Summary
The mechanisms whereby chromatin structure and cell cycle progression are restored after DNA repair are largely unknown. We show that chromatin is reassembled following double-strand break (DSB) repair and that this requires the histone chaperone Asf1. Absence of Asf1 causes persistent activation of the DNA damage checkpoint after DSB repair as a consequence of defective checkpoint recovery, leading to cell death. The contribution of Asf1 towards chromatin assembly after DSB repair is due to its role in promoting acetylation of free histone H3 on lysine 56 (K56) by the histone acetyl transferase Rtt109, because mimicking acetylation of K56 bypasses the requirement for Asf1 for chromatin reassembly and checkpoint recovery after repair, while mutations that prevent K56 acetylation block chromatin reassembly after repair. These results indicate that restoration of the chromatin following DSB repair is driven by acetylated H3 K56 and that this is a signal for the completion of repair.
Introduction
Cell survival and maintenance of genomic integrity are dependent on the efficient and accurate repair of DNA double-strand breaks (DSBs). DSBs occur during DNA replication, in response to exogenous DNA damaging agents, or as a programmed event during growth or development (Aguilera and Gomez-Gonzalez, 2008). Although our knowledge of the pathways that repair DSBs and the cell cycle checkpoints that respond to DNA damage is rapidly growing, we still know very little about how DSB repair occurs in the natural context in the cell, that is, chromatin. The basic repeating unit of chromatin, termed the nucleosome, is made up of two molecules each of histone H2A, H2B, H3, and H4 with approximately 147 base pairs of DNA wrapped around it (Luger et al., 1997). By its very nature, chromatin provides a formidable obstacle to the repair machinery gaining access to the DNA lesion. Accordingly many recent studies have discovered that the chromatin structure around a DNA lesion is altered by the action of chromatin remodelers (for recent reviews see (Altaf et al., 2007; Osley et al., 2007)). However, we know very little about how the chromatin structure is reinstated after double-strand DNA repair.
Cells employ two major pathways to repair a DSB; homologous recombination and non-homologous end joining (NHEJ). An early event during both of these repair pathways is the 5’ to 3’ resection of the DNA ends by the Mre11-Rad50-Xrs2 (MRX) complex to yield 3’ single-stranded (ssDNA) tails that enable subsequent annealing or strand invasion (Williams et al., 2007). Single-strand annealing (SSA) is a form of homologous recombination that involves annealing of ssDNA tails at complementary sequences on both sides of the DSB and removal of the intervening DNA (Prado et al.,2003). By contrast, repair of a DSB by NHEJ requires no sequence homology (Dudasova et al., 2004).
The repair of DSBs usually depends on the DNA damage checkpoint that detects and signals the presence of DNA damage and arrests cell cycle progression until the damage is repaired (Qin and Li, 2003). The DNA damage checkpoint in budding yeast is initiated by the recruitment of multiple checkpoint components to the DSB, including the PI3-family kinase ATR homolog Mec1 and its binding partner, the ATRIP homolog Ddc2 (Melo and Toczyski, 2002). Once recruited to the DNA, Mec1 phosphorylates various targets including histone H2A on serine 129 (or serine 139 on the histone variant H2AX in mammals) in the chromatin flanking the lesion (Downs et al., 2000; Rogakou et al., 1998). Mec1-mediated phosphorylation also activates the Rad9 adaptor protein, which couples the upstream Mec1 kinase with Rad53 phosphorylation (Gilbert et al., 2001). Rad53 is important for maintaining nucleotide levels necessary for DNA synthesis and arresting the cell cycle at the metaphase to anaphase transition (Pasero et al., 2003). Once DSB repair is complete, the DNA damage checkpoint signals are reversed so that cells can resume cell cycle progression, by a process that has been dubbed “checkpoint recovery” (Bartek and Lukas, 2007). The mechanism of checkpoint recovery is virtually unknown, and only a few proteins have been identified to play a role in this process so far, including the helicase Srs2 (Vaze et al., 2002). Cells can also turn off their DNA damage checkpoint in the absence of DNA repair by the process called “adaptation” (Bartek and Lukas, 2007). The mechanism for this is also not clear, but requires several proteins including the NHEJ protein, Ku (Lee et al., 1998). Although checkpoint recovery and adaptation are most highly studied in budding yeast,their molecular mechanisms appear to be conserved in multicellular organisms (Lupardus and Cimprich, 2004; van Vugt and Medema, 2004; van Vugt and Medema, 2005).
Chromatin is taken apart and reassembled during DNA replication and transcription by chromatin assembly factors including histone chaperones, and this is also likely to the be the case during double-strand DNA repair (Groth et al., 2007). The histone chaperone Anti-silencing Function 1 (Asf1) was identified biochemically by its ability to deposit histones H3 and H4 onto newly-replicated DNA in vitro (Tyler et al., 1999). Yeast deleted for ASF1 are highly sensitive to DNA damaging agents (Le et al., 1997; Tyler et al., 1999), which is likely to reflect a direct role for Asf1 in modulating chromatin structure during repair. Indeed, human Asf1 is required for the assembly of nucleosomes following nucleotide excision repair in vitro (Mello et al., 2002). Furthermore, yeast asf1 mutants have elevated rates of genomic instability (Myung et al., 2003; Ramey et al., 2004). Furthermore, there exists a dynamic interaction between Asf1 and the Rad53 DNA damage checkpoint kinase, which suggests that activation of Asf1 may be an important cellular response to DNA damage (Emili et al., 2001; Hu et al., 2001). In addition to its role in chromatin assembly and disassembly, Asf1 is also essential for stimulating the acetylation of free histone H3 on lysine 56 (K56) by the histone acetyl transferase (HAT) Rtt109 (Recht et al., 2006; Tsubota et al., 2007). Despite its occurrence in eukaryotes from yeast to humans, the molecular function of acetylation of H3 K56 remains unknown.
Although chromatin disassembly has been previously documented at a site of double-strand DNA damage (Tsukuda et al., 2005), chromatin reassembly following double-strand DNA repair has not been reported. In this work, we set out to discover why the Asf1 histone chaperone is required for rapid growth after DSB repair. In addition to finding a role for Asf1 in chromatin reassembly following DSB repair, we have also discovered a role for Asf1 in recovery and adaptation to the DNA damage checkpoint following repair, explaining why asf1 mutant yeast die after DNA repair. These roles for Asf1 can be bypassed by a mimic of permanent acetylation of histone H3 on lysine 56, while deletion of the gene encoding the K56 histone acetyl transferase, RTT109, also leads to persistent DNA damage checkpoint activation following DNA repair. As such, acetylated K56 on H3 is required to reinstate the chromatin structure over the repaired DNA, which in turn is a critical signal for turning off the DNA damage checkpoint, allowing cell cycle re-entry following DNA repair.
Results
Chromatin disassembly is tightly coupled to DNA resection
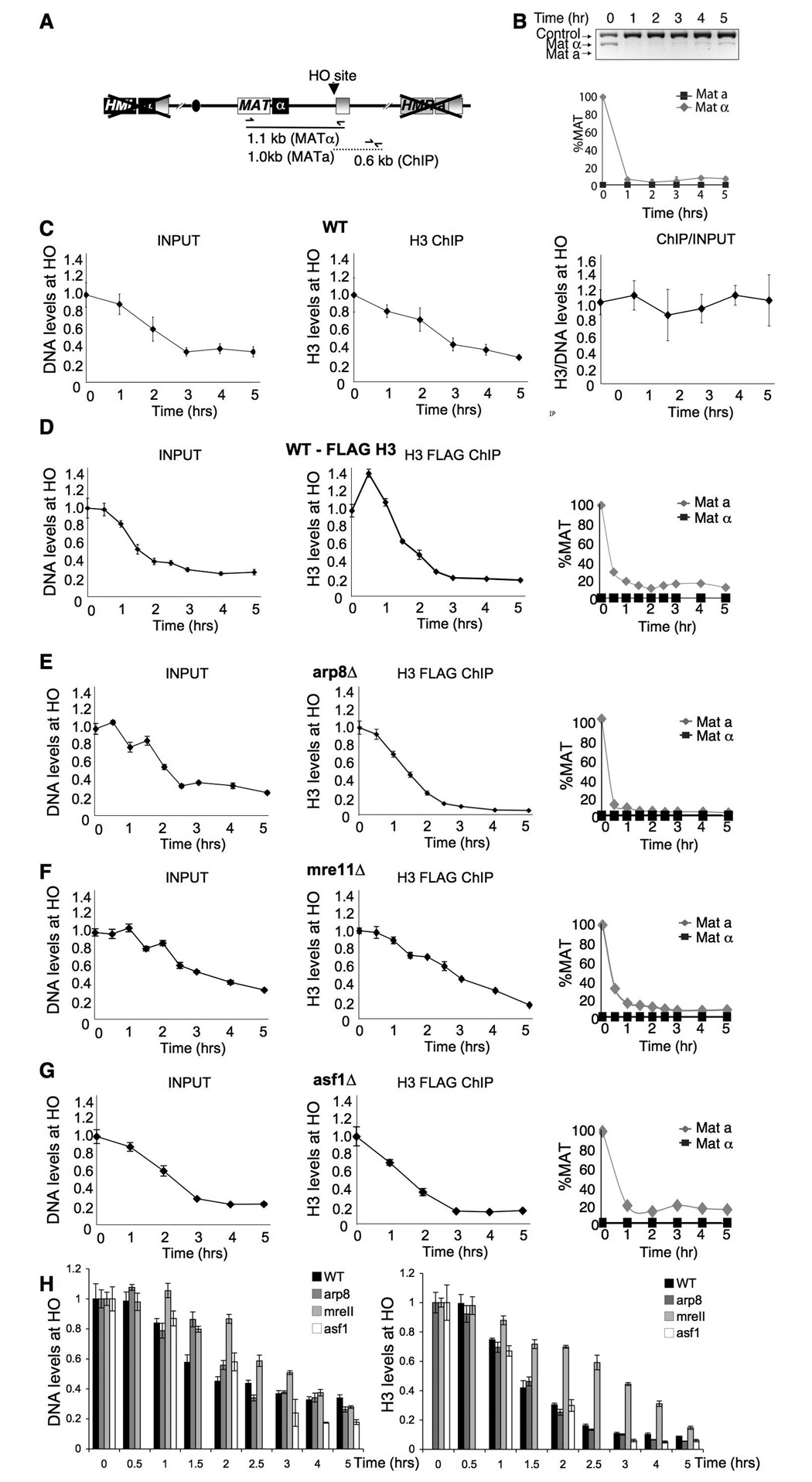
Recent chromatin immunoprecipitation (ChIP) analyses have shown that histone occupancy drops flanking an unrepairable HO endonuclease-induced DNA break in budding yeast (Shim et al., 2007; Tsukuda et al., 2005). This observation was interpreted as demonstrating that chromatin is disassembled flanking a DSB. When we had performed the same studies in the identical yeast strain (Fig. 1A,B) we observed a loss of input DNA with increasing time after induction of the DNA break as a consequence of resection to single-stranded DNA, which results in a reduced PCR signal (Fig. 1C left panel). As such, the kinetics of the loss of input DNA in the region flanking the DSB can be used as a measure of DNA resection rate. Using an antibody to the C-terminus of histone H3 that is not affected by histone modifications, we found that the kinetics of loss of histones flanking the DSB by ChIP analysis closely mimics the loss of DNA due to resection (Fig. 1C, middle panel), apparent by the horizontal line that is obtained when we normalize the histone ChIP signal to the input signal for each time point (Fig. 1C, right panel), as we and another group had previously reported (Shroff et al., 2004; Tamburini and Tyler, 2005). By contrast, the reports that interpreted their data as showing chromatin disassembly at a DSB had either normalized all their histone ChIP data to the input signal prior to inducing the DSB, rather than using the corresponding input for each time point (Tsukuda et al., 2005), or had not normalized their histone ChIP data to the input signal at all (Shim et al., 2007). By failing to normalize to, or acknowledge, the loss of input DNA as a consequence of DNA resection that occurs with an unrepairable break, we suspected that the apparent chromatin disassembly flanking the lesion reported by these groups was merely a consequence of DNA resection and the inability of histones to bind to the resulting single-stranded DNA.
Figure 1. DNA resection drives chromatin disassembly around a DSB.
A. Schematic of mating type loci in strain JKM179 and positions of primers used for DNA repair and ChIP analyses below. The HMR and HML donor loci are deleted in strain JKM179. B. PCR assay of DNA cutting and failure to repair following induction of the HO endonuclease by addition of galactose at Time = 0hr. Below is quantitation of three independent experiments, after normalization to the control. C. Analysis of DNA levels and H3 levels flanking the double-strand break. The left panel shows input DNA used for the ChIP analysis 0.6kb from the HO site normalized to a distal SMC2 site. The middle panel shows the amount of DNA 0.6kb from the HO site from the H3 ChIP analysis (“H3 IP”), normalized to a distal SMC2 site. The right panel shows the normalization of the H3 IP to input signals. D. As for C, but in the wild type strain carrying H3-FLAG (YTT035). In addition, the right panel shows quantitation of cutting and repair, determined as in B. E. As for D, but in the arp8Δ strain carrying H3-FLAG (BAT058). F.. As for D, but in the mre11Δ strain carrying H3-FLAG (BAT061). G. As for D, but in the asf1Δ strain carrying H3-FLAG (BAT062). H. The left panel shows a plot of all the input DNAs from panels D–G, while the right panel shows a plot of all the DNA from the H3 ChIPs from panels D–G. Note that the wild type H3 ChIP data was normalized to 1 at time 0.5 hrs rather than time 0 hrs to better enable comparison between the strains.
To further verify that the histone loss flanking the DSB is coupled to DNA resection, we repeated the ChIP analyses of histone occupancy around a break using the FLAG-tagged H3 construct that had been used in the previous report (Tsukuda et al., 2005). As with the H3 antibody, we observed DNA resection as loss of the input DNA at increasing times after inducing the DSB that mirrored the loss of histones from the DNA over time (Fig. 1D). It had previously been reported that the Arp8 component of the ATP-dependent chromatin remodeling complex INO80 mediates chromatin disassembly, because deletion of ARP8 delayed the loss of histones flanking a DSB (Tsukuda et al., 2005). However, we suspected this result might have been an indirect consequence of the delayed DNA resection that occurs in the arp8 mutant (van Attikum et al., 2004). To address this possibility, we examined the DNA resection and histone H3 loss from around the DSB in an arp8 mutant, by looking at the input signal and H3 ChIP signal respectively, at increasing times after inducing the DNA lesion (Fig. 1E). The delay in DNA resection in the arp8 mutant was subtle (Fig. 1E), in comparison to the delay observed upon deletion of MRE11, which encodes a protein whose inactivation is known to delay DNA resection (Tsubouchi and Ogawa, 1998) (Fig. 1F, H). Accordingly, the mre11 mutant has a clear delay in chromatin disassembly, while there is no significant delay in chromatin disassembly seen in the absence of Arp8 (Fig. 1H). As such, the rate of chromatin disassembly from around the DSB is tightly coupled to the rate of DNA resection.
Towards elucidating which proteins mediate chromatin disassembly flanking a DSB, we examined yeast mutant for the histone chaperone Asf1. Asf1 contributes to chromatin disassembly from promoter regions during transcriptional induction (Adkins et al., 2004; Schwabish and Struhl, 2006). By contrast, we observed no significant defect in either DNA resection or chromatin disassembly in the absence of Asf1 (Fig. 1G). As such, Asf1 is not required for chromatin disassembly flanking a DSB.
Asf1 is required for chromatin reassembly following DSB repair
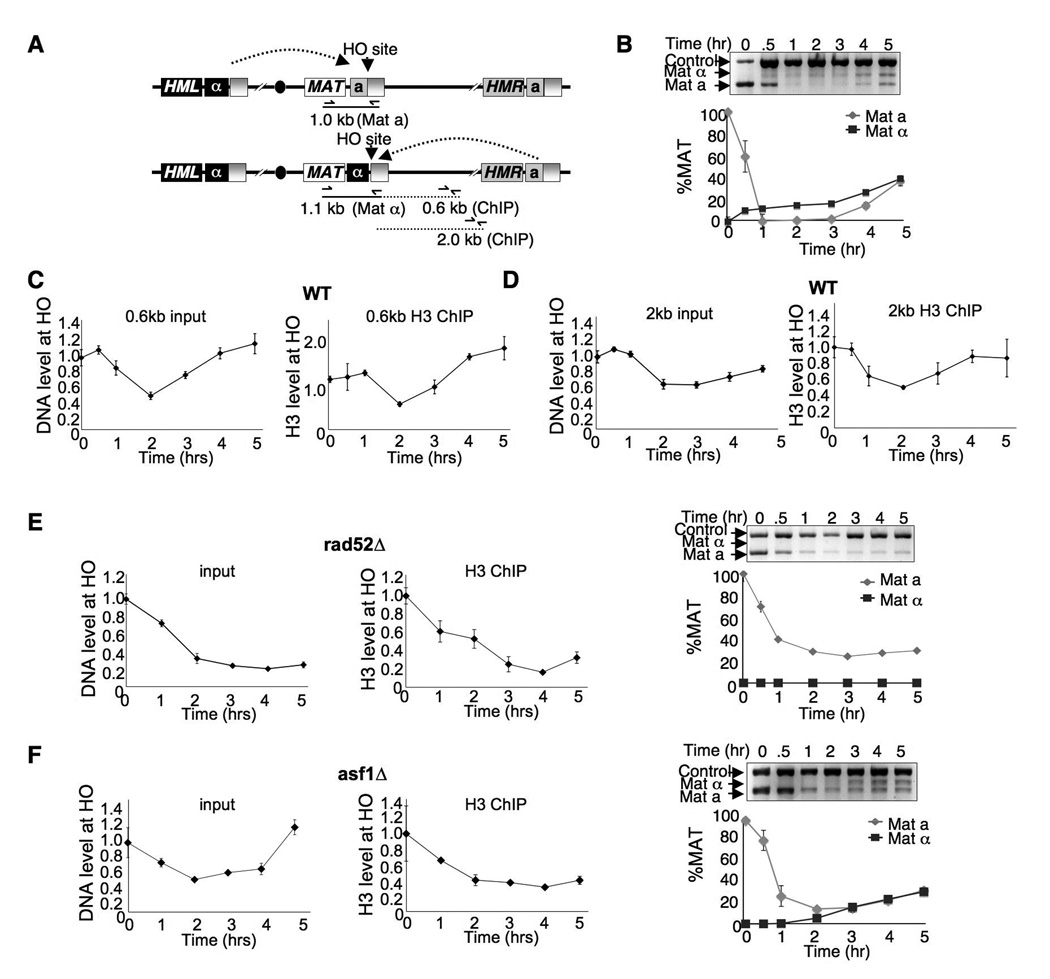
In order to determine whether chromatin is reassembled after DSB repair, we used an inducible HO lesion that was repairable using the donor sequences at HML or HMR (Fig. 2A, B). The ChIP input DNA samples show clear resection and repair of the DNA at 0.6kb from the HO lesion in a wild type strain (Fig. 2C), occurring with slightly delayed kinetics at 2.0kb from the HO lesion (Fig. 2D). The histone H3 ChIP showed a decrease and then an increase in histone levels flanking the HO site that closely followed the time course of DNA resection and DNA repair (Fig. 2C and D). The increase in histone levels around the HO site following DNA repair is dependent on repair, as it failed to occur in a rad52 mutant that cannot repair the HO lesion (Fig. 2E). It was not clear whether the increase in histone levels during repair in the wild type strain really was chromatin reassembly or whether it is just reflecting the increased amount of input DNA due to repair in the ChIP samples. Therefore, we repeated the analysis in yeast deleted for the histone chaperone ASF1. In the asf1 mutant, we observed resynthesis of DNA at 0.6kb from the HO site, albeit with slower kinetics (delayed by about 1 hour) than in wild type cells (Fig. 2F). However, we failed to see an increase in histone levels around the HO site following DNA repair in the asf1 mutant, indicating that Asf1 is required for chromatin reassembly following DNA repair.
Figure 2. Asf1-dependent reassembly of chromatin following DSB repair.
A. Schematic of mating type loci, showing positions of PCR primers generating MATa and MATα products for assaying DNA cutting and repair, and positions of primer pairs used for histone ChIP. B. Gel and quantitation of DNA cutting and repair in a wild type strain (BAT009), as described in Fig. 1. Galactose was added at time 0 to induce HO endonuclease and glucose added at 2 hours to allow repair using the donor sequences at HMR and HML. C. Chromatin disassembly and reassembly during DNA repair in wild type yeast (BAT009) at 0.6kb from the HO site. The input DNA is shown in the left panel. Quantitation of the ChIP (“H3 IP”) analysis of histone H3 is shown in the right panel, normalized as described in Fig. 1. D. As for C but at 2.0 kb from the HO site. E. As for C but in a strain deleted for RAD52 (JLY075). The right panel shows the HO cutting and repair analysis from the same time course. F. As for E but in a strain deleted for ASF1 (BAT063).
Asf1 contributes to recovery from the DNA damage checkpoint after repair
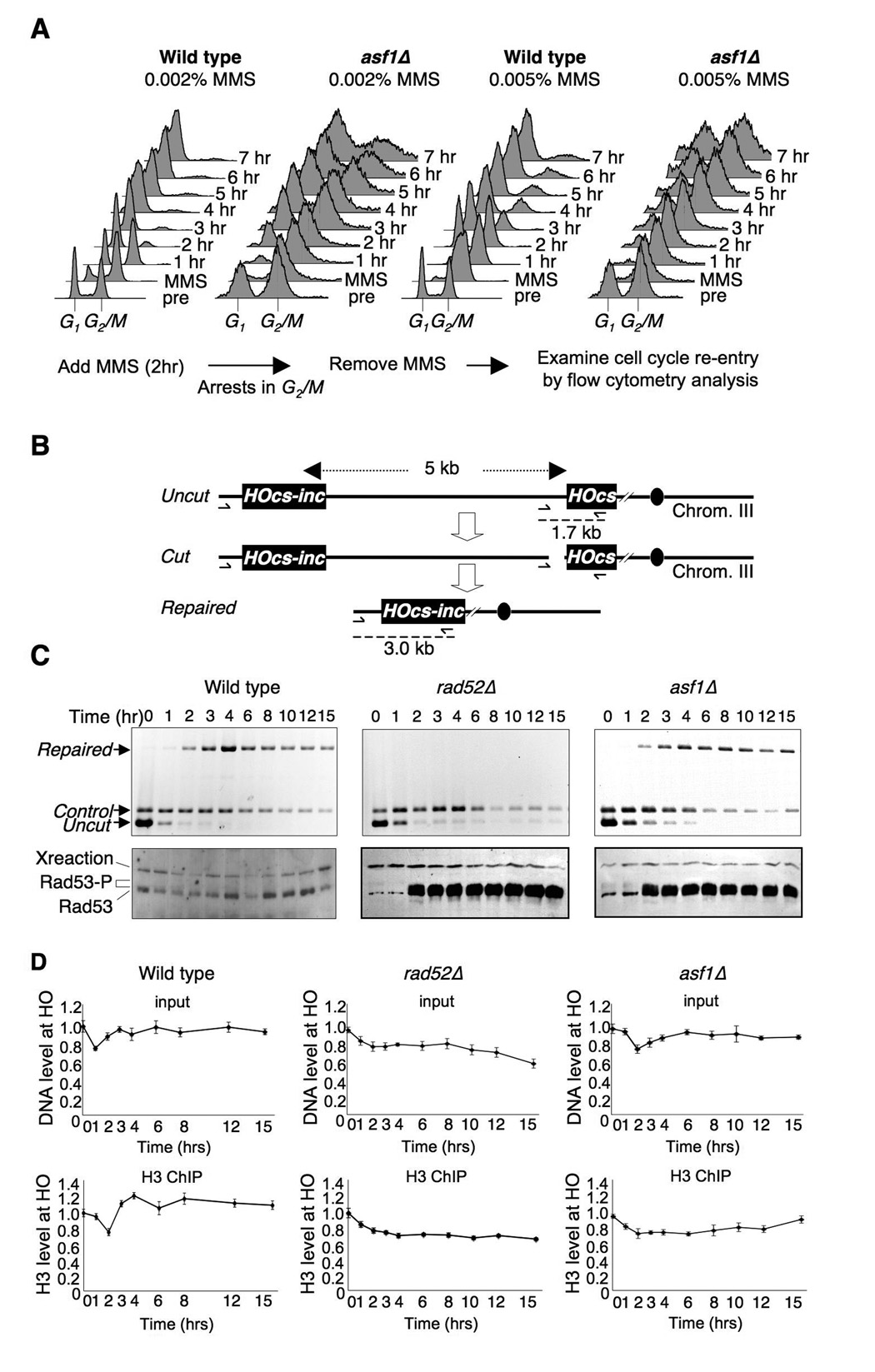
Yeast lacking Asf1 are relatively sensitive to agents that generate DSBs (Tyler et al., 1999). However, this cell death is unlikely due to a defect in DNA repair, as asf1 mutants are competent for all pathways of DSB repair (Ramey et al., 2004) (Fig. 2F). This finding led us to investigate why asf1 mutant yeast die or recover slowly even though they have already repaired their DNA lesions. We found that low doses of exposure to the alkylating agent methyl methane sulfonate (MMS) that results in single and double-strand DNA breaks led to accumulation of cells with a G2/M DNA content, presumably due to activation of the DNA damage checkpoint (Fig. 3A). Upon removal of the MMS, wild type yeast re-enter the cell cycle after 1–2 hours. By contrast, the asf1 mutant is delayed 2–5 hours (depending on the dose of MMS) from re-entering the cell cycle (Fig. 3A). This result suggests that asf1 mutants maintain an active DNA damage checkpoint for longer than normal following DNA repair, suggesting that Asf1 may contribute to recovery from the DNA damage checkpoint.
Figure 3. Delayed cell cycle re-entry in asf1 mutants following DSB repair.
A. Asynchronous cultures of WT (JKT010) and asf1Δ (JKT018) yeast were exposed to MMS for 2 hours, which causes a G2/M accumulation. Flow cytometry analysis of DNA content was used to follow the cell cycle distribution after washing out the MMS. B. Schematic for system used to measure SSA. Repair of the HO lesion at the HO-cs site requires 5 kb of resection back to the uncleavable HOcs-inc site. The position of PCR primers and products used to measure repair in panel C are shown. C. The Rad53 kinase remains activated in asf1 mutants. The DSB was induced at time 0 by addition of galactose to wild type (YMV045), rad52Δ (YMV046) and asf1Δ strains (JKT200). The top panels show analysis of the repair of the HO lesion (note that the cut DNA gives no product). The lower panels show western analyses of Rad53 at the same time points. “Xreaction” refers to a cross-reacting protein that serves as a normalization control for loading. D. Chromatin disassembly and reassembly analysis using the identical strains and time course shown in C. The top panels show the input, and the lower panels show the ChIP analysis of H3, normalized as in Fig. 1.
Because the sequence-independent nature of MMS-induced damage makes it difficult to follow DSB repair, we switched to the inducible HO endonuclease system to follow the kinetics of DNA repair and activation of the DNA damage checkpoint. Repair of the HO lesion at MAT is unusual in that it does not normally require activation of the DNA damage checkpoint for its repair (Pellicioli et al., 2001). Therefore, we used Jim Haber’s SSA system that requires 5 kilobases of DNA resection in order to repair the HO site, resulting in activation of the DNA damage checkpoint (Vaze et al., 2002). Using a set of three PCR primers to measure DNA damage and repair (Fig. 3B), the asf1 mutant showed no defect in DSB repair as compared to yeast deleted for RAD52 that failed to repair the HO lesion (Fig. 3C). Activation of the DNA damage checkpoint was followed by western blotting analysis to measure the induction and hyperphosphorylation of the Rad53 DNA damage checkpoint kinase. In the wild type strain, the DNA damage checkpoint was turned on and off too rapidly to be detected in our analysis. By contrast, in the rad52 mutant that cannot repair the DSB, the DNA damage checkpoint was activated by 2 hours and remained active for at least a further 13 hours (Fig. 3C). Strikingly, in the asf1 mutant that does repair the DSB, the DNA damage checkpoint was activated by 2 hours and remained active for at least a further 13 hours (Fig. 3C). This result demonstrates that the DNA damage checkpoint remains active in the asf1 mutants even after DNA repair, and is reminiscent of a defect in recovery from the DNA damage checkpoint. The profound delay in inactivation of Rad53 in the asf1 mutant after DNA repair closely mimicked the profound delay in chromatin reassembly proximal to the HO site in the asf1 mutant in the same strain (Fig. 3D).
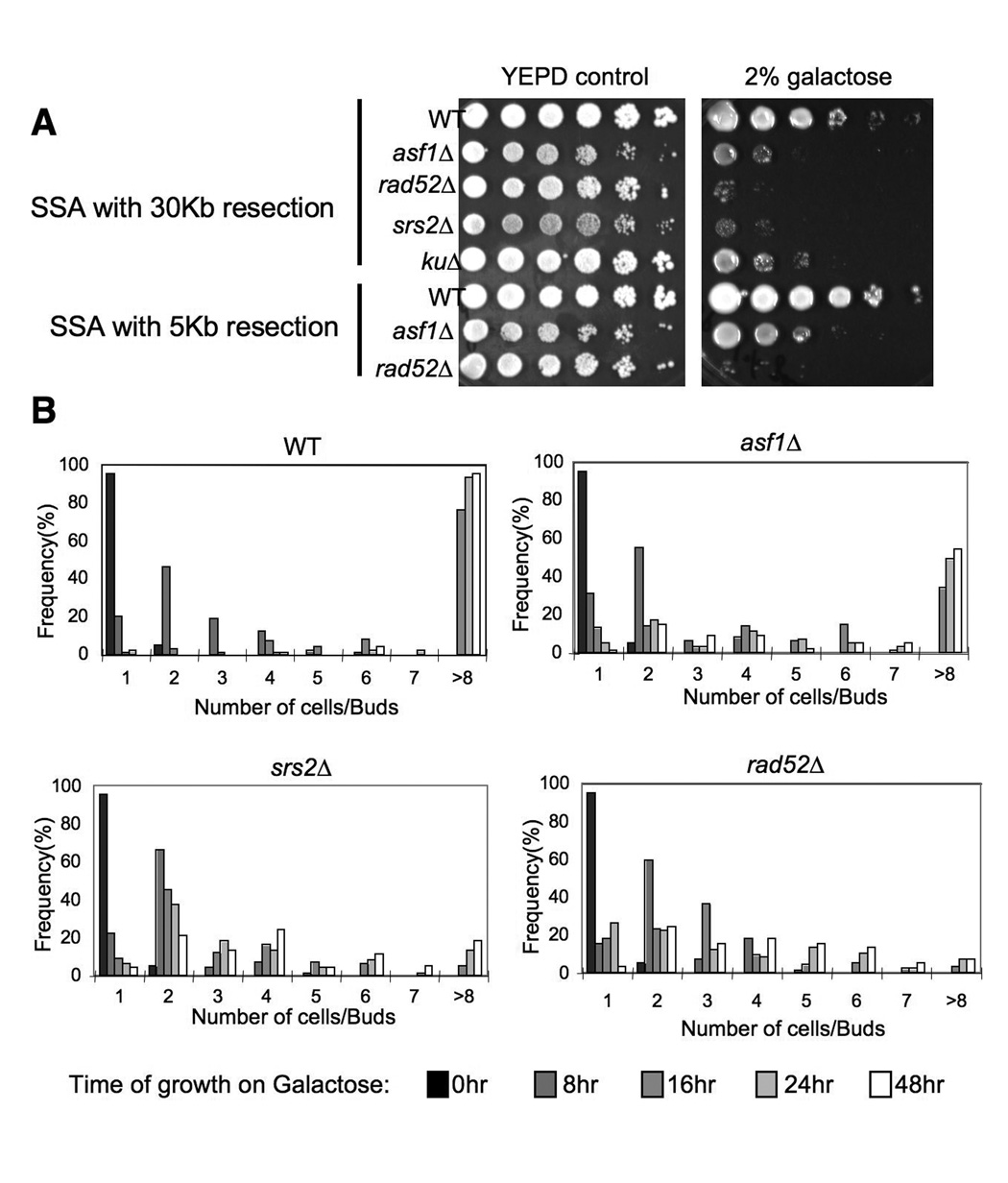
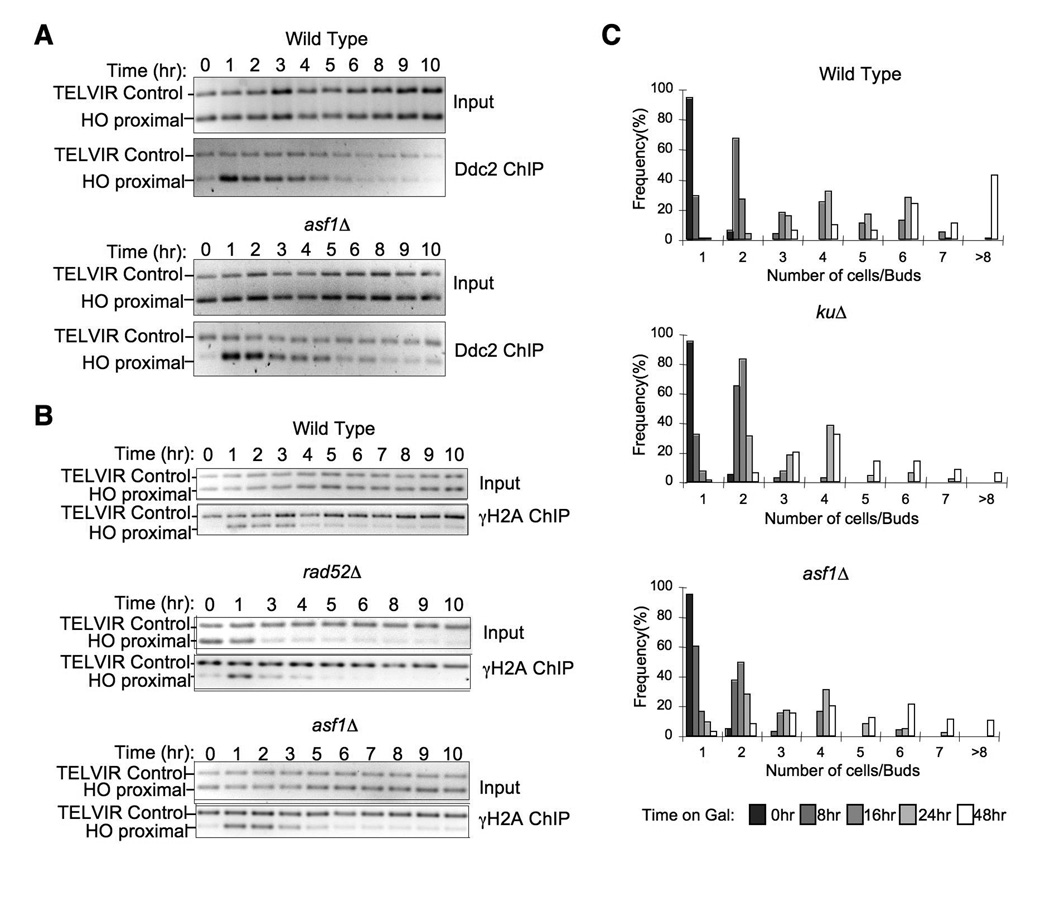
To investigate the consequence, if any, of induction of a single DNA break on viability of an asf1 mutant, we plated the SSA strains onto galactose to induce the HO endonuclease. As a control we included a strain deleted for SRS2, whose gene product is known to be required for checkpoint recovery (Vaze et al., 2002). We found that the asf1 mutants are as sensitive to this single DNA break as srs2 mutants (Fig. 4A). Furthermore, the degree of sensitivity depended on the length of resection required for SSA, with a 5kb resection causing less sensitivity of an asf1 mutant as compared to 30kb resection (Fig.4A). To determine unequivocally whether Asf1 is required for recovery from the DNA damage checkpoint, we examined the ability of asf1 mutant cells to resume cell division after DNA damage. To do this, we measured the effect of inducing the HO lesion on the ability of single cells to divide over time, as previously described (Vaze et al., 2002) (Fig. 4B). As a control we included the srs2 mutant that has a known role in checkpoint recovery (Vaze et al., 2002). Following induction of the DSB, 90% of the wild type cells divided twice by 48 hours, while around 20% of the srs2 mutant had divided twice by 48 hours. The asf1 mutant had an intermediate defect, with about 50% of the cells having divided twice by 48 hours (Fig. 4B). Importantly, this result was not due to the asf1 mutant having a growth defect on galactose plates, because colony size analysis of the asf1 mutant resembled wild type when the HO site was not cleaved (Suppl. Fig. 1). These results demonstrate that Asf1 contributes to recovery from the DNA damage checkpoint.
Figure 4. asf1 mutants have a defect in recovery from the DNA damage checkpoint.
A. 10-fold serial dilution analysis of the indicated strains, showing that asf1 mutants are sensitive to a galactose-induced unique HO endonuclease cut. Strains WT (YMV002), asf1Δ (JCY001), rad52Δ (YMV037), srs2Δ (YMV057), and kuΔ (YMV2-1) required 30kb of resection during repair by SSA, while strains WT (YMV045), asf1Δ (JKT200), and rad52Δ (YMV046) required 5kb of resection during repair by SSA. B. Quantitation of colony size formation from single cells following the indicated length of times of growth on galactose-containing plates, in WT (YMV002), asf1Δ (JCY001), rad52Δ (YMV037), and srs2Δ (YMV057) strains.
Asf1 is not required for removal of Ddc2 or phosphorylated histone H2A from the vicinity of the DNA lesion
Recruitment of the central checkpoint kinase Mec1 to a DSB activates the DNA damage checkpoint (Dubrana et al., 2007), and conversely removal of Mec1 from the DNA after repair is likely to be critical for recovery from the DNA damage checkpoint. To determine whether the checkpoint recovery defect in the asf1 mutant reflects a requirement for chromatin reassembly in order to displace the Mec1-Ddc2 complex, we examined the removal of Mec1-Ddc2 from the region flanking the HO lesion following DNA repair. By ChIP analysis of Ddc2 levels in the vicinity of the HO lesion, the kinetics of Ddc2 recruitment and removal were very similar in wild type and asf1 mutant strains during SSA repair (Fig. 5A). As such, chromatin assembly is not required for displacement of the DNA damage checkpoint protein Ddc2. Because Ddc2 tethers Mec1 to DNA (Dubrana et al., 2007), this result suggests that Asf1 is also not required for the displacement of Mec1 after DNA repair. Consistent with this observation, we saw no defect in the loss of phosphorylated H2A from the vicinity of the HO lesion in the absence of Asf1 (Fig. 5B).
Figure 5. Chromatin reassembly is not required for removal of Mec1-Ddc2 or phosphorylated H2A after DNA repair, but is required for checkpoint adaptation.
A. Removal of Ddc2 from the site of DNA repair does not require chromatin assembly. The HO lesion was induced in strains WT (JFY016) and asf1Δ (JFY017) at time 0 by addition of galactose. The level of Ddc2 flanking the HO lesion during SSA repair was measured by ChIP analysis. B. Loss of phosphorylated H2A from chromatin does not require Asf1. The HO lesion was induced in strains WT (YMV045), asf1Δ (JKT200), and rad52Δ (YMV046) at time 0 by addition of galactose. The level of H2A phosphorylated on serine 129 flanking the HO site in strains undergoing HO repair by SSA was measured by ChIP analysis. C. Asf1 contributes to checkpoint adaptation. Colony formation was assessed at the indicated times after placing single unbudded cells onto galactose plates to induce the unrepairable DSB in isogenic WT, kuΔ and asf1Δ strains derived from JKM179.
Asf1 contributes to checkpoint adaptation
Given the defect in checkpoint recovery, we examined whether asf1 mutants also have a defect in the related process of checkpoint adaptation. We performed colony size analyses on single cells carrying an unrepairable HO lesion (Fig. 5C). For comparison, we included a mutant of the yeast counterpart of Ku70, which has an established role in checkpoint adaptation (Vaze et al., 2002). By 48 hours, most of the wild type cells had divided 4 or more times when the unrepairable break was induced by growth on galactose. By contrast, the ku mutant yielded much smaller colonies as a consequence of its role in checkpoint adaptation. We found that the asf1 mutant had a checkpoint adaptation defect almost as severe as that of the ku mutant (Fig. 5C). Importantly, this result was not due to the asf1 mutant having a growth defect on galactose plates, because the colony size analysis of the asf1 mutant resembled wild type in a strain where the HO site was not cleaved (Suppl. Fig. 2). As such, Asf1 contributes to both adaptation and recovery from the DNA damage checkpoint.
Acetylated K56 on histone H3 drives chromatin assembly and checkpoint recovery after repair
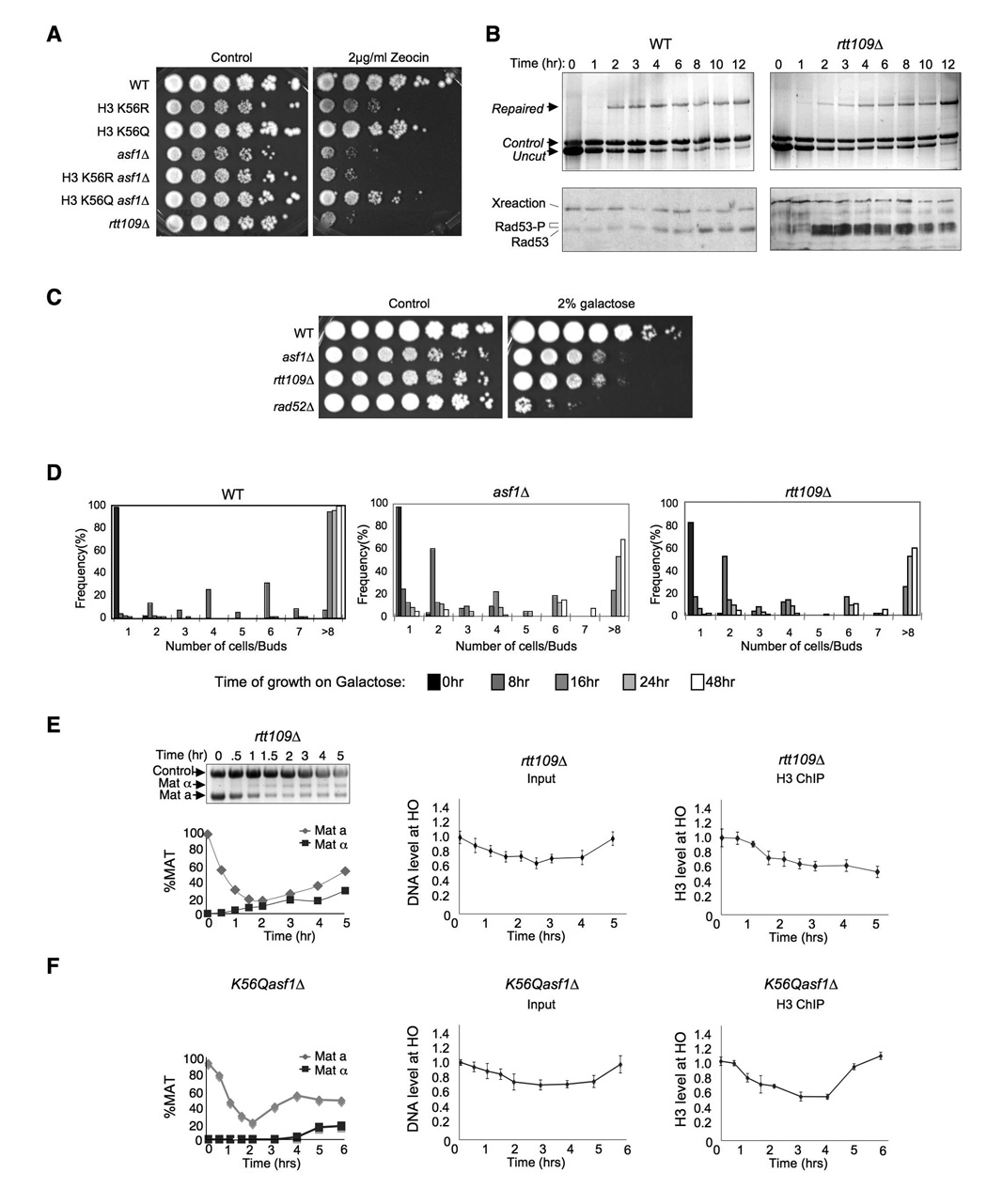
The histone chaperone Asf1 is essential for achieving acetylation of newly-synthesized histone H3 on lysines 9 and 56 (Adkins et al., 2007; Recht et al., 2006). Because the role of Asf1 in providing resistance to replicational stress can be partially bypassed by mutation of lysine 56 to Q to mimic permanent acetylation (Recht et al., 2006), we asked whether the K56Q mutation could also bypass the role of Asf1 in resistance to double-strand DNA damaging agents. Upon exposure to the radiomimetic zeocin that generates double-strand DNA breaks, the K56Q asf1Δ double mutant was significantly more resistant to zeocin than the asf1Δ mutant (Fig. 6A). This result indicates that mimicking acetylation of K56Ac can mostly bypass the role of Asf1 in protecting against double-strand DNA damaging agents. As such, an important role of Asf1 in promoting survival following double-strand DNA repair is to achieve acetylation of K56 on histone H3.
Figure 6. Acetylated H3 K56 is required for chromatin assembly and DNA damage checkpoint recovery after DNA repair.
A. Mimicking H3 K56 acetylation can bypass the requirement for Asf1 for resistance to double-strand DNA damage. 10-fold serial dilution analysis of the indicated isogenic strains. B. Rtt109 is required for recovery from the DNA damage checkpoint. The same analysis presented in Fig. 3 was performed on WT (YMV045) and rtt109Δ (JFY013) strains undergoing SSA with 5kb of resection. C. Rtt109 is required for viability after repair of the HO site. 10 fold serial dilution analysis of WT (YMV045), asf1Δ (JKT200), rad52Δ (YMV046) and rtt109Δ (JFY013) strains was performed as described in Fig. 4A. D. Rtt109 is required for recovery from the DNA damage checkpoint. Colony formation analysis of WT (YMV045), asf1Δ (JKT200) and rtt109Δ (JFY013) strains as described in Fig. 4B. E. Rtt109 is required for chromatin reassembly after DNA repair. Analysis of cutting / repair and chromatin assembly and disassembly was performed on strain rtt109Δ (JFY013) as described in Fig. 2. F. A mimic of permanent H3 K56 acetylation bypasses the requirement for Asf1 for chromatin reassembly after DNA repair. Analysis of cutting / repair and chromatin assembly and disassembly was performed on strain asf1ΔK56Q as described in Fig. 2.
Given that mimicking acetylation of K56 restores double-strand damage resistance to asf1Δ cells (Fig. 6A), it is likely that K56 acetylation per se is required for turning off the DNA damage checkpoint after DNA repair. As such, we predicted that the HAT Rtt109 that mediates acetylation of K56 (Driscoll et al., 2007; Han et al., 2007) would be required for recovery from the DNA damage checkpoint. It is known that yeast lacking Rtt109 are sensitive to double-strand DNA damaging agents (Driscoll et al., 2007) (Fig. 6A), but it has never been examined whether rtt109Δ cells are capable of repairing DNA damage. Accordingly, we found that yeast lacking Rtt109 are fully competent for double-strand DNA repair (Fig. 6B,E). Despite the repair of the HO lesion in the absence of Rtt109, it is apparent that the DNA damage checkpoint protein Rad53 was persistently activated for up to 12 hours after induction of the HO endonuclease (Fig. 6B). Consistent with the persistent activation of the DNA damage checkpoint after repair in the rtt109 mutants, loss of Rtt109 lead to a reduction of cell viability following repair of the single HO site equivalent to that due to loss of Asf1 (Fig. 6C). Analysis of the ability of single cells to form colonies demonstrated that the rtt109 mutant had a checkpoint recovery defect that is equivalent to the recovery defect in the asf1 mutant (Fig. 6D). To further investigate the relationship between chromatin assembly and checkpoint recovery, we examined whether the rtt109 mutant was capable of reassembling chromatin following DNA repair. This analysis revealed that chromatin reassembly does not occur after DNA repair in the absence of Rtt109 (Fig. 6E) nor in a H3 K56R mutant that mimics unacetylated K56 (Suppl. Fig. 10). Finally, we tested whether acetylation of H3 K56 was sufficient for chromatin reassembly after DNA repair even in the absence of the histone chaperone Asf1. We found that a yeast strain carrying both the K56Q mutant that mimics K56 acetylation and deletion of ASF1 was fully competent for chromatin reassembly following DNA repair (Fig. 6F). Taken together, these data indicate that acetylation of H3 K56 by Asf1 and Rtt109 per se is driving chromatin reassembly after DNA repair, leading to inactivation of the DNA damage checkpoint and cell survival (Fig. 7).
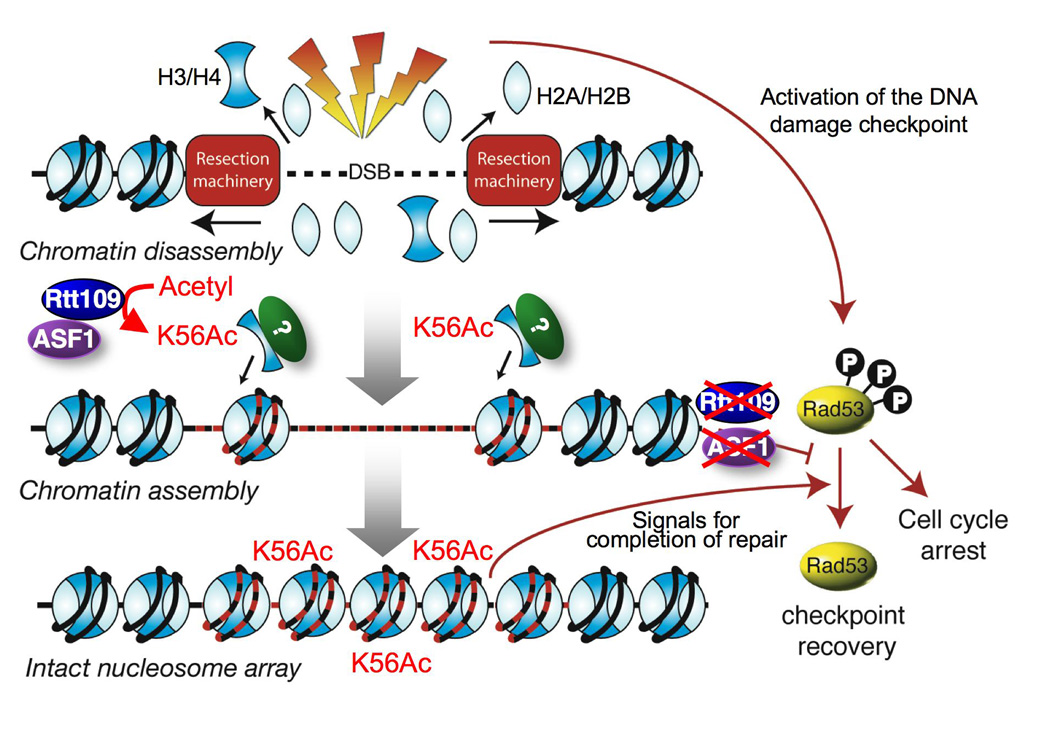
Figure 7. Model for the role of chromatin in deactivation of the DNA damage checkpoint.
Discussion
It is well appreciated that alterations to the chromatin structure in the vicinity of double-strand DNA breaks provide access for the machinery that mediates DNA repair and activation of the DNA damage checkpoint. However, many questions remain as to the events that occur after the DNA is repaired. Is the chromatin structure reestablished following double-strand DNA repair? If so, how is the chromatin reassembled following DSB repair? How does the cell know when DNA repair is complete? How is the DNA damage checkpoint turned off after repair, or in the absence of DNA repair? In this study we show that chromatin disassembly at the DNA lesion occurs concomitant with DNA resection and that chromatin reassembly is tightly coupled to DNA repair. The Asf1 and Rtt109-dependent acetylation of H3 K56 drives chromatin reassembly following DSB repair and its absence leads to persistent checkpoint activation after repair, leading to cell death. Our results indicate that reinstating the chromatin structure carrying H3 K56 over the site of the DNA lesion signals to the DNA damage checkpoint that the repair process is complete.
DNA resection drives chromatin disassembly at the DNA lesion
In every mutation that we have examined, the kinetics of chromatin disassembly in the vicinity of the DNA break closely mimic the kinetics of DNA resection. Furthermore, we have not been able to separate the two processes, i.e. to achieve DNA resection in the absence of chromatin disassembly using any of the mutants of the histone chaperones that we have examined (Fig. 1G; data not shown). Mutants that slow down DNA resection also result in slower chromatin disassembly (Fig. 1F). Indeed the only mutation that has been reported to kinetically separate the processes of DNA resection and chromatin disassembly at a DSB is inactivation of the Arp8 subunit of the INO80 ATP-dependent chromatin remodeler (Tsukuda et al., 2005). This study showed slower chromatin disassembly in the arp8 mutant as compared to wild type yeast in the vicinity of a DSB (Tsukuda et al., 2005). However, this study did not show the input DNA to the histone ChIPs to enable comparison of the rate of chromatin disassembly to the DNA resection rate, instead, reporting that there was no resection defect using a different assay. The resection defect of the arp8 mutant is apparent in other assays (van Attikum et al., 2007; van Attikum et al., 2004). As such, it is likely that the slightly delayed chromatin disassembly that is seen in response to a DSB in an arp8 mutant (Tsukuda et al., 2005) is merely a reflection of the slightly delayed DNA resection in this mutant. Indeed, recent evidence demonstrates that the INO80 complex promotes recruitment of Mre11 which itself contributes to DNA resection (van Attikum et al., 2007). All the evidence taken together indicates that although chromatin remodeling is required for recruitment of the resection machinery, DNA resection itself is sufficient to disassemble the chromatin from the vicinity of a DSB (Fig. 7).
Chromatin reassembly following repair is driven by K56 acetylation on histone H3
We have discovered that chromatin is reassembled following DSB repair. Normally, the kinetics of chromatin reassembly closely mimic the kinetics of DSB repair, but the two processes can be uncoupled by deletion of ASF1 or RTT109, where the DNA is repaired but the chromatin is not reassembled (Fig. 2F, 6E). The fact that the K56Q mimic of acetylation can bypass the requirement for Asf1 for chromatin reassembly after repair (Fig. 6F) implies that Asf1 is not the histone chaperone that is physically depositing the histones onto the DNA after DSB repair, but instead is promoting chromatin assembly indirectly by helping Rtt109 acetylate H3 K56. We do not know how acetylation of free histones on H3 K56 is required for chromatin assembly after repair, but it is possible that this acetylation mark promotes their recruitment to the repaired DNA, perhaps by increasing their binding affinity to a repair-specific histone chaperone.
Why does the DNA damage checkpoint remain active after DNA repair in cells that fail to assemble chromatin carrying the H3K56 mark?
Activation of the checkpoint kinase Rad53 is a critical response to DNA damage that leads to delayed entry to mitosis. Activation of Rad53 is well understood and involves phosphorylation by the protein kinase Mec1 following its recruitment to the DNA lesion. However, deactivation of Rad53, which must occur to allow the cell to recover and adapt from checkpoint arrest, is not well understood. The phosphatases Pph3, Psy2, (O'Neill et al., 2007) Ptc2 and Ptc3 (Leroy et al., 2003) play a role in dephosphorylation of Rad53, but what signals for dephosphorylation of Rad53 by these phosphatases is unclear. Noteworthy, the continued phosphorylation of Rad53 in the absence of Asf1 is not due to an indirect transcriptional role for Asf1 in regulating expression of these phosphatases, as the levels of the Pph3, Psy2, Ptc2 and Ptc3 transcripts are not significantly altered upon deletion of ASF1 (Zabaronick and Tyler, 2005).
Strikingly, the repair of the DNA lesion itself is not sufficient to signal for turning off the DNA damage checkpoint, as yeast lacking Asf1 and Rtt109 repair their DSBs but maintain persistent activation of the DNA damage checkpoint. Because our DNA repair assay utilized PCR not Southern blotting, we can also rule out the possibility that the continued activation of the DNA damage checkpoint following DNA repair in the asf1 and rtt109 mutants is due to a gap or nick remaining at the site of the DNA lesion, because this would prevent production of the repair PCR product.
Asf1 may play a direct role in inactivation of the DNA damage checkpoint via its dynamic physical interaction with Rad53 (Emili et al., 2001; Hu et al., 2001). All inactive Rad53 in the cell is bound to Asf1 (Hu et al., 2001), mediated via interaction with the Rad53 FHA1 domain (Schwartz et al., 2003). Upon activation of the DNA damage checkpoint, Rad53 becomes phosphorylated and Asf1 is released (Emili et al., 2001; Hu et al., 2001). It is tempting to speculate that Asf1 turns off the DNA damage checkpoint by binding to the transiently dephosphorylated FHA1 domain of Rad53 to block re-phosphorylation by Mec1. In such a model, there is nothing to physically block the continuous re-phosphorylation of the FHA1 domain of Rad53 by Mec1 in the absence of Asf1, leading to an inability to inactivate Rad53 during checkpoint recovery and adaptation. However, this model would predict that the initial activation of Rad53 by Mec1 would also be physically blocked by Asf1, which is clearly not the case. Furthermore, this model cannot explain why acetylation of histone H3 on K56 is sufficient to turn off the DNA damage checkpoint even in the absence of Asf1 (Fig. 6F).
Asf1 is required for turning off the DNA damage checkpoint during both adaptation and recovery. Although, many proteins involved in adaptation are not required for recovery, there are examples of proteins that are needed for both processes. The phosphatases Ptc2 and Ptc3 that dephosphorylate Rad53 are required for both adaptation and recovery (Leroy et al., 2003). One further example is the DNA helicase Srs2 (Vaze et al., 2002). It has been proposed that Srs2 may physically remove DNA repair or checkpoint proteins from the DNA template after DNA repair, in order to achieve inactivation of the DNA damage checkpoint (Vaze et al., 2002). Accordingly, Srs2 can displace Rad51 from DNA templates in vitro (Krejci et al., 2004; Krejci et al., 2003). It is possible that in a similar manner to Srs2, chromatin reassembly is required in order to displace or compete with the repair or checkpoint machinery for occupancy of the DNA at the site of the repaired lesion. However, chromatin reassembly after repair does not play a role in displacement of Rad51, because we observed no recruitment of Rad51 to the vicinity of the HO lesion during SSA (data not shown), consistent with the lack of genetic requirement for RAD51 during SSA (Vaze et al., 2002). We also see no requirement for chromatin reassembly in order to remove phosphorylated H2A or Mec1-Ddc2 from the vicinity of the DNA lesion (Fig. 5A,B), although it is still possible that chromatin reassembly may be required for displacement of the Ddc1-Rad17-Mec3 checkpoint complex. Following removal of phosphorylated H2A from sites of DNA damage, its dephosphorylation is required for recovery from the DNA damage checkpoint (Keogh et al., 2006). It was not possible to confirm whether the disassembled histone H2A was dephosphorylated following SSA repair in the asf1 mutant due to the high background level of H2A phosphorylation that is present in asf1 mutants even in the absence of DNA damaging agents (data not shown) (Prado et al., 2004).
Our favored model is that the altered chromatin structure itself is sensed by the DNA damage checkpoint (Fig. 7). Precedent for this idea comes from a report from the Kastan lab in which perturbations that disrupt the chromatin structure but do not damage DNA resulted in activation of the DNA damage checkpoint in human cells (Bakkenist and Kastan, 2003). Specifically, we propose that it is the reassembly of new histones carrying the H3K56 acetylation mark onto the repaired DNA that signals to the DNA damage checkpoint that DNA repair is complete (Fig. 7). Seemingly all newly-synthesized histone H3 is acetylated on lysine K56 (Masumoto et al., 2005), but this K56 acetyl mark is usually rapidly removed after replication-dependent histone deposition by the Hst3/Hst4 deacetylases (Celic et al., 2006; Maas et al., 2006). However, following incorporation of the K56 acetylated histone H3 onto newly-repaired DNA the K56 acetyl mark will persist as a consequence of the transcriptional repression and degradation of the histone deacetylase Hst3 by the activated DNA damage checkpoint (Maas et al., 2006; Thaminy et al., 2007). Consistent with this model, histone H3 with acetylated K56 is enriched on chromatin fractions undergoing DNA repair (Masumoto et al., 2005). We propose that the patch of chromatin bearing the K56Ac mark locally signals for the nearby checkpoint machinery to be deactivated (Fig. 7). Indeed, deletion of HST3/HST4 (and hence global over-acetylation of H3 K56 on chromatin), causes a defective DNA damage checkpoint response (Thaminy et al., 2007) which is consistent with the idea that acetylated K56 signals for the DNA damage checkpoint to be deactivated. In our model, after the K56 acetyl mark on the chromatin has signaled for deactivation of the DNA damage checkpoint, this would allow the subsequent restabilization of Hst3 in order to deacetylate K56 to enable future activation of the DNA damage checkpoint as necessary.
A role for histone acetylation in turning off the DNA damage checkpoint may not be specific to K56 acetylation. We previously observed that acetylation of the N-termini of histones H3 and H4 in the vicinity of a DNA lesion only occurs after the strand invasion step of homologous recombination is complete, and the absence of these marks leads to cell death even though DNA repair per se was complete (Tamburini and Tyler, 2005). Taken together, our studies indicate that reinstating chromatin with acetylation mark(s) over the repaired DNA is essential for deactivating the DNA damage checkpoint. Future studies should reveal how acetylation marks on chromatin communicate with the DNA damage checkpoint in order to allow cell cycle reentry after repair.
Methods
Chromatin Immunoprecipitation analyses
ChIP analyses were performed as previously described (Tamburini and Tyler, 2005). Plotted are the average and standard error of the mean of three independent cultures for each experiment.
DNA damage and repair quantitation of the HO site at MAT
Cutting, repair and mating type switching of the HO lesion at MAT was measured by PCR amplification of genomic DNA templates taken from the time courses described above, using primers flanking the HO site in the MAT locus, as described previously (Ramey et al., 2004).
Analysis of SSA repair and Rad53 activation
Cutting and repair of the HO site in the single strand annealing strains was performed using the three primers indicated in Fig. 3B. Protein samples were prepared by TCA precipitation as previously described (Keogh et al., 2006). Samples were resolved on 8% SDS PAGE gels and Rad53 was detected by western blotting using an antibody against Rad53 at 1:200 dilution (Santa Cruz, sc-6749).
Microscopic recovery and adaptation analyses
Individual unbudded G1 cells were generated by starvation and sonication (as described by (Vaze et al., 2002)). These cells were then spread onto plates containing YERP+2% galactose to induce the HO lesion, and the number of cells/buds of the same region was assayed using a dissection microscope at the times indicated in the figures.
Supplementary Material
Acknowledgements
Thanks to Seth Noone and Candice Wike for careful reading of the manuscript. We are very grateful for the generous gift of yeast strains from Jim Haber, David Allis and Mary-Ann Osley. We gratefully acknowledge William Bonner, John Diffley and Patrick Sung for antisera. We thank Steve Kron for helpful discussions. We gratefully acknowledge the University of Colorado Cancer Center Flow Cytometry Core facility and the Real Time PCR Core facility, especially Umarani Pugazhenthi and Bryan Haugen. This work was supported by a grant from the NCI to Jessica Tyler. Jessica Tyler is a Leukemia and Lymphoma Society Scholar.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adkins MW, Carson JJ, English CM, Ramey CJ, Tyler JK. The histone chaperone anti-silencing function 1 stimulates the acetylation of newly synthesized histone H3 in S-phase. J Biol Chem. 2007;282:1334–1340. doi: 10.1074/jbc.M608025200. [DOI] [PubMed] [Google Scholar]
- Adkins MW, Howar SR, Tyler JK. Chromatin disassembly mediated by the histone chaperone Asf1 is essential for transcriptional activation of the yeast PHO5 and PHO8 genes. Mol Cell. 2004;14:657–666. doi: 10.1016/j.molcel.2004.05.016. [DOI] [PubMed] [Google Scholar]
- Aguilera A, Gomez-Gonzalez B. Genome instability: a mechanistic view of its causes and consequences. Nat Rev Genet. 2008;9:204–217. doi: 10.1038/nrg2268. [DOI] [PubMed] [Google Scholar]
- Altaf M, Saksouk N, Cote J. Histone modifications in response to DNA damage. Mutat Res. 2007 doi: 10.1016/j.mrfmmm.2006.09.009. [DOI] [PubMed] [Google Scholar]
- Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421:499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- Bartek J, Lukas J. DNA damage checkpoints: from initiation to recovery or adaptation. Curr Opin Cell Biol. 2007 doi: 10.1016/j.ceb.2007.02.009. [DOI] [PubMed] [Google Scholar]
- Celic I, Masumoto H, Griffith WP, Meluh P, Cotter RJ, Boeke JD, Verreault A. The sirtuins hst3 and Hst4p preserve genome integrity by controlling histone h3 lysine 56 deacetylation. Curr Biol. 2006;16:1280–1289. doi: 10.1016/j.cub.2006.06.023. [DOI] [PubMed] [Google Scholar]
- Downs JA, Lowndes NF, Jackson SP. A role for Saccharomyces cerevisiae histone H2A in DNA repair. Nature. 2000;408:1001–1004. doi: 10.1038/35050000. [DOI] [PubMed] [Google Scholar]
- Driscoll R, Hudson A, Jackson SP. Yeast Rtt109 promotes genome stability by acetylating histone H3 on lysine 56. Science. 2007;315:649–652. doi: 10.1126/science.1135862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubrana K, van Attikum H, Hediger F, Gasser SM. The processing of double-strand breaks and binding of single-strand-binding proteins RPA and Rad51 modulate the formation of ATR-kinase foci in yeast. J Cell Sci. 2007 doi: 10.1242/jcs.018366. [DOI] [PubMed] [Google Scholar]
- Dudasova Z, Dudas A, Chovanec M. Non-homologous end-joining factors of Saccharomyces cerevisiae. FEMS Microbiol Rev. 2004;28:581–601. doi: 10.1016/j.femsre.2004.06.001. [DOI] [PubMed] [Google Scholar]
- Emili A, Schieltz DM, Yates JR, 3rd, Hartwell LH. Dynamic interaction of DNA damage checkpoint protein Rad53 with chromatin assembly factor Asf1. Mol Cell. 2001;7:13–20. doi: 10.1016/s1097-2765(01)00150-2. [DOI] [PubMed] [Google Scholar]
- Gilbert CS, Green CM, Lowndes NF. Budding yeast Rad9 is an ATP-dependent Rad53 activating machine. Mol Cell. 2001;8:129–136. doi: 10.1016/s1097-2765(01)00267-2. [DOI] [PubMed] [Google Scholar]
- Groth A, Rocha W, Verreault A, Almouzni G. Chromatin challenges during DNA replication and repair. Cell. 2007;128:721–733. doi: 10.1016/j.cell.2007.01.030. [DOI] [PubMed] [Google Scholar]
- Han J, Zhou H, Horazdovsky B, Zhang K, Xu RM, Zhang Z. Rtt109 acetylates histone H3 lysine 56 and functions in DNA replication. Science. 2007;315:653–655. doi: 10.1126/science.1133234. [DOI] [PubMed] [Google Scholar]
- Hu F, Alcasabas AA, Elledge SJ. Asf1 links Rad53 to control of chromatin assembly. Genes Dev. 2001;15:1061–1066. doi: 10.1101/gad.873201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keogh MC, Mennella TA, Sawa C, Berthelet S, Krogan NJ, Wolek A, Podolny V, Carpenter LR, Greenblatt JF, Baetz K, Buratowski S. The Saccharomyces cerevisiae histone H2A variant Htz1 is acetylated by NuA4. Genes Dev. 2006;20:660–665. doi: 10.1101/gad.1388106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krejci L, Macris M, Li Y, Van Komen S, Villemain J, Ellenberger T, Klein H, Sung P. Role of ATP hydrolysis in the antirecombinase function of Saccharomyces cerevisiae Srs2 protein. J Biol Chem. 2004;279:23193–23199. doi: 10.1074/jbc.M402586200. [DOI] [PubMed] [Google Scholar]
- Krejci L, Van Komen S, Li Y, Villemain J, Reddy MS, Klein H, Ellenberger T, Sung P. DNA helicase Srs2 disrupts the Rad51 presynaptic filament. Nature. 2003;423:305–309. doi: 10.1038/nature01577. [DOI] [PubMed] [Google Scholar]
- Le S, Davis C, Konopka JB, Sternglanz R. Two new S-phase-specific genes from Saccharomyces cerevisiae. Yeast. 1997;13:1029–1042. doi: 10.1002/(SICI)1097-0061(19970915)13:11<1029::AID-YEA160>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- Lee SE, Moore JK, Holmes A, Umezu K, Kolodner RD, Haber JE. Saccharomyces Ku70, mre11/rad50 and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell. 1998;94:399–409. doi: 10.1016/s0092-8674(00)81482-8. [DOI] [PubMed] [Google Scholar]
- Leroy C, Lee SE, Vaze MB, Ochsenbien F, Guerois R, Haber JE, Marsolier-Kergoat MC. PP2C phosphatases Ptc2 and Ptc3 are required for DNA checkpoint inactivation after a double-strand break. Mol Cell. 2003;11:827–835. doi: 10.1016/s1097-2765(03)00058-3. [DOI] [PubMed] [Google Scholar]
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- Lupardus PJ, Cimprich KA. Checkpoint adaptation; molecular mechanisms uncovered. Cell. 2004;117:555–556. doi: 10.1016/j.cell.2004.05.005. [DOI] [PubMed] [Google Scholar]
- Maas NL, Miller KM, DeFazio LG, Toczyski DP. Cell cycle and checkpoint regulation of histone H3 K56 acetylation by Hst3 and Hst4. Mol Cell. 2006;23:109–119. doi: 10.1016/j.molcel.2006.06.006. [DOI] [PubMed] [Google Scholar]
- Masumoto H, Hawke D, Kobayashi R, Verreault A. A role for cell-cycle-regulated histone H3 lysine 56 acetylation in the DNA damage response. Nature. 2005;436:294–298. doi: 10.1038/nature03714. [DOI] [PubMed] [Google Scholar]
- Mello JA, Sillje HH, Roche DM, Kirschner DB, Nigg EA, Almouzni G. Human Asf1 and CAF-1 interact and synergize in a repair-coupled nucleosome assembly pathway. EMBO Rep. 2002;3:329–334. doi: 10.1093/embo-reports/kvf068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo J, Toczyski D. A unified view of the DNA-damage checkpoint. Curr Opin Cell Biol. 2002;14:237–245. doi: 10.1016/s0955-0674(02)00312-5. [DOI] [PubMed] [Google Scholar]
- Myung K, Pennaneach V, Kats ES, Kolodner RD. Saccharomyces cerevisiae chromatin-assembly factors that act during DNA replication function in the maintenance of genome stability. Proc Natl Acad Sci U S A. 2003;100:6640–6645. doi: 10.1073/pnas.1232239100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill BM, Szyjka SJ, Lis ET, Bailey AO, Yates JR, 3rd, Aparicio OM, Romesberg FE. Pph3-Psy2 is a phosphatase complex required for Rad53 dephosphorylation and replication fork restart during recovery from DNA damage. Proc Natl Acad Sci U S A. 2007;104:9290–9295. doi: 10.1073/pnas.0703252104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osley MA, Tsukuda T, Nickoloff JA. ATP-dependent chromatin remodeling factors and DNA damage repair. Mutat Res. 2007 doi: 10.1016/j.mrfmmm.2006.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasero P, Shimada K, Duncker BP. Multiple roles of replication forks in S phase checkpoints: sensors, effectors and targets. Cell Cycle. 2003;2:568–572. [PubMed] [Google Scholar]
- Pellicioli A, Lee SE, Lucca C, Foiani M, Haber JE. Regulation of Saccharomyces Rad53 checkpoint kinase during adaptation from DNA damage-induced G2/M arrest. Mol Cell. 2001;7:293–300. doi: 10.1016/s1097-2765(01)00177-0. [DOI] [PubMed] [Google Scholar]
- Prado F, Cortes-Ledesma F, Aguilera A. The absence of the yeast chromatin assembly factor Asf1 increases genomic instability and sister chromatid exchange. EMBO Rep. 2004;5:497–502. doi: 10.1038/sj.embor.7400128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prado F, Cortes-Ledesma F, Huertas P, Aguilera A. Mitotic recombination in Saccharomyces cerevisiae. Curr Genet. 2003;42:185–198. doi: 10.1007/s00294-002-0346-3. [DOI] [PubMed] [Google Scholar]
- Qin J, Li L. Molecular anatomy of the DNA damage and replication checkpoints. Radiat Res. 2003;159:139–148. doi: 10.1667/0033-7587(2003)159[0139:maotdd]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Ramey CJ, Howar S, Adkins M, Linger J, Spicer J, Tyler JK. Activation of the DNA damage checkpoint in yeast lacking the histone chaperone anti-silencing function 1. Mol Cell Biol. 2004;24:10313–10327. doi: 10.1128/MCB.24.23.10313-10327.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Recht J, Tsubota T, Tanny JC, Diaz RL, Berger JM, Zhang X, Garcia BA, Shabanowitz J, Burlingame AL, Hunt DF, et al. Histone chaperone Asf1 is required for histone H3 lysine 56 acetylation, a modification associated with S phase in mitosis and meiosis. Proc Natl Acad Sci U S A. 2006;103:6988–6993. doi: 10.1073/pnas.0601676103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- Schwabish MA, Struhl K. Asf1 mediates histone eviction and deposition during elongation by RNA polymerase II. Mol Cell. 2006;22:415–422. doi: 10.1016/j.molcel.2006.03.014. [DOI] [PubMed] [Google Scholar]
- Schwartz MF, Lee SJ, Duong JK, Eminaga S, Stern DF. FHA domain-mediated DNA checkpoint regulation of Rad53. Cell Cycle. 2003;2:384–396. [PubMed] [Google Scholar]
- Shim EY, Hong SJ, Oum JH, Yanez Y, Zhang Y, Lee SE. RSC mobilizes nucleosomes to improve accessibility of repair machinery to the damaged chromatin. Mol Cell Biol. 2007;27:1602–1613. doi: 10.1128/MCB.01956-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shroff R, Arbel-Eden A, Pilch D, Ira G, Bonner WM, Petrini JH, Haber JE, Lichten M. Distribution and dynamics of chromatin modification induced by a defined DNA double-strand break. Curr Biol. 2004;14:1703–1711. doi: 10.1016/j.cub.2004.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamburini BA, Tyler JK. Localized histone acetylation and deacetylation triggered by the homologous recombination pathway of double-strand DNA repair. Mol Cell Biol. 2005;25:4903–4913. doi: 10.1128/MCB.25.12.4903-4913.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaminy S, Newcomb B, Kim J, Gatbonton T, Foss E, Simon J, Bedalov A. HST3 is regulated by MEC1-dependent proteolysis and controls the S phase checkpoint and sister chromatid cohesion by deacetylating histone H3 at lysine 56. J Biol Chem. 2007 doi: 10.1074/jbc.M706384200. [DOI] [PubMed] [Google Scholar]
- Tsubota T, Berndsen CE, Erkmann JA, Smith CL, Yang L, Freitas MA, Denu JM, Kaufman PD. Histone H3-K56 acetylation is catalyzed by histone chaperone-dependent complexes. Mol Cell. 2007;25:703–712. doi: 10.1016/j.molcel.2007.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsubouchi H, Ogawa H. A novel mre11 mutation impairs processing of double-strand breaks of DNA during both mitosis and meiosis. Mol Cell Biol. 1998;18:260–268. doi: 10.1128/mcb.18.1.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukuda T, Fleming AB, Nickoloff JA, Osley MA. Chromatin remodeling at a DNA double-strand break site in Saccharomyces cerevisiae. Nature. 2005;438:379–383. doi: 10.1038/nature04148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler JK, Adams CR, Chen SR, Kobayashi R, Kamakaka RT, Kadonaga JT. The RCAF complex mediates chromatin assembly during DNA replication and repair. Nature. 1999;402:555–560. doi: 10.1038/990147. [DOI] [PubMed] [Google Scholar]
- van Attikum H, Fritsch O, Gasser SM. Distinct roles for SWR1 and INO80 chromatin remodeling complexes at chromosomal double-strand breaks. Embo J. 2007;26:4113–4125. doi: 10.1038/sj.emboj.7601835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Attikum H, Fritsch O, Hohn B, Gasser SM. Recruitment of the INO80 complex by H2A phosphorylation links ATP-dependent chromatin remodeling with DNA double-strand break repair. Cell. 2004;119:777–788. doi: 10.1016/j.cell.2004.11.033. [DOI] [PubMed] [Google Scholar]
- van Vugt MA, Medema RH. Checkpoint adaptation and recovery: back with Polo after the break. Cell Cycle. 2004;3:1383–1386. doi: 10.4161/cc.3.11.1248. [DOI] [PubMed] [Google Scholar]
- van Vugt MA, Medema RH. Getting in and out of mitosis with Polo-like kinase-1. Oncogene. 2005;24:2844–2859. doi: 10.1038/sj.onc.1208617. [DOI] [PubMed] [Google Scholar]
- Vaze MB, Pellicioli A, Lee SE, Ira G, Liberi G, Arbel-Eden A, Foiani M, Haber JE. Recovery from checkpoint-mediated arrest after repair of a double-strand break requires Srs2 helicase. Mol Cell. 2002;10:373–385. doi: 10.1016/s1097-2765(02)00593-2. [DOI] [PubMed] [Google Scholar]
- Williams RS, Williams JS, Tainer JA. Mre11-Rad50-Nbs1 is a keystone complex connecting DNA repair machinery, double-strand break signaling, and the chromatin template. Biochem Cell Biol. 2007;85:509–520. doi: 10.1139/O07-069. [DOI] [PubMed] [Google Scholar]
- Zabaronick SR, Tyler JK. The histone chaperone anti-silencing function 1 is a global regulator of transcription independent of passage through S phase. Mol Cell Biol. 2005;25:652–660. doi: 10.1128/MCB.25.2.652-660.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.