Abstract
PTEN is a tumor suppressor gene associated with multiple tumor types. PTEN function is essential for early embryonic development and is involved in the regulation of cell size, number, and survival. By dephosphorylating PIP3, PTEN normally acts to inhibit the PI3-Kinase/AKT pathway. Here we have identified two zebrafish orthologs, ptena and ptenb, of the single mammalian PTEN gene and analyzed the role of these genes in zebrafish development. ptena transcripts were expressed throughout the embryo at early somitogenesis. By 24 hpf, expression was predominant in the central nervous system, axial vasculature, retina, branchial arches, ear, lateral line primordium, and pectoral fin bud. ptenb was also ubiquitously expressed early in somitogenesis, but transcripts became more restricted to the somites and central nervous system as development progressed. By 48 hpf, ptena and ptenb were expressed predominantly in the central nervous system, branchial arches, pectoral fins, and eye. Antisense morpholinos were used to knock down translation of ptena and ptenb mRNA in zebrafish embryos. Knockdown of either pten gene caused increased levels of phosphorylated Akt in morphant embryos, indicating that Ptena and Ptenb each possess PIP3 lipid phosphatase activity. ptena morphants had irregularities in notochord shape (73%), vasculogenesis (83%), head shape (72%), and inner ear development (59%). The most noticeable defects in ptenb morphants were upward hooked tails (73%), domed heads (83%), and reduced yolk extensions (90%). These results indicate that ptena and ptenb encode functional enzymes and that each pten gene plays a distinct role during zebrafish embryogenesis.
Keywords: Zebrafish, PTEN, mRNA expression, development, morpholino knockdown
INTRODUCTION
PTEN (phosphatase and tensin homolog deleted on chromosome 10) is a tumor suppressor gene that is mutated in a number of human cancers including glioblastoma, advanced prostate cancer, breast cancer, endometrial carcinoma, and melanoma (Li and Sun, 1997; Li et al., 1997; Steck et al., 1997; Podsypanina et al., 1999; Birck et al., 2000; Zhou et al., 2002). Mutations in the PTEN gene are also responsible for the tumor predisposition syndromes Cowden disease (Liaw et al., 1997), Bannayan-Zonana (Marsh et al., 1997), and Lhermitte-Duclose disease (Zhou et al., 2003). PTEN has been shown to have protein and lipid phosphatase activity in vitro, although only the lipid phosphatase activity has been demonstrated in vivo (Maehama and Dixon, 1998). PTEN acts as a negative regulator of the PI3-Kinase/AKT pathway via its ability to dephosphorylate phosphatidylinositol-3,4,5-triphosphate (PIP3). The PI3-Kinase/AKT pathway is involved in regulating cell growth, motility, and survival (Thompson and Thompson, 2004). Mutations or deletions of PTEN lead to overactivation of the PI3-Kinase/AKT pathway which contributes to tumorigenesis (Wu et al., 1998; Stahl et al., 2003).
PTEN also plays an essential role in early animal development, although the precise function of PTEN in embryogenesis remains enigmatic. Homozygous knockouts of Pten in mice have been found to cause embryonic lethality, although the mutant phenotypes differed between various studies (Di Cristofano et al., 1998; Suzuki et al., 1998; Podsypanina et al., 1999). Since Pten is normally expressed ubiquitously during early murine development, embryonic lethality in Pten null mice has been interpreted to result from abnormal development of multiple organs and tissues (Stiles et al., 2004). Tissue-specific knockout of Pten in mice suggests a possible role for PTEN in the regulation of cell size and growth (Kishimoto et al., 2003). PTEN has also been shown to be essential for Drosophila development as dPTEN null mutants die at early larval stages (Goberdhan et al., 1999; Huang et al., 1999; Gao et al., 2000). PTEN appears to regulate the size of cells and organs as well as cell survival in Drosophila development. These properties of PTEN have been attributed to PTEN regulation of the PI3-Kinase/AKT pathway (Goberdhan et al., 1999).
To analyze the role of PTEN in vertebrate development, we took advantage of the powerful reverse genetic tools available in zebrafish. Antisense morpholino (MO) gene knockdown provides a means to produce hypomorphs for any gene in the zebrafish genome (Nasevicius and Ekker, 2000). Graded knockdowns with MOs in zebrafish make it possible to study the developmental function of essential genes ex utero, which is extremely difficult in mice. For example, homozygous looptail (Ltap) mutants (Kibar et al., 2001) or knockouts of VEGF (Ferrara et al., 1996) in mice result in embryonic lethality. Alternatively, MO knockdown of the Ltap ortholog strabismus/Van Gogh (Park and Moon, 2002) or VEGF (Nasevicius et al., 2000) in zebrafish provided further insight into the developmental function of these genes.
We have identified two paralogous PTEN genes in zebrafish, ptena and ptenb. These genes exhibit distinct but overlapping expression patterns early in embryogenesis. By 48 hpf, both genes show similar expression in the CNS, branchial arches, pectoral fin, and eye. ptena and ptenb appear to function as PIP3 lipid phosphatases based on their ability to decrease phosphorylation of Akt. Knockdown of ptena caused irregularities in notochord and head shape, vasculogenesis, and ear development, while knockdown of ptenb caused hooked tails, domed heads, and reduced yolk extensions. Since morpholino knockdown of ptena and ptenb mRNA translation in zebrafish produced unique phenotypes, it is likely that these two highly related genes play distinct roles in the developing zebrafish embryo.
RESULTS
Identification and Characterization of Zebrafish pten Genes
Zebrafish pten genes were identified by BLAST searches of the zebrafish EST database. These searches revealed two distinct classes of cDNAs, one class encoding ptena and the other ptenb. Primers were designed to amplify full-length pten ORFs using RT-PCR. Sequencing of the initial amplification products revealed the presence of two splice variants of the ptena and ptenb genes. The short splice variants were found to contain the same intron/exon organization as the mammalian PTEN gene (Fig. 1). The long splice variant of both ptena and ptenb contained a 69 base pair (bp) insertion corresponding to the boundary of the 6th and 7th exons of the mammalian PTEN gene (GenBank accession no. AH005966). The inserted 69 bp exon was found in zebrafish genomic DNA, and in the RT-PCR products amplified from embryonic and adult zebrafish mRNA. The additional exon found in the zebrafish ptena and ptenb genes is also present in the pten genes of Fugu, Tetraodon, and medaka (not shown), but not in chicken, human, or other tetrapod PTEN genes.
Figure 1.
Comparison of human PTEN and zebrafish Pten polypeptides. Human (hPTEN) and zebrafish (zfPtena and zfPtenb) polypeptides were aligned using CLUSTALW. Identical amino acids are highlighted in black, and conserved amino acids are highlighted in gray. Amino acids are numbered to the right of each line. Arrowheads indicate the location of introns and are flanked by the corresponding exon numbers. The asterisk indicates the extra exon found in the deduced zebrafish amino acid sequence. The region encompassing the C2 domain is underlined.
The complete ORFs for the ptena long and short splice variants encode polypeptides of 454 and 431 amino acids. The complete ORFs for the ptenb splice variants encode polypeptides 422 and 399 amino acids in length. An amino acid sequence alignment of the zebrafish Ptena, Ptenb, and human PTEN polypeptides is shown in Fig. 1. Zebrafish Ptena and Ptenb polypeptides exhibit 88% and 86% identity, respectively, with human PTEN. The phosphatase motif (amino acids 123–130 of human PTEN) shows complete identity between the human and zebrafish PTEN polypeptides. The region of greatest divergence, including the alternatively spliced region of Ptena and Ptenb and a 32 amino acid insertion found in Ptena, occurs within the C2 domain, a region proposed to have a potential role in membrane localization (Lee et al., 1999).
BLAST searches of the zebrafish genome assembly (Version 4) available from the Zebrafish Sequencing Group indicate that the zebrafish ptena gene is located on linkage group (LG) 17 (Zv4_scaffold1476.4), and the ptenb gene maps to LG 12 (GenBank accession nos. AL731788.8 and BX548001.4). These linkage groups have significant synteny with human chromosome 10, the location of the human PTEN gene (Woods et al., 2000). These results suggest that the zebrafish ptena and ptenb genes most likely arose as a result of the genome-wide duplication that occurred in teleost fish (Amores et al., 1998; Postlethwait et al., 1998).
Expression of ptena and ptenb Genes in Zebrafish Embryos
We used an RT-PCR approach to analyze stage-specific expression of the zebrafish pten splice variants. Primers specific for ptena were found to amplify both the long and short ptena splice variants from all stages of embryonic development tested (6 hpf-72 hpf) as well as the adult (Fig. 2A). Similarly, long and short splice variants of ptenb were also detected at equivalent developmental stages (Fig. 3A). It should be noted that the RT-PCR approach used was not quantitative, and therefore the relative abundance of reaction products may not accurately reflect ptena and ptenb mRNA levels in vivo.
Figure 2.
Expression of zebrafish ptena mRNA during embryogenesis. Expression of ptena was analyzed by RT-PCR and whole-mount in situ hybridization. (A) Expression of ptena splice variants during zebrafish development determined by RT-PCR. Bands of 305 bp and 236 bp represent fragments of the long and short splice variants, respectively. (B–K) Expression of ptena determined by whole-mount in situ hybridization. (B) Early somitogenesis, lateral view. (C) Mid-somitogenesis, lateral view. (D) 24 hpf, lateral view. (E) 24 hpf, lateral view of head. (F) 24 hpf, lateral view of tail. (G) 36 hpf, lateral view of otic vesicle. (H) 24 hpf, transverse section of trunk. (I) 48 hpf, lateral view. (J) 48 hpf, transverse section of trunk. (K) 5 dpf, lateral view. A, aorta wall; AV, axial vasculature; BA, branchial arches; END, endoderm; FP, floor plate; LLP, lateral line primordium; NC, notochord; OV, otic vesicle; PF, pectoral fin; R, retina; SC, spinal cord; TEL, telencephalon.
Figure 3.
Expression of zebrafish ptenb mRNA during embryogenesis. Expression of ptenb was analyzed by RT-PCR and whole-mount in situ hybridization. (A) Expression of ptenb splice variants during zebrafish development determined by RT-PCR. Bands of 298 bp and 229 bp represent fragments of the long and short splice variants, respectively. (B–K) Expression of ptenb determined by whole-mount in situ hybridization. (B) Early somitogenesis, lateral view. (C–H) Mid-somitogenesis. (C) 10-somite stage, lateral view. (D) 10-somite stage, dorsal view of somites obtained with differential interference contrast microscopy. (E) 10-somite stage, transverse section. (F) 18-somite stage, lateral view. (G) 18-somite stage, dorsal view of tail. (H) 18-somite stage, dorsal view of head. (I) 24 hpf, lateral view. (J) 24 hpf, lateral view of head. (K) 36 hpf, lateral view of otic vesicle. (L) 48 hpf, lateral view. (M) 5 dpf, lateral view. AC, adaxial cells; BA, branchial arches; CG, cranial ganglia; D, diencephalon; EPI, epiphysis; E, eye; NC, notochord; OV, otic vesicle; PF, pectoral fin; S, somite; SC, spinal cord; TEL, telencephalon; TEG, tegmentum..
Whole-mount in situ hybridization was used to analyze the expression patterns of ptena and ptenb during zebrafish embryogenesis. As shown in Fig. 2B–C, ptena mRNA was broadly expressed throughout the embryo at early and mid-somitogenesis. At 24 hpf, transcripts of ptena were more abundant in the CNS, axial vasculature, retina, branchial arches and pectoral fin bud (Fig 2 D–F). In the CNS, ptena was expressed predominantly in the telencephlon and the spinal cord (with the exception of the floor plate). Additional expression of ptena in the wall of the aorta and in the lateral line primordium was observed in transverse sections of 24 hpf embryos (Fig. 2H). Between 24 hpf and 48 hpf, low levels of ptena expression were detected in the otic vesicle (Fig. 2E, G). From 48 hpf through 5 dpf, robust ptena expression was detected in the eye, CNS, branchial arches, endoderm, and pectoral fin buds (Fig. 2I–K).
From early somitogenesis through 24 hpf, low levels of ptenb mRNA were detected throughout the embryo with higher levels present in the caudal somites (Fig. 3B–H). At the 10-somite stage, expression of ptenb was restricted to the posterior and lateral compartments of the somites, which are destined to become fast muscle fibers (Fig. 3D–E). At the 18-somite stage, ptenb expression became restricted to the ventral portion of the somites, while transcripts of ptenb were detectable in neurons of the spinal cord and the cranial ganglia, particularly those in the trigeminal placode (Fig. 3F–H). At 24 hpf, ptenb expression was observed in the spinal cord, the brain, and in the ventral portion of the somites (Fig. 3I–J). In brain, ptenb transcripts were particularly abundant in nuclei of the diencephalon, telencephalon, and tegmentum (Fig. 3J). Between 48 hpf and 5 dpf, ptenb transcripts were most abundant in the central nervous system, branchial arches, pectoral fin, and eye (Fig. 3L–M). In contrast to ptena, there was no apparent enrichment of ptenb transcripts in the 36 hpf otic vesicle (Fig. 3K).
Antisense Morpholino Knockdown of ptena and ptenb Expression
We have utilized antisense morpholinos (MOs) to analyze the role of the ptena and ptenb genes in zebrafish development. Two independent non-overlapping antisense MOs were designed to target the 5’UTR of each pten gene. One of each pair of MOs was used in an in vitro translation assay (Blasiole et al., 2005) to confirm the specificity of the MOs for their respective target sequences. The ptena MO (ptena-MO1) decreased the translation of ptena mRNA by 83% compared to ptena mRNA translated in the absence of MO (Fig. 4A). Translation of ptena mRNA was not inhibited by a ptenb MO (ptenb-MO1) or an MO targeted to the zebrafish Na,K-ATPase α1a.1 subunit (atp1a1a.1-MO). Compared to the in vitro translation of ptenb mRNA in the absence of MO, translation of ptenb mRNA was reduced 90% by ptenb-MO1, but was not decreased in the presence of either ptena-MO1 or the non-specific atp1a1a.1-MO (Fig. 4B). These results strongly suggest that the ptena and ptenb MOs specifically block translation of their target mRNA sequences.
Figure 4.
Specificity of pten antisense morpholinos. ptena and ptenb mRNAs were separately translated in vitro in the presence or absence of ptena or ptenb MOs (ptena-MO1 or ptenb-MO1), respectively. An MO targeted to the Na,K-ATPase α1a.1 subunit (atp1a1a.1-MO) was used as a non-specific control. (A) In vitro translation of ptena. (B) In vitro translation of ptenb. [35S]-labeled proteins were separated on an SDS-containing 10% polyacrylramide gel. The gel was dried and exposed to X-ray film. The ~60 kDa bands corresponding to Ptena and Ptenb polypeptides are shown at the bottom of each panel. Quantitation of bands by laser densitometry is shown at the top of each panel. Bars indicate the levels of [35S]-labeled Ptena or Ptenb in the presence of MOs compared to levels of [35S]-labeled Ptena or Ptenb in the absence of MOs.
We examined the effect of ptena-MO1 and ptenb-MO1 on zebrafish embryogenesis. At 15 hpf, high doses of ptena-MO1 (4 ng/embryo) produced extensive tissue disorganization, necrosis, and death of all injected embryos (n=273; data not shown). At a dose of 1 ng of ptena-MO1, >90% of injected embryos survived to 4 dpf (n=250). At 24 hpf, a variety of developmental defects were observed in ptena morphants (Fig. 5, Fig 6, and Table 1). The most conspicuous morphological defects observed were smaller eyes, curved tails, and a shortened body axis. As outlined in Table 1, microscopic examination of ptena morphants (n=100) revealed defects in notocord shape (73%), ear morphology (59%), head shape (72%), and intersegmental blood flow (83%). By 4 dpf, the heads of ptena morphants failed to properly straighten, and a high percentage (>60%) of morphants displayed cardiac edema (Fig. 4P).
Figure 5.
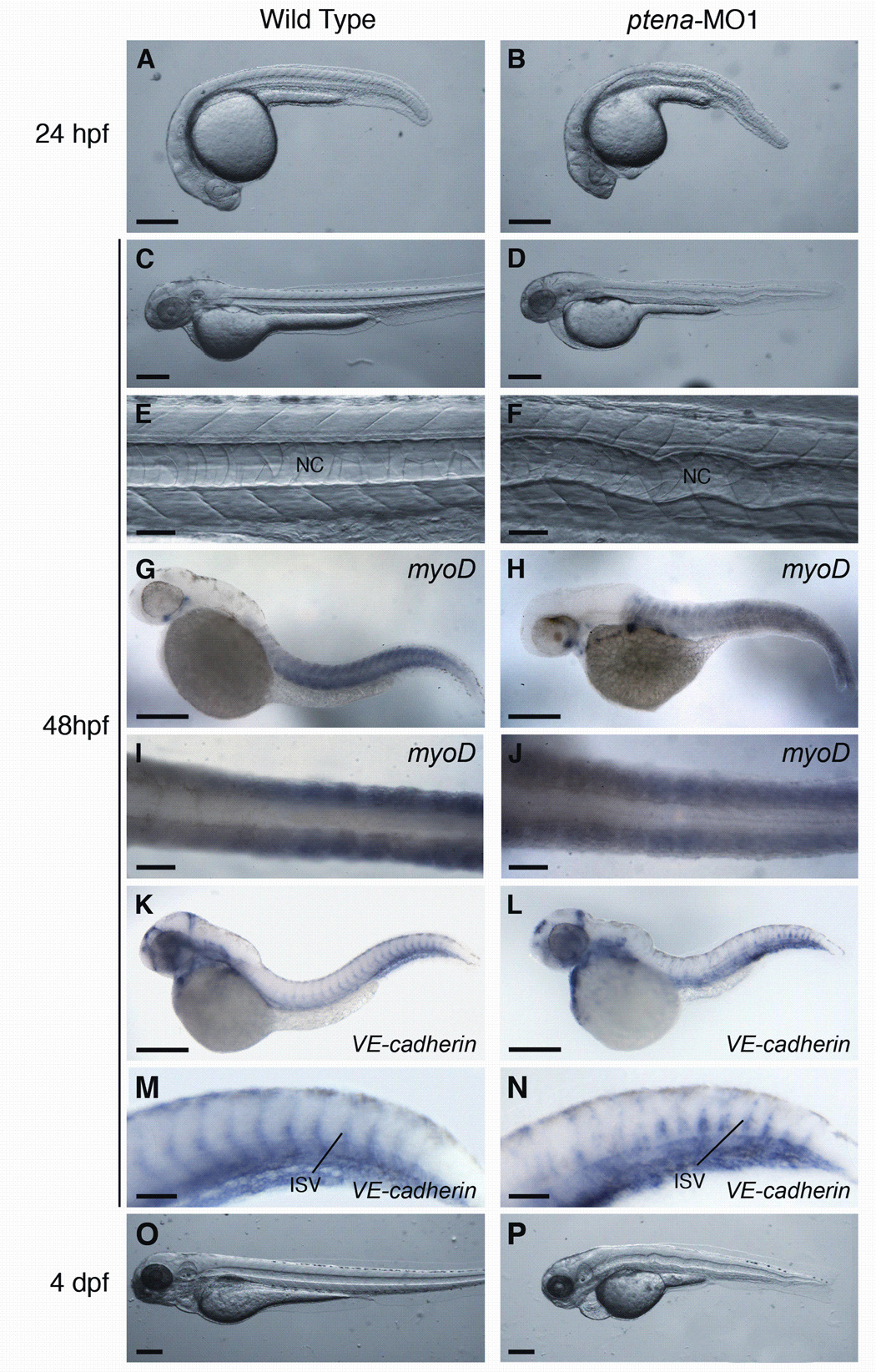
Effect of ptena-MO1 morpholino on zebrafish development. All panels (except I and J) are lateral views with anterior to the left. (A) 24 hpf, wild type embryo. (B) 24 hpf, ptena morphant. (C) 48 hpf, wild type embryo. (D) 48 hpf, ptena morphant. (E) 48 hpf, trunk of wild type embryo. (F) 48 hpf, trunk of ptena morphant. (G–J) 48 hpf, in situ hybridization using myoD probe. (G) Wild type embryo. (H) ptena morphant. (I) Tail of wild type embryo, dorsal view. (J) Tail of ptena morphant, dorsal view. (K–P) 48 hpf, in situ hybridization using VE-cadherin probe. (K) Wild type embryo. (L) ptena morphant. (M) Tail of wild type embryo. (N) Tail of ptena morphant. (O) 4 dpf, wild type embryo. (P) 4 dpf, ptena morphant. Scale bars: A–D, G, H, K, L, O, P = 250 µm; E, F, I, J, M, N = 50 µm.
Figure 6.
Effect of ptena-MO1 morpholino on zebrafish inner ear development. All panels are lateral views of the otic vesicle with anterior to the left. (A) 24 hpf, wild type embryo. (B–D) 24 hpf, ptena morphants. (E) 48 hpf, wild type embryo. (F) 48 hpf, ptena morphant. (G) 4 dpf, wild type embryo. (H–J) 4 dpf, ptena morphants. (K–T) 48 hpf, in situ hybridization using ear-specific markers. (K) pax2a, wild type embryo. (L) pax2a, ptena morphant. (M) otx1, wild type embryo. (N) otx1, ptena morphant. (O) starmaker, wild type embryo. (P) starmaker, ptena morphant. (Q) dfna5, wild type embryo. (R) dfna5, ptena morphant. (S) ncs-1a, wild type embryo. (T) ncs1-a, ptena morphant. Scale bars: A–J = 25 µm. Arrowhead indicates site where epithelial pillars fail to fuse. Arrows point to abnormal tissue masses.
Table 1.
Occurrence of morphant phenotypes at 48 hpf.
| wildtype | ptena morphants (ptena-MO1) | ptenb morphants (ptenb-MO1) | |
|---|---|---|---|
| Abnormal Otoliths | 0 | 59 | 5 |
| Domed Head | 0 | 72 | 83 |
| Hooked Tail | 0 | 0 | 73 |
| Wavy Notochord | 0 | 73 | 19 |
| Lacking Axial Blood flow | 0 | 19 | 22 |
| Lacking Intersegmental Blood Flow | 7 | 83 | 28 |
| Reduced Yolk Extension | 0 | 0 | 90 |
| Total Number of Fish Assayed | 100 | 100 | 100 |
A prominent defect in ptena morphants (73%) was the appearance of an irregularly shaped (wavy) notocord (Fig. 5F). To analyze whether the notochord defect might be caused by alterations in the surrounding tissue, we examined the expression of myoD in the adjacent somites. As shown in Fig 5, G-J, expression of myoD in wild type embryos and ptena morphants appeared very similar, suggesting that the defect in notochord morphology was not due to disorganization of the surrounding somites. At a dose of 2 ng of ptenA-MO1, >75% of injected embryos survived through 4 dpf (n=250). We observed a lack of intersegmental blood flow in 83% of these ptena morphants. Analysis of ptena morphants with a VE-cadherin riboprobe, a marker of vascular endothelial tissue (Larson et al., 2004) revealed that the intersegmental vessels were irregularly formed and did not follow the well-defined chevron shape characteristic of intersegmental vessels seen in wild type embryos (Fig. 5K–N). In addition, some of the intersegmental vessels in the morphants did not appear to completely traverse the trunk or form connections with the dorsal longitudinal anastomotic vessel (Fig. 5N). These results suggest that ptena may play a role in vascular development.
Knockdown of the ptena gene produced a noticeable effect on ear development in zebrafish embryos. At 24 hpf, wild type otic vesicles contained two characteristic rounded otoliths (Fig. 6A). In contrast, ptena morphants exhibited globular-shaped otoliths (Fig. 6B), and/or a single otolith (Fig. 6C, D). At 48 hpf, 59% (n=100) of ptena morphants contained only one otolith (Fig. 6F). By 4 dpf, ptena morphants containing a single otolith failed to generate a second otolith (100%; n = 59). In addition, 22% of ptena morphants containing one otolith also exhibited a defect in semicircular canal formation (Fig. 6H, J). Aberrant semicircular canal morphologies included failure of the epithelial pillars to fuse (Fig 6H), as well as the presence of disorganized epithelial cell masses in the ear (Fig. 6J). Using whole-mount in situ hybridization, we analyzed expression of a number of ear markers involved in ear patterning (pax2a, otx1), otolith formation (starmaker), and semicircular canal development (dfna5, ncs-1a). We did not observe differences in mRNA expression between ptena morphants and wild type controls for any of the markers analyzed (Fig. 6K–T), suggesting that the inner ear defects caused by ptena-MO1 are not due to abnormal patterning of the otic vesicle or disrupted expression of genes involved in otolith formation or semicircular canal development.
To confirm the role of ptena in zebrafish development, we analyzed the effects of a second non-overlapping morpholino (ptena-MO2; 1.5 ng) on zebrafish embryogenesis. At 24 hpf, ptena-MO2 phenocopied the otolith and notochord defects in 45% and 46% of injected embryos (n=58), respectively. At 48 hpf, defects in intersegmental blood flow and head shape were observed in 47% and 35% (n=43) of injected embryos, respectively. None of the ptena-MO2 morphants exhibited defects in semicircular canal formation by 4dpf. These results are consistent with the view that in zebrafish, ptena plays an essential role in a variety of developmental processes including head and notochord formation as well as ear and vascular system development.
We next analyzed the effect of knocking down translation of ptenb mRNA in zebrafish embryos using the ptenb MOs. High doses of ptenb-MO1 (8 ng) or ptenb-MO2 (8 ng) severely affected embryogenesis, such that all tissues were extremely disorganized, the body axis was shortened, and all embryos died within 36 hpf (n=157; data not shown). At a dose of 4 ng of ptenb-MO1, 72% of ptenb morphants were viable at 48 hpf. At this stage, the principal defects we observed in ptenb morphants (n=100) were an upward hooked tail that occurred in 73% of the embryos, a domed head that occurred in 83% of the embryos, and a reduction in the yolk extension that occurred in 90% of the embryos (Fig. 7 and Table 1). Compared to ptena morphants, <20% exhibited defects in the shape of the notochord, and only 28% lacked intersegmental blood flow. By 24 hpf, >95% of ptenb morphants developed normal otoliths. Microinjection of ptenb-MO2 at a dose of 6 ng phenocopied the curved tail defect only. Taken together, these results suggest that ptena and ptenb play distinct roles in zebrafish development.
Figure 7.
Effect of ptenb-MO1 morpholino on zebrafish development. All panels are lateral views of 48 hpf embryos with anterior to the left. (A) Wild type embryo. (B) ptenb morphant. (C) Tail of wild type embryo. (D) Tail of ptenb morphant. Scale bars = 250 µm.
Biological Activity of Ptena and Ptenb
PTEN lipid phosphatase activity has previously been shown to negatively regulate phosphorylation of Akt by virtue of its ability to dephosphorylate PIP3 (Maehama and Dixon, 1998). Because knockdown of ptena and ptenb expression gave distinct developmental phenotypes, we asked whether the PIP3 lipid phosphatase activity of PTEN was conserved in the Ptena and Ptenb enzymes of zebrafish. To do this, we compared the level of activated (phosphorylated) Akt (pAkt) in lysates prepared from wild type zebrafish and ptena and ptenb morphant embryos. Expression of Ptena and Ptenb was knocked down using the ptena-MO1 and ptenb-MO1 morpholinos, and lysates were prepared from wild type and morphant embryos at 48 hpf. Western blot analysis was then used to compare the relative amount of pAkt to total Akt (tAkt) in whole fish lysates. As shown in Fig. 8, MO knockdown of Ptena expression produced a significant increase in the relative amount of pAkt compared to uninjected control embryos. A similar effect on pAkt levels was also observed in lysates prepared from ptenb morphants (Fig 8). These results indicate that zebrafish Ptena and Ptenb both exhibit PIP3 lipid phosphatase activity and function to negatively regulate the PI3-Kinase/AKT pathway.
Figure 8.
Lipid phosphatase activity of zebrafish pten genes. Lysates were prepared from wild type embryos as well as ptena and ptenb morphants at 48 hpf (30 embryos/group). Proteins were separated by SDS-PAGE and transferred to a nitrocellulose filter. Blots were probed with an anti-Akt antibody, stripped, and reprobed with an anti-phospho-Akt (pAkt) antibody. (A) Western blot. (B) Quantitation of bands by laser densitometry. The bars for ptena (n=6 separate experiments) and ptenb (n=4 separate experiments) represent the relative ratio of pAkt to total Akt (tAkt) compared to wild type embryos. The error bars represent the standard error of the mean. The asterisk indicates a statistically significant increase in the ratio of pAkt to tAkt (p<.05) as calculated by an unpaired Students t-test.
DISCUSSION
Studies of mouse knockouts and Drosophila mutants indicate that the PTEN tumor suppressor gene plays an essential role in early development. However, since homozygous null mutations in mice cause embryonic lethality, it has been difficult to gain a better understanding of the precise developmental function of PTEN in vertebrates. Here we have studied PTEN function using zebrafish as a model vertebrate developmental organism. Zebrafish offer many advantages for studying complex developmental processes. In particular, MO-based antisense knockdowns offer the possibility of creating hypomorphs of specific genes. Thus intermediate phenotypes may become manifest without the embryonic lethality associated with null mutations in species such as mouse. In zebrafish, we identified two paralogous pten genes, ptena and ptenb, that exhibit unique but overlapping expression patterns through development. Each gene encodes an enzyme that appears to negatively regulate Akt phosphorylation and is required for embryogenesis. However, our observation that knockdowns of ptena and ptenb expression produce distinct morphant phenotypes suggests that these two pten genes have different functional roles in zebrafish development.
Gene mapping data show that the zebrafish ptena and ptenb genes map to LGs 17 and12, respectively, while the human PTEN gene has been localized to chromosome 10 (Li et al., 1997). Both LG17 and LG 12 contain multiple orthologs of human genes located on chromosome 10 (Woods et al., 2000). The fact that these linkage groups share significant synteny with human chromosome 10 is consistent with the view that these are duplicate chromosome segments. Our studies indicate that both Ptena and Ptenb negatively regulate the PI3-Kinase/AKT pathway via their lipid phosphatase activity, and that each enzyme is required to support embryogenesis. However, it is of interest that knockdown of each pten gene produces a distinct morphant phenotype. This result suggests that the two genes are not able to compensate for one another in early embryogenesis. The duplication-degeneration-complementation model has been proposed to account for the retention of duplicated genes in the genome (Force et al., 1999). This model proposes that retention of duplicated genes in the genome is accompanied by changes in localization or function such that the duplicates together retain the original functions of the single ancestral gene. It will now be of considerable interest to determine whether the two zebrafish pten genes together perform the function of the single mammalian PTEN gene, or whether either of these duplicates have evolved new functional properties. This question can be addressed using morpholino-based knockdown coupled with mRNA rescue approaches that are now possible in zebrafish.
Mammals contain a single functional PTEN gene, and to date no splice variants of this gene have been identified. Both pten genes in zebrafish exhibit alternative splicing, with short and long transcripts of ptena and ptenb expressed throughout embryogenesis and in adult fish. The presence of the alternatively spliced exon in both fish genes indicates that it predates the duplication in the teleost lineage. However, available data are insufficient to determine whether this exon was present in an ancestral vertebrate and lost in the tetrapod lineage, or whether it arose in the teleost lineage. The long splice variant of each zebrafish Pten polypeptide contains a 23 amino acid insertion within one of the three loops that form the putative membrane association region of the pten C2 domain (Lee et al., 1999). The location of the insertion suggests the possibility that the zebrafish pten splice variants may exhibit differences in membrane targeting, binding, or function.
The expression patterns of the zebrafish pten genes during embryonic development closely resemble those of the orthologous mammalian genes. In zebrafish, ptena and ptenb are expressed ubiquitously during early somitogenesis. At the 18-somite stage, ptenb shows strong expression in the cranial ganglia and brain nuclei. As development proceeds, ptena and ptenb genes are predominantly expressed in the central nervous system. In early mouse development, Pten expression is not spatially restricted at E7 (Luukko et al., 1999), whereas by E15, Pten transcripts are abundant in the central and peripheral nervous system (especially in the spinal cord and peripheral nerve ganglia). During embryogenesis, the mouse and human PTEN genes are expressed in the vasculature, lung, kidney, thymus, thyroid gland, skin, and gastrointestinal tract (Luukko et al., 1999; Gimm et al., 2000). In zebrafish, we detected expression of pten genes in the axial vasculature (ptena), endoderm (ptena), and branchial arches (ptena and ptenb). Although expression of pten genes was detected in ear (ptena), somites (ptenb) and eye (ptena and ptenb) of zebrafish embryos, it is not yet clear whether PTEN genes are expressed in these tissues during mouse or human embryogenesis.
Knockdown of ptena mRNA translation resulted in a number of morphological phenotypes including defects in the formation of intersegmental blood vessels. Previous studies have demonstrated that VEGF (vascular endothelial growth factor) signals through the Akt pathway to regulate vasculogenesis and angiogenesis in zebrafish (Chan et al., 2002). Our data indicates that knockdown of ptena causes an increase in the levels of pAkt, and that elevated pAkt levels may lead to abnormal vasculogenesis in zebrafish. Our results thus provide the first indication that ptena may play a role in vascular development. ptena morphants also exhibited defects in notochord morphology. It is possible that the defect in notochord shape may arise early in development when ptena is ubiquitously expressed. It is of further interest that knockdown of ptena also produced a defect in otolith formation. Rather than the two rounded otoliths characteristic of 48 hpf wild type embryos, ptena morphants had abnormal numbers and/or morphology of otoliths. To our knowledge, a role for PTEN in ear development has not been previously reported. It will clearly be of interest to determine whether ptena regulation of the Akt pathway may serve to mediate aspects of inner ear morphogenesis.
Studies of mouse Pten knockouts and Drosophila PTEN mutants have shown that during development, PTEN plays a role in regulating cell and organ size as well as cell proliferation (Kishimoto et al., 2003; Goberdhan et al., 1999; Huang et al., 1999; Gao et al., 2000). We did not observe gross increase in organ size in either ptena or ptenb morphants. While it is possible that pten knockdown may have caused changes in cell size, detailed histological analysis will be required to resolve this issue. We did observe that both ptena and ptenb morphants had heads that were abnormally shaped compared to wild type embryos. This domed appearance could possibly reflect an increase in cell number or cell size within the brain. Results from neuron-specific knockouts of Pten in mice showed that altering Pten expression caused an increase in neuron size leading to an overall increase in brain size (Kishimoto et al., 2003). It will be of interest to determine if similar changes in cell size contribute to the domed head phenotype observed in pten morphant zebrafish.
In summary, we have identified two pten genes in zebrafish that have overlapping expression patterns. Using MO knockdown we showed that ptena and ptenb play distinct roles in embryonic development of zebrafish, while both have the ability to anatagonize the PI3-Kinase/AKT pathway. It will be important to determine whether the developmental roles of ptena and ptenb are solely dependent on the PI3-Kinase/AKT pathway. In addition, it will be interesting to see whether the additional exon in the long spice variants of zebrafish pten genes affects their function.
EXPERIMENTAL PROCEDURES
Identification and Characterization of Zebrafish pten Genes
A BLAST search of the GenBank EST database revealed numerous zebrafish cDNAs with a high degree of similarity to mammalian PTEN. Assembly of overlapping sequences (ESTs fm55g11, fv35h02, faa38e03) yielded a full-length composite sequence corresponding to ptena. Two additional non-overlapping ESTs (fy71d09, fl55h02) encoded the 5' and 3' ends of a distinct pten gene (ptenb). PCR primer pairs were used to amplify the full-length ptena and ptenb cDNAs; ptena forward primer: 5’- GCTGTCATGGCAATGAC -3’, ptena reverse primer: 5’- TCAGACTTTTGTAATCTGTGCG -3’, ptenb forward primer: 5’- GACTCCTGTCACAGCCATGGCTGCG -3’, ptenb reverse primer: 5’- CTTCCCATAAAAATATTTCAAC -3’. BLAST searches of the zebrafish genome assembly (Version 4) available from the Zebrafish Sequencing Group (http://www.ensembl.org/) were used to assign linkage groups.
mRNA Expression Analysis
For RT-PCR, zebrafish embryos at various stages of development and ~ 6 month old adults were collected and homogenized in TRIzol Reagent (Invitrogen; Carlsbad, CA). Total RNA was extracted according to the method of Chomczynski and Sacchi (1987), and developmental stage-specific RNA (0.5 µg) was used as template to generate single stranded cDNA using the SuperScript First Strand Synthesis kit (Invitrogen). PCR was carried out with REDTaq DNA polymerase (Sigma; St. Louis, MO) using a RoboCycler Gradient Temperature Cycler (Stratagene; La Jolla, CA) and primers that flank the alternatively spliced exons of ptena (forward primer 5’-CCAGCCAGCGCAGGTATGTGTA-3’ and reverse primer 5’- GCGGCTGAGGAAACTCGAAGATC-3’) and ptenb (forward primer 5’-GCTACCTTCTGAGGAATAAGCTGG-3’ and reverse primer 5’-CTTGATGTCCCCACACACAGGC-3’). PCR products were analyzed by electrophoresis on a 1.5% agarose gel. The PCR products from each primer pair were verified by DNA sequencing.
Whole-mount in situ hybridization analysis was performed as described by Thisse et al. (1999; http://zfin.org/cgi-bin/webdriver?MIval=aa-pubview2.apg&OID=ZDB-PUB-010810-1). The following antisense probes were utilized to characterize pten expression: ptena (GenBank accession no. AY398669, nucleotides 1–1371); ptenb (GenBank accession no. AY398670, nucleotides 1–1302). Additional antisense RNA probes include dfna5 (Busch-Nentwich et al., 2004), myoD (Weinberg et al., 1996), otx1 (from E. Weinberg), ncs-1a (Blasiole et al., 2005) and starmaker (Sollner et al., 2003), VE-cadherin (cdh5) (Larson et al., 2004).
Antisense Morpholino Knockdowns
Antisense morpholino oligonucleotides (MOs) (Gene Tools LLC; Philomath, OR) were designed to target the 5’UTR of each zebrafish pten gene. ptena-MO1 (5’-CCTCGCTCACCCTTGACTGTGTATG-3’); ptena-MO2 (5’- CAGTTTTATTCCGGTTTATTGTCAG-3’); ptenb-MO1(5’-CTTTCGGACGGTC-GGTCGTCTTTA-3’); ptenb-MO2 (5’-GGCTGTGACAGGAGTCTTTAGGGTT-3’). The MOs were resuspended in 1x Danieau buffer (58 mM NaCl, 0.7 mM KCl, 0.4 mM MgSO4, 0.6 mM Ca(NO3)2, 5 mM HEPES, pH 7.6) and microinjected into the yolk of single cell embryos.
The specificity of the pten MOs was tested using an in vitro translation assay as previously described (Blasiole et al., 2005). All experiments were performed with the short splice forms of the pten genes. ptena (GenBank accession no. AY398668) and ptenb (GenBank accession no. AY398671) cDNAs were subcloned into pBluescript II KS+ (Stratagene). Capped ptena and ptenb mRNAs were synthesized using the mMESSAGE mMACHINE (Ambion) transcription kit, and 0.5 µg of mRNA was used to program the synthesis of [35S]-labeled proteins using an in vitro rabbit reticulocyte lysate translation kit (Ambion) under conditions described by the manufacturer. The translation of mRNA was tested in the presence of 4 µg of antisense MOs (ptena-MO1 and ptenb-MO1). An antisense MO targeted against the zebrafish Na,K-ATPase α1a.1 gene (atp1a1a.1) (5’-GCCTTCTCCTCGTCCCATTTTGCTG-3) (Shu et al., 2003) was used as a non-specific control. The entire reaction mixture (20 µl) of each in vitro translation assay was separated by SDS-PAGE. The gels were dried, exposed to X-ray film, and the relative intensity of the bands quantitated by laser densitometry (Molecular Dynamics; Sunnyvale, CA) and analyzed using the Quantity One software package (PDI, Inc.; Huntington Station, NY).
Phosphorylated Akt Assay
Pten activity was assayed in wild type and morphant embryos as follows. Wild type embryos, ptena morphants (injected with 6 ng ptena-MO1), and ptenb morphants (injected with 6 ng ptenb-MO1) were collected at 48 hpf and homogenized in lysis buffer (Blasiole et al., 2005). Thirty embryos were collected in each group. Lysates were incubated at 4°C for one hour, then centrifuged at 10,000 rpm for 10 minutes at 4°C. Supernatants were normalized for total protein content, fractionated by SDS-PAGE (50 µg protein/lane), then transferred to a nitrocellulose filter. Immunoblots were probed with a rabbit anti-human Akt antibody (1:1000 dilution; Cell Signalling; Beverly, MA). Blots were stripped and reprobed with a rabbit anti-human phospho-Akt (Ser 473) antibody (1:1000 dilution, Cell Signalling). Peroxidase-conjugated secondary antibodies were from Jackson ImmunoResearch (West Grove, PA). Immunoreactivity was visualized by enhanced chemiluminescence (ECL) using an ECL Plus kit (Amersham Pharmacia; Piscataway, NJ). Immunoblots were quantitated by laser densitometry (Molecular Dynamics) and analyzed using the Quantity One software package (PDI, Inc.). Statistical analyses of the data were performed using an unpaired one-tailed Student’s t-test.
ACKNOWLEDGEMENTS
This work was supported by grants from the NIH (RL, KC, CT and BT).
Footnotes
WEBSITE REFERENCES
http://www.ensembl.org/; Zebrafish Sequencing Group
http://zfin.org/cgi-bin/webdriver?MIval=aa-pubview2.apg&OID=ZDB-PUB-010810-1; Thisse, B., Pflumio, S., Fürthauer, M., Loppin, B., Heyer, V., Degrave, A., Woehl, R., Lux, A., Steffan, T., Charbonnier, X.Q. and Thisse, C. Expression of the zebrafish genome during embryogenesis.
REFERENCES
- Amores A, Force A, Yan YL, Joly L, Amemiya C, Fritz A, Ho RK, Langeland J, Prince V, Wang YL, Westerfield M, Ekker M, Postlethwait JH. Zebrafish hox clusters and vertebrate genome evolution. Science. 1998;282:1711–1714. doi: 10.1126/science.282.5394.1711. [DOI] [PubMed] [Google Scholar]
- Birck A, Ahrenkiel V, Zeuthen J, Hou-Jensen K, Guldberg P. Mutation and allelic loss of the PTEN/MMAC1 gene in primary and metastatic melanoma biopsies. J Invest Dermatol. 2000;114:277–280. doi: 10.1046/j.1523-1747.2000.00877.x. [DOI] [PubMed] [Google Scholar]
- Blasiole B, Kabbani N, Boehmler W, Thisse B, Thisse C, Canfield V, Levenson R. Neuronal calcium sensor-1 gene ncs-1a is essential for semicircular canal formation in zebrafish inner ear. J Neurobiol. 2005;64:285–297. doi: 10.1002/neu.20138. [DOI] [PubMed] [Google Scholar]
- Busch-Nentwich E, Sollner C, Roehl H, Nicolson T. The deafness gene dfna5 is crucial for ugdh expression and HA production in the developing ear in zebrafish. Development. 2004;131:943–951. doi: 10.1242/dev.00961. [DOI] [PubMed] [Google Scholar]
- Chan J, Bayliss PE, Wood JM, Roberts TM. Dissection of angiogenic signaling in zebrafish using a chemical genetic approach. Cancer Cell. 2002;1:257–267. doi: 10.1016/s1535-6108(02)00042-9. [DOI] [PubMed] [Google Scholar]
- Di Cristofano A, Pesce B, Cordon-Cardo C, Pandolfi PP. Pten is essential for embryonic development and tumour suppression. Nat Genet. 1998;19:348–355. doi: 10.1038/1235. [DOI] [PubMed] [Google Scholar]
- Ferrara N, Carver-Moore K, Chen H, Dowd M, Lu L, O'Shea KS, Powell-Braxton L, Hillan KJ, Moore MW. Heterozygous embryonic lethality induced by targeted inactivation of the VEGF gene. Nature. 1996;380:439–442. doi: 10.1038/380439a0. [DOI] [PubMed] [Google Scholar]
- Force A, Lynch M, Pickett FB, Amores A, Yan YL, Postlethwait J. Preservation of duplicate genes by complementary, degenerative mutations. Genetics. 1999;151:1531–1545. doi: 10.1093/genetics/151.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Neufeld TP, Pan D. Drosophila PTEN regulates cell growth and proliferation through PI3K-dependent and -independent pathways. Dev Biol. 2000;221:404–418. doi: 10.1006/dbio.2000.9680. [DOI] [PubMed] [Google Scholar]
- Gimm O, Attie-Bitach T, Lees JA, Vekemans M, Eng C. Expression of the PTEN tumour suppressor protein during human development. Hum Mol Genet. 2000;9:1633–1639. doi: 10.1093/hmg/9.11.1633. [DOI] [PubMed] [Google Scholar]
- Goberdhan DC, Paricio N, Goodman EC, Mlodzik M, Wilson C. Drosophila tumor suppressor PTEN controls cell size and number by antagonizing the Chico/PI3-kinase signaling pathway. Genes Dev. 1999;13:3244–3258. doi: 10.1101/gad.13.24.3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Potter CJ, Tao W, Li DM, Brogiolo W, Hafen E, Sun H, Xu T. PTEN affects cell size, cell proliferation and apoptosis during Drosophila eye development. Development. 1999;126:5365–5372. doi: 10.1242/dev.126.23.5365. [DOI] [PubMed] [Google Scholar]
- Kibar Z, Vogan KJ, Groulx N, Justice MJ, Underhill DA, Gros P. Ltap, a mammalian homolog of Drosophila Strabismus/Van Gogh, is altered in the mouse neural tube mutant Loop-tail. Nat Genet. 2001;28:251–255. doi: 10.1038/90081. [DOI] [PubMed] [Google Scholar]
- Kishimoto H, Hamada K, Saunders M, Backman S, Sasaki T, Nakano T, Mak TW, Suzuki A. Physiological functions of Pten in mouse tissues. Cell Struct Funct. 2003;28:11–12. doi: 10.1247/csf.28.11. [DOI] [PubMed] [Google Scholar]
- Larson JD, Wadman SA, Chen E, Kerley L, Clark KJ, Eide M, Lippert S, Nasevicius A, Ekker SC, Hackett PB, Essner JJ. Expression of VE-cadherin in zebrafish embryos: a new tool to evaluate vascular development. Dev Dyn. 2004;231:204–213. doi: 10.1002/dvdy.20102. [DOI] [PubMed] [Google Scholar]
- Lee JO, Yang H, Georgescu MM, Di Cristofano A, Maehama T, Shi Y, Dixon JE, Pandolfi P, Pavletich NP. Crystal structure of the PTEN tumor suppressor: implications for its phosphoinositide phosphatase activity and membrane association. Cell. 1999;99:323–334. doi: 10.1016/s0092-8674(00)81663-3. [DOI] [PubMed] [Google Scholar]
- Li DM, Sun H. TEP1, encoded by a candidate tumor suppressor locus, is a novel protein tyrosine phosphatase regulated by transforming growth factor beta. Cancer Res. 1997;57:2124–2129. [PubMed] [Google Scholar]
- Li J, Yen C, Liaw D, Podsypanina K, Bose S, Wang SI, Puc J, Miliaresis C, Rodgers L, McCombie R, Bigner SH, Giovanella BC, Ittmann M, Tycko B, Hibshoosh H, Wigler MH, Parsons R. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breastand prostate cancer. Science. 1997;275:1943–1947. doi: 10.1126/science.275.5308.1943. [DOI] [PubMed] [Google Scholar]
- Liaw D, Marsh DJ, Li J, Dahia PL, Wang SI, Zheng Z, Bose S, Call KM, Tsou HC, Peacocke M, Eng C, Parsons R. Germline mutations of the PTEN gene in Cowden disease, an inherited breast and thyroid cancer syndrome. Nat Genet. 1997;16:64–67. doi: 10.1038/ng0597-64. [DOI] [PubMed] [Google Scholar]
- Luukko K, Ylikorkala A, Tiainen M, Makela TP. Expression of LKB1 and PTEN tumor suppressor genes during mouse embryonic development. Mech Dev. 1999;83:187–190. doi: 10.1016/s0925-4773(99)00050-7. [DOI] [PubMed] [Google Scholar]
- Maehama T, Dixon JE. The tumor suppressor, PTEN/MMAC1, dephosphorylates the lipid second messenger, phosphatidylinositol 3,4,5-trisphosphate. J Biol Chem. 1998;273:13375–13378. doi: 10.1074/jbc.273.22.13375. [DOI] [PubMed] [Google Scholar]
- Marsh DJ, Dahia PL, Zheng Z, Liaw D, Parsons R, Gorlin RJ, Eng C. Germline mutations in PTEN are present in Bannayan-Zonana syndrome. Nat Genet. 1997;16:333–334. doi: 10.1038/ng0897-333. [DOI] [PubMed] [Google Scholar]
- Nasevicius A, Ekker SC. Effective targeted gene 'knockdown' in zebrafish. Nat Genet. 2000;26:216–220. doi: 10.1038/79951. [DOI] [PubMed] [Google Scholar]
- Nasevicius A, Larson J, Ekker SC. Distinct requirements for zebrafish angiogenesis revealed by a VEGF-A morphant. Yeast. 2000;17:294–301. doi: 10.1002/1097-0061(200012)17:4<294::AID-YEA54>3.0.CO;2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park M, Moon RT. The planar cell-polarity gene stbm regulates cell behaviour and cell fate in vertebrate embryos. Nat Cell Biol. 2002;4:20–25. doi: 10.1038/ncb716. [DOI] [PubMed] [Google Scholar]
- Podsypanina K, Ellenson LH, Nemes A, Gu J, Tamura M, Yamada KM, Cordon-Cardo C, Catoretti G, Fisher PE, Parsons R. Mutation of Pten/Mmac1 in mice causes neoplasia in multiple organ systems. Proc Natl Acad Sci U S A. 1999;96:1563–1568. doi: 10.1073/pnas.96.4.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postlethwait JH, Yan YL, Gates MA, Horne S, Amores A, Brownlie A, Donovan A, Egan ES, Force A, Gong Z, Goutel C, Fritz A, Kelsh R, Knapik E, Liao E, Paw B, Ransom D, Singer A, Thomson M, Abduljabbar TS, Yelick P, Beier D, Joly JS, Larhammar D, Rosa F, et al. Vertebrate genome evolution and the zebrafish gene map. Nat Genet. 1998;18:345–349. doi: 10.1038/ng0498-345. [DOI] [PubMed] [Google Scholar]
- Shu X, Cheng K, Patel N, Chen F, Joseph E, Tsai HJ, Chen JN. Na,K-ATPase is essential for embryonic heart development in the zebrafish. Development. 2003;130:6165–6173. doi: 10.1242/dev.00844. [DOI] [PubMed] [Google Scholar]
- Sollner C, Burghammer M, Busch-Nentwich E, Berger J, Schwarz H, Riekel C, Nicolson T. Control of crystal size and lattice formation by starmaker in otolith biomineralization. Science. 2003;302:282–286. doi: 10.1126/science.1088443. [DOI] [PubMed] [Google Scholar]
- Stahl JM, Cheung M, Sharma A, Trivedi NR, Shanmugam S, Robertson GP. Loss of PTEN promotes tumor development in malignant melanoma. Cancer Res. 2003;63:2881–2890. [PubMed] [Google Scholar]
- Steck PA, Pershouse MA, Jasser SA, Yung WK, Lin H, Ligon AH, Langford LA, Baumgard ML, Hattier T, Davis T, Frye C, Hu R, Swedlund B, Teng DH, Tavtigian SV. Identification of a candidate tumour suppressor gene, MMAC1, at chromosome 10q23.3 that is mutated in multiple advanced cancers. Nat Genet. 1997;15:356–362. doi: 10.1038/ng0497-356. [DOI] [PubMed] [Google Scholar]
- Stiles B, Groszer M, Wang S, Jiao J, Wu H. PTENless means more. Dev Biol. 2004;273:175–184. doi: 10.1016/j.ydbio.2004.06.008. [DOI] [PubMed] [Google Scholar]
- Suzuki A, de la Pompa JL, Stambolic V, Elia AJ, Sasaki T, del Barco Barrantes I, Ho A, Wakeham A, Itie A, Khoo W, Fukumoto M, Mak TW. High cancer susceptibility and embryonic lethality associated with mutation of the PTEN tumor suppressor gene in mice. Curr Biol. 1998;8:1169–1178. doi: 10.1016/s0960-9822(07)00488-5. [DOI] [PubMed] [Google Scholar]
- Thompson JE, Thompson CB. Putting the rap on Akt. J Clin Oncol. 2004;22:4217–4226. doi: 10.1200/JCO.2004.01.103. [DOI] [PubMed] [Google Scholar]
- Weinberg ES, Allende ML, Kelly CS, Abdelhamid A, Murakami T, Andermann P, Doerre OG, Grunwald DJ, Riggleman B. Developmental regulation of zebrafish MyoD in wild-type, no tail and spadetail embryos. Development. 1996;122:271–280. doi: 10.1242/dev.122.1.271. [DOI] [PubMed] [Google Scholar]
- Woods IG, Kelly PD, Chu F, Ngo-Hazelett P, Yan YL, Huang H, Postlethwait JH, Talbot WS. A comparative map of the zebrafish genome. Genome Res. 2000;10:1903–1914. doi: 10.1101/gr.10.12.1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Senechal K, Neshat MS, Whang YE, Sawyers CL. The PTEN/MMAC1 tumor suppressor phosphatase functions as a negative regulator of the phosphoinositide 3-kinase/Akt pathway. Proc Natl Acad Sci U S A. 1998;95:15587–15591. doi: 10.1073/pnas.95.26.15587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou XP, Kuismanen S, Nystrom-Lahti M, Peltomaki P, Eng C. Distinct PTEN mutational spectra in hereditary non-polyposis colon cancer syndrome-related endometrial carcinomas compared to sporadic microsatellite unstable tumors. Hum Mol Genet. 2002;11:445–450. doi: 10.1093/hmg/11.4.445. [DOI] [PubMed] [Google Scholar]
- Zhou XP, Marsh DJ, Morrison CD, Chaudhury AR, Maxwell M, Reifenberger G, Eng C. Germline inactivation of PTEN and dysregulation of the phosphoinositol-3-kinase/Akt pathway cause human Lhermitte-Duclos disease in adults. Am J Hum Genet. 2003;73:1191–1198. doi: 10.1086/379382. [DOI] [PMC free article] [PubMed] [Google Scholar]