12.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J Chem Phys. 1983;79:926–935. [Google Scholar]
- Kanyo ZF, Scolnick LR, Ash DE, Christianson DW. Structure of a unique binuclear manganese cluster in arginase. Nature. 1996;383:554–557. doi: 10.1038/383554a0. [DOI] [PubMed] [Google Scholar]
- Khvorova A, Lescoute A, Westhof E, Jayasena SD. Sequence elements outside the hammerhead ribozyme catalytic core enable intracellular activity. Nat Struct Biol. 2003;10:708–712. doi: 10.1038/nsb959. [DOI] [PubMed] [Google Scholar]
- Kim NK, Murali A, DeRose VJ. Separate metal requirements for loop interactions and catalysis in the extended hammerhead ribozyme. J Am Chem Soc. 2005;127:14134–14135. doi: 10.1021/ja0541027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kisseleva N, Khvorova A, Westhof E, Schiemann O. Binding of manganese(II) to a tertiary stabilized hammerhead ribozyme as studied by electron paramagnetic resonance spectroscopy. RNA. 2005;11:1–6. doi: 10.1261/rna.7127105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leclerc F, Karplus M. Two-metal-ion mechanism for hammerhead-ribozyme catalysis. J Phys Chem B. 2006;110:3395–3409. doi: 10.1021/jp053835a. [DOI] [PubMed] [Google Scholar]
- Lee TS, Silva-Lopez C, Martick M, Scott WG, York DM. Insight into the role of Mg2+ in hammerhead ribozyme catalysis from X-ray crystallography and molecular dynamics simulation. J Chem Theory Comput. 2007;3:325–327. doi: 10.1021/ct6003142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TS, Silva-Lopez C, Giambasu GM, Martick M, Scott WG, York DM. Role of Mg2+ in hammerhead ribozyme catalysis from molecular simulation. J Am Chem Soc. 2008;130:3053–3064. doi: 10.1021/ja076529e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley DMJ. Structure, folding and mechanisms of ribozymes. Curr Opin Struct Biol. 2005;15:313–323. doi: 10.1016/j.sbi.2005.05.002. [DOI] [PubMed] [Google Scholar]
- Martick M, Scott WG. Tertiary contacts distant from the active site prime a ribozyme for catalysis. Cell. 2006;126:309–320. doi: 10.1016/j.cell.2006.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayaan E, Range K, York DM. Structure and binding of Mg(II) ions and di-metal bridge complexes with biological phosphates and phosphoranes. J Biol Inorg Chem. 2004;9:807–817. doi: 10.1007/s00775-004-0583-7. [DOI] [PubMed] [Google Scholar]
- Mayaan E, Moser A, Mackerell AD, Jr, York DM. CHARMM force field parameters for simulation of reactive intermediates in native and thio-substituted ribozymes. J Comput Chem. 2007;28:495–507. doi: 10.1002/jcc.20474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray JB, Seyhan AA, Walter NG, Burke JM, Scott WG. The hammerhead, hairpin and VS ribozymes are catalytically proficient in monovalent cations alone. Chem Biol. 1998;5:587–595. doi: 10.1016/s1074-5521(98)90116-8. [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Nelson JA, Uhlenbeck OC. When to believe what you see. Mol Cell. 2006;23:447–450. doi: 10.1016/j.molcel.2006.08.001. [DOI] [PubMed] [Google Scholar]
- Nelson JA, Uhlenbeck OC. Minimal and extended hammerheads utilize a similar dynamic reaction mechanism for catalysis. RNA. 2007;14:43–54. doi: 10.1261/rna.717908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne EM, Schaak JE, Derose VJ. Characterization of a native hammerhead ribozyme derived from schistosomes. RNA. 2005;11:187–196. doi: 10.1261/rna.7950605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penedo JC, Wilson TJ, Jayasena SD, Khvorova A, Lilley DMJ. Folding of the natural hammerhead ribozyme is enhanced by interaction of auxiliary elements. RNA. 2004;10:880–888. doi: 10.1261/rna.5268404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kalé L, Schulten K. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pley HW, Flaherty KM, McKay DB. Three-dimensional structure of a hammerhead ribozyme. Nature. 1994;372:68–74. doi: 10.1038/372068a0. [DOI] [PubMed] [Google Scholar]
- Prody GA, Bakos JT, Buzayan JM, Schneider IR, Breuning G. Autolytic processing of dimeric plant virus satellite RNA. Science. 1986;231:1577–1580. doi: 10.1126/science.231.4745.1577. [DOI] [PubMed] [Google Scholar]
- Przybilski R, Hammann C. The tolerance to exchanges of the Watson Crick base pair in the hammerhead ribozyme core is determined by surrounding elements. RNA. 2007;13:1625–1630. doi: 10.1261/rna.631207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyle AM. Ribozymes: a distinct class of metalloenzymes. Science. 1993;261:709–714. doi: 10.1126/science.7688142. [DOI] [PubMed] [Google Scholar]
- Rhodes MM, Reblova K, Sponer J, Walter NG. Trapped water molecules are essential to structural dynamics and function of a ribozyme. Proc Natl Acad Sci USA. 2006;103:13380–13385. doi: 10.1073/pnas.0605090103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roychowdhury-Saha M, Burke DH. Extraordinary rates of transition metal ion-mediated ribozyme catalysis. RNA. 2006;12:1846–1852. doi: 10.1261/rna.128906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruffner DE, Stormo GD, Uhlenbeck OC. Sequence requirements of the hammerhead RNA self-cleavage reaction. Biochemistry. 1990;29:10695–10702. doi: 10.1021/bi00499a018. [DOI] [PubMed] [Google Scholar]
- Ryckaert JP, Ciccotti G, Berendsen HJC. Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J Comp Physiol. 1977;23:327–341. [Google Scholar]
- Schiemann O, Fritscher J, Kisseleva N, Sigurdsson ST, Prisner TF. Structural investigation of a high-affinity MnII binding site in the hammerhead ribozyme by EPR spectroscopy and DFT calculations. Effects of neomycin B on metal-ion binding. ChemBioChem. 2003;4:1057–1065. doi: 10.1002/cbic.200300653. [DOI] [PubMed] [Google Scholar]
- Scott WG. RNA structure, metal ions, and catalysis. Curr Opin Chem Biol. 1999;3:705–709. doi: 10.1016/s1367-5931(99)00029-0. [DOI] [PubMed] [Google Scholar]
- Scott WG, Murray JB, Arnold JR, Stoddard BL, Klug A. Capturing the structure of a catalytic RNA intermediate: the hammerhead ribozyme. Science. 1996;274:2065–2069. doi: 10.1126/science.274.5295.2065. [DOI] [PubMed] [Google Scholar]
- Uhlenbeck OC. A small catalytic oligoribonucleotide. Nature. 1987;328:596–600. doi: 10.1038/328596a0. [DOI] [PubMed] [Google Scholar]
- Wang S, Karbstein K, Peracchi A, Beigelman L, Herschlag D. Identification of the hammerhead ribozyme metal ion binding site responsible for rescue of the deleterious effect of a cleavage site phosphorothioate. Biochemistry. 1999;38:14363–14378. doi: 10.1021/bi9913202. [DOI] [PubMed] [Google Scholar]
- Wedekind JE, McKay DB. Crystallographic structures of the hammerhead ribozyme: relationship to ribozyme folding and catalysis. Annu Rev Biophys Biomol Struct. 1998;27:475–502. doi: 10.1146/annurev.biophys.27.1.475. [DOI] [PubMed] [Google Scholar]
- Winn MD. An overview of the CCP4 project in protein crystallography: an example of a collaborative project. J Synchrotron Radiat. 2003;10:23–25. doi: 10.1107/s0909049502017235. [DOI] [PubMed] [Google Scholar]
- Zhou JM, Zhou DM, Takagi Y, Kasai Y, Inoue A, Baba T, Taira K. Existence of efficient divalent metal ion-catalyzed and inefficient divalent metal ion-independent channels in reactions catalyzed by a hammerhead ribozyme. Nucleic Acids Res. 2002;30:2374–2382. doi: 10.1093/nar/30.11.2374. [DOI] [PMC free article] [PubMed] [Google Scholar]