Abstract
High levels of the intracellular signaling molecule cyclic diguanylate (c-di-GMP) supress motility and activate exopolysaccharide (EPS) production in a variety of bacterial species. In many bacteria part of the effect of c-di-GMP is on gene expression, but the mechanism involved is not known for any species. We have identified the protein FleQ as a c-di-GMP-responsive transcriptional regulator in Pseudomonas aeruginosa. FleQ is known to activate expression of flagella biosynthesis genes. Here we show that it also represses transcription of genes including the pel operon involved in EPS biosynthesis, and that this repression is relieved by c-di-GMP. Our in vivo data indicate that FleQ represses pel transcription and that pel transcription is not repressed when intracellular c-di-GMP levels are high. FleN, a known antiactivator of FleQ also participates in control of pel expression. In in vitro experiments we found that FleQ binds to pel promoter DNA and that this binding is inhibited by c-di-GMP. FleQ binds radiolabeled c-di-GMP in vitro. FleQ does not have amino acid motifs that resemble previously defined c-di-GMP binding domains. Our results show that FleQ is a new type of c-di-GMP binding protein that controls the transcriptional regulation of EPS biosynthesis genes in P. aeruginosa.
Introduction
Biofilms are surface-associated communities of bacteria that are encased in exopolysaccharides (EPS) (Costerton, 1999; Kolter and Greenberg, 2006). Cells of the opportunistic pathogen Pseudomonas aeruginosa form biofilms in a variety of situations including on indwelling medical devices as well as in the lungs of cystic fibrosis patients (Costerton et al., 1999; Costerton et al., 2003; Hoiby, 2002; Singh et al., 2000; Yahr and Greenberg, 2004). Biofilm infections are difficult to treat because bacteria in biofilms tend to be highly resistant to antimicrobial treatment and are able to more easily evade immune responses than planktonic cells (Hoiby, 2002; Stewart and Costerton, 2001). Because of this there is significant interest in understanding factors important for biofilm formation and maintenance. One such factor is the intracellular signaling molecule cyclic diguanylate (c-di-GMP). Originally discovered as an allosteric effector of cellulose synthesis in Gluconacetobacter xylinus (Ross et al., 1987), c-di-GMP is now recognized as playing a central role in modulating the transition between planktonic and biofilm lifestyles in a large and growing number of bacterial species, including P. aeruginosa, P. fluorescens, Salmonella typhimurium, Escherichia coli, and Vibrio cholerae (Cotter and Stibitz, 2007; Goymer et al., 2006; Hickman et al., 2005; Kulasakara et al., 2006; Romling et al., 2005; Romling and Amikam, 2006; Ross et al., 1991; Simm et al., 2004; Tamayo et al., 2007; Thormann et al., 2006; Tischler and Camilli, 2004; Wolfe and Visick, 2008).
In general, increased intracellular c-di-GMP stimulates EPS production and inhibits flagella- and pilus-mediated motility, whereas low intracellular c-di-GMP promotes motility and inhibits EPS synthesis (Aldridge et al., 2003; Boles and McCarter, 2002; Garcia et al., 2004; Hickman et al., 2005; Kader et al., 2006; Lim et al., 2007; Simm et al., 2004; Tischler and Camilli, 2004). Accumulating evidence indicates that c-di-GMP has effects both on enzyme activities and protein assembly as well as on the expression of genes for EPS synthesis, flagella biogenesis, and virulence in a variety of bacteria (Beyhan et al., 2006; Ferreira et al., 2008; Hickman et al., 2005; Lee et al., 2007; Mendez-Ortiz et al., 2006). Recently a number of c-di-GMP binding proteins have been identified that affect the activities of protein complexes including alginate synthase, cellulose synthase, flagella, and pili (Christen et al., 2007; Lee et al., 2007; Merighi et al., 2007; Pratt et al., 2007; Ryjenkov et al., 2006). To date no c-di-GMP binding proteins that influence gene expression have been reported.
We set out to identify the mechanism by which c-di-GMP affects gene expression in P. aeruginosa. The observation that flagella biosynthesis genes are expressed at lower levels in cells with high intracellular c-di-GMP prompted us to investigate if c-di-GMP might affect the activity of one of the transcription factors known to regulate flagella gene expression. One candidate is FleQ (PA1097), the master regulator of flagella gene expression in P. aeruginosa (Arora et al., 1997). FleQ sits at the top of a four-tiered transcription hierarchy that controls flagellar gene expression (Dasgupta et al., 2003). It is an enhancer binding protein that contains an N-terminal FleQ domain, an AAA σ54 -interaction domain, and a helix-turn-helix DNA binding domain. FleQ activates expression of the two component regulatory genes fleSR as well as genes that are necessary for the assembly of the flagella export apparatus and initiation of flagella basal body-hook formation in conjunction with the alternative RNA polymerase sigma factor, σ54 (Jyot et al., 2002). A fleQ mutant is nonmotile (Dasgupta et al., 2000; Dasgupta and Ramphal, 2001). A transcriptome analysis of a fleQ mutant of P. aeruginosa strain PAK showed that not only was FleQ involved in activating the transcription of flagella genes, but FleQ also negatively regulated the expression of a number of genes that are now known to specify EPS synthesis (Dasgupta et al., 2003). The expression levels of these genes are also now known to be increased in cells with high intracellular c-di-GMP (Hickman et al., 2005).
Here we show that c-di-GMP acts directly on FleQ to cause it to derepress the expression of the Pel EPS synthesis operon and other genes. We demonstrate that full length FleQ, as well as a truncated version of FleQ that lacks the N-terminal FleQ domain, bind c-di-GMP and that the ability of FleQ to bind to the pel promoter is inhibited by c-di-GMP. Our results indicate that c-di-GMP functions to relieve transcriptional repression by FleQ of EPS genes necessary for biofilm formation in P. aeruginosa.
Results
A fleQ mutant forms wrinkly colonies
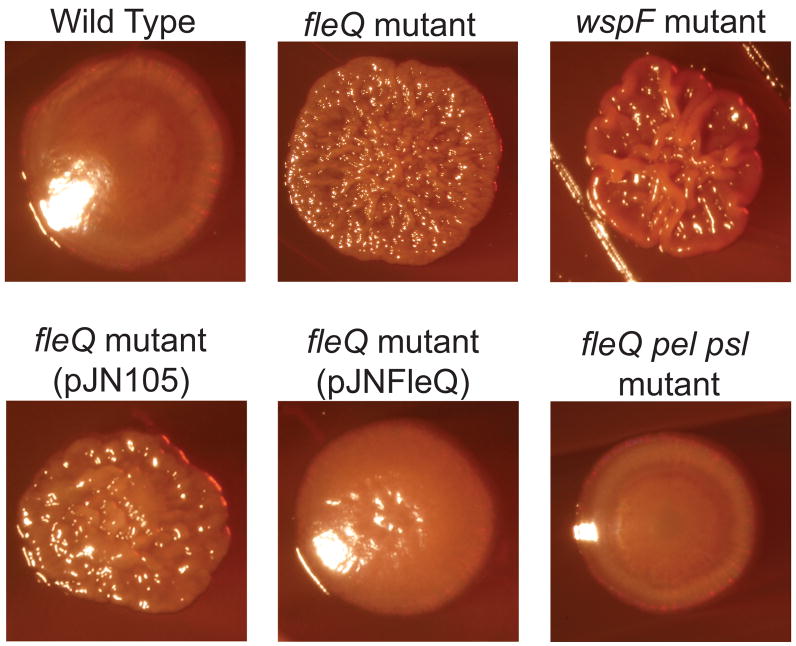
A fleQ::Tn5 mutant of P. aeruginosa strain PAO1 was nonmotile, as expected, and the motility defect was complemented in trans with a wild-type copy of fleQ. In addition to having a motility defect, we found that the fleQ mutant formed colonies that were wrinkled in appearance compared to those of the wild type on tryptone agar at room temperature (Figure 1). The fleQ mutant colonies were slightly less wrinkly than wspF mutant colonies (Figure 1). wspF mutations cause the diguanylate cyclase WspR to be activated, resulting in the accumulation of high intracellular concentrations of c-di-GMP relative to wild type cells (Goymer et al., 2006; Hickman et al., 2005). The colony morphology phenotype of a fleQ mutant on tryptone agar was reverted by complementation with the fleQ gene in trans (Figure 1). Several studies have shown that increased production of Pel or Psl EPS by P. aeruginosa results in a wrinkled colony morphology (Friedman and Kolter, 2004a, 2004b; Hickman et al., 2005; Sakuragi and Kolter, 2007). When we generated deletions in the pelA and pslBCD genes in a fleQ mutant background the wrinkled colony morphology typical of the fleQ mutant changed to smooth (Figure 1), indicating that Pel and Psl EPS are responsible for the wrinkled colony phenotype.
Figure 1. A fleQ mutant has a wrinkly colony morphology.
P. aeruginosa wild type (strain PAO1), fleQ mutant, wspF mutant, fleQ mutant containing fleQ on a plasmid (pJNFleQ) or control vector (pJN105), and a fleQ pel psl mutant were streaked on tryptone agar containing congo red and colonies were imaged after 5 days at 25°C.
A fleQ mutation and high intracellular c-di-GMP have nonadditive effects in causing elevated EPS gene expression
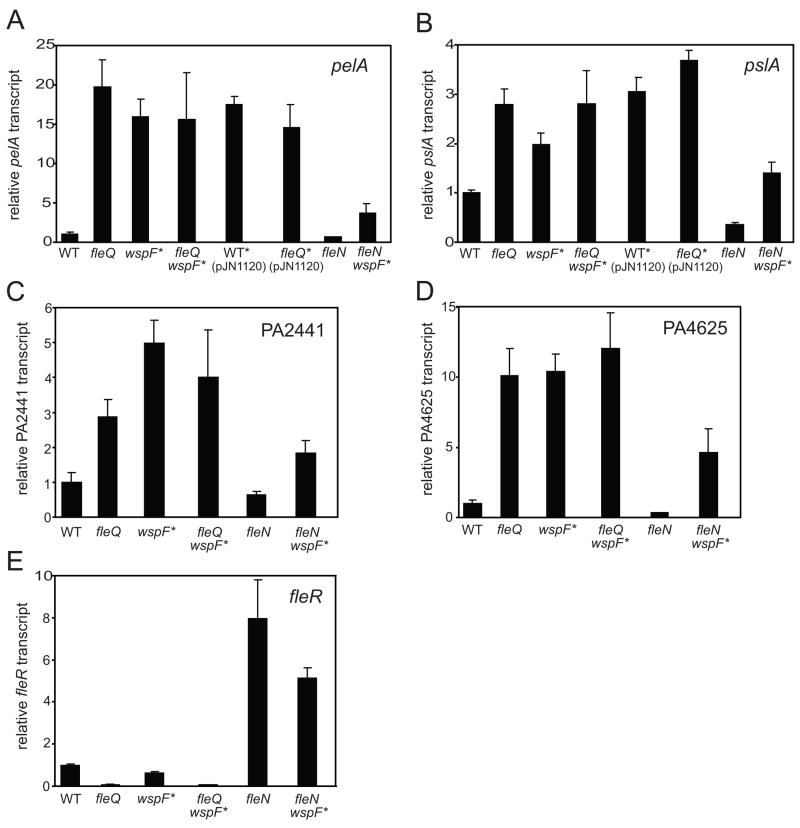
We have previously shown that high intracellular c-di-GMP results in elevated expression of a number of genes in P. aeruginosa PAO1, including genes for EPS biosynthesis (Hickman et al., 2005). An earlier study by Dasgupta et al. (2003) showed that a fleQ mutant of P. aeruginosa strain PAK had increased expression of many of the same genes that we found to be elevated in high c-di-GMP cells. To confirm and extend these findings in P. aeruginosa strain PAO1, we used reverse transcriptase PCR to compare levels of the EPS biosynthesis genes pelA (PA3064) and pslA (PA2231), as well as levels of hypothetical genes PA2441 and PA4625 in a fleQ mutant to those of the wild type and we also quantitated transcript levels in a wspF mutant. In our previous work we found that expression of pelA, pslA, PA2441, and PA4625 were elevated in the wspF mutant (Hickman et al., 2005). We found that the fleQ mutant had a 20-fold increased level of pelA transcript and a 2.8-fold increased level pslA transcript compared to wild type (Figure 2A and 2B). The levels of PA2441 and PA4625 transcription were also higher in a fleQ mutant compared to wild type (Figure 2C and 2D). A wspF strain with high intracellular c-di-GMP had levels of these transcripts that were comparable to those observed with the fleQ mutant. We also generated elevated levels of intracellular c-di-GMP in wild type and fleQ mutant cells by overexpressing the diguanylate cyclase encoded by PA1120 (Kulasakara et al., 2006). This also resulted in elevated transcript levels of pelA and pslA (Figure 2 A and B). Thus FleQ appears to negatively regulate transcription of pel, psl, PA2441, and PA4625 whereas high c-di-GMP appears to somehow stimulate transcription of these genes.
Figure 2. FleQ is a negative regulator of genes that are also expressed at high c-di-GMP levels.
Relative transcript levels for pelA (A), pslA (B), PA2441(C), PA4625 (D) and fleR (E) as assayed by quantitative RT-PCR are shown for wild type (PAO1), wspF mutant (high c-di-GMP), a fleQ mutant, a fleQ wspF mutant (high c-di-GMP), wild type expressing PA1120 from a plasmid (pJN1120), a fleQ mutant expressing PA1120, fleN mutant, and a fleN wspF mutant (high c-di-GMP). Strains with an asterisk indicate that these strains have high c-di-GMP levels and display phenotypes consistent with elevated c-di-GMP. Data points are the average of three independent biological samples. Error bars represent the standard deviations between replicates.
If FleQ and c-di-GMP influence gene transcription at the same point, then the effects of a fleQ mutation and high c-di-GMP on transcription should not be additive. Consistent with this, when we quantified transcript levels of pelA, pslA, PA2441, and PA4625 in a wspF fleQ double mutant we saw no increase in expression compared to either single wpsF or fleQ mutants (Figure 2).
FleN, a known antagonist of FleQ activation of flagella gene expression, also modulates pel and psl transcription
The activity of FleQ is known to be modulated by FleN (PA1454), which has been reported to bind to FleQ and dampen its ability to activate gene transcription (Dasgupta et al., 2000; Dasgupta and Ramphal, 2001). Strains lacking FleN have increased expression of flagella biosynthesis genes and fleN mutants are multiflagellated (Dasgupta et al., 2000). If FleN also dampens the ability of FleQ to repress transcription of EPS biosynthesis and other genes, then fleN mutants would be predicted to show decreased expression of genes that are negatively regulated by FleQ. In keeping with this, the transcript levels of pelA, pslA, PA2441, and PA4625 were 1.5, 2.8, 3.0, and 1.6 fold lower in a fleN mutant compared to wild type (Figure 2). The transcript levels of our target genes were lower in a wspF fleN double mutant than in a wspF mutant (a strain with elevated c-di-GMP) (Figure 2). However, the levels of the target gene transcripts were higher in the wspF fleN double mutant than in the fleN single mutant, indicating that the double mutant strain responded to c-di-GMP to modulate transcription. These results are consistent with a model where FleQ acts as a repressor of pel and psl gene transcription and FleN interacts with FleQ to antagonize its repressor activity.
Elevated c-di-GMP levels have small effects on transcription of flagella biosynthesis genes
Consistent with previously published results showing that FleQ is an important positive activator of flagella gene expression (Arora et al., 1997), we observed 20-fold lower levels of fleR transcript in the fleQ mutant compared to wild type (Figure 2E). The fleR transcript levels were 1.6 fold lower in a wspF mutant compared to wild type, indicating that elevated c-di-GMP has a much smaller effect on expression of flagella genes than on EPS biosynthesis genes. A wspF fleQ double mutant had the same low levels of fleR transcript as did a fleQ mutant alone. Loss of fleN resulted in an eight-fold increase in fleR transcript level (Figure 2E), consistent with previous reports (Dasgupta et al., 2000). A wspF fleN double mutant showed a 1.6 fold decrease in fleR transcript levels compared with the fleN mutant alone, indicating that the small effect of c-di-GMP on fleR transcript levels is observed in the presence or absence of the fleN gene (Figure 2E). Similar effects of elevated c-di-GMP on transcript levels of the flhA gene were also observed (data not shown). Since the effect of elevated c-di-GMP on the expression of flagella genes was small we decided to focus on the effects of c-di-GMP and FleQ on the pel operon.
fleQ or fleN mutations have minimal effects on c-di-GMP concentrations
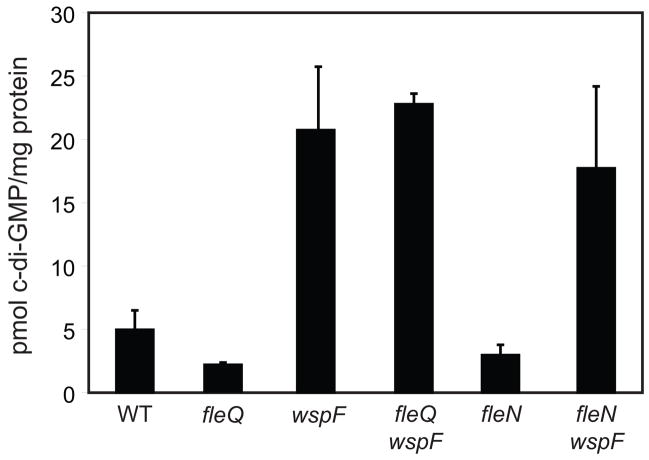
One explanation for the effects of FleQ and FleN on expression of the pel and psl genes is that loss of FleQ or FleN affects c-di-GMP levels, which then influence gene transcription. In order to rule out this possibility we quantified c-di-GMP levels in strains lacking FleQ or FleN and compared them to the levels in wild-type cells. We measured c-di-GMP by liquid chromatography-mass spectrometry using a modification of a previously reported method (Thormann et al., 2006). We estimate from our measurements that the wild type strain PAO1 has an average intracellular concentration of 0.7 μM c-di-GMP and the wspF mutant has an intracellular concentration of c-di-GMP of about 3.0 μM. The fleQ and fleN mutants had intracellular c-di-GMP levels that were slightly lower than those of the wild-type strain (Figure 3). The intracellular concentration of c-di-GMP in the fleQ wspF double mutant was essentially the same as those measured in a strain with only a wspF mutation. Levels of c-di-GMP in the fleN wspF double mutant were slightly lower than in the wspF mutant alone. These small changes in c-di-GMP levels likely do not cause the large changes in gene expression that we observed in the fleQ and fleN mutants. This indicates that the effects of the fleQ mutation in causing elevated pel, psl, PA2441, and PA4625 transcription are not due to a secondary effect of this mutation causing increases in intracellular c-di-GMP.
Figure 3. Loss of FleQ or FleN does not cause elevated c-di-GMP levels.
C-di-GMP was extracted from whole cells and levels were measured as described in the Experimental Procedures. Strains are wild type (PAO1), a fleQ mutant, a wspF mutant, a fleQ wspF mutant, a fleN mutant, and a fleN wspF mutant. Data represent averages of three independent cultures. Error bars are the standard deviations between independent cultures.
FleQ binds to pel promoter DNA
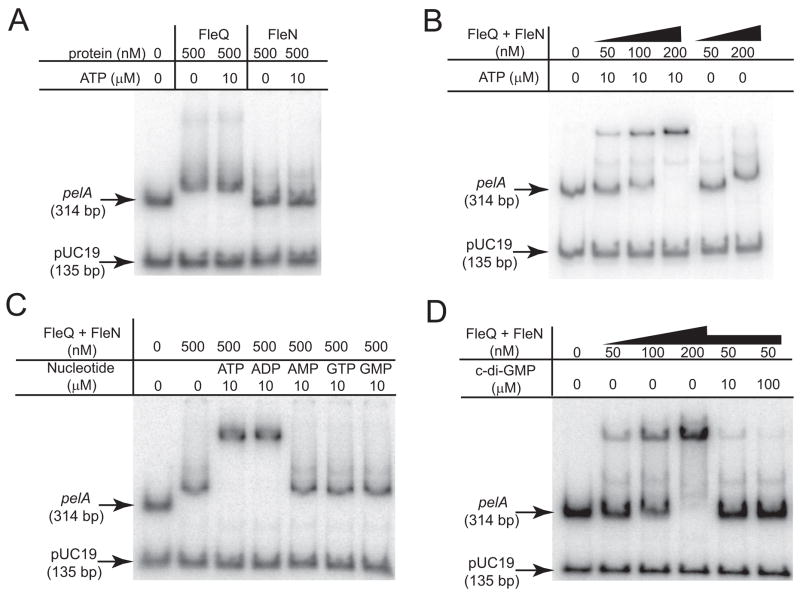
Although there are relatively few examples of bacterial enhancer binding proteins like FleQ directly repressing transcription, it is not unprecedented for members this class of protein to act as transcriptional repressors (Chang et al., 2007; North et al., 1996; Weiss et al., 1991). To test the possibility that FleQ directly represses transcription, we purified FleQ and assayed its ability to bind to pelA promoter DNA in an electrophoretic mobility shift assay (EMSA). We incubated purified FleQ with a fragment of DNA spanning −284 to +30 base pairs relative to translational start of pelA. The addition of FleQ to reaction mixtures caused a shift in the mobility of the pel promoter DNA fragment (Figure 4A). FleQ did not affect the mobility of a 135 base pair fragment from the plasmid pUC19 that we included as a negative control (Figure 4A). Although FleQ was able to bind specifically to the pel promoter region, relatively large amounts of protein (500 nM) were required to see even a slight shift in DNA mobility, suggesting that FleQ is a relatively poor DNA binding protein. This is in agreement with previous reports showing that purified FleQ bound poorly to the promoters of several genes for flagella biosynthesis (Dasgupta and Ramphal, 2001; Jyot et al., 2002).
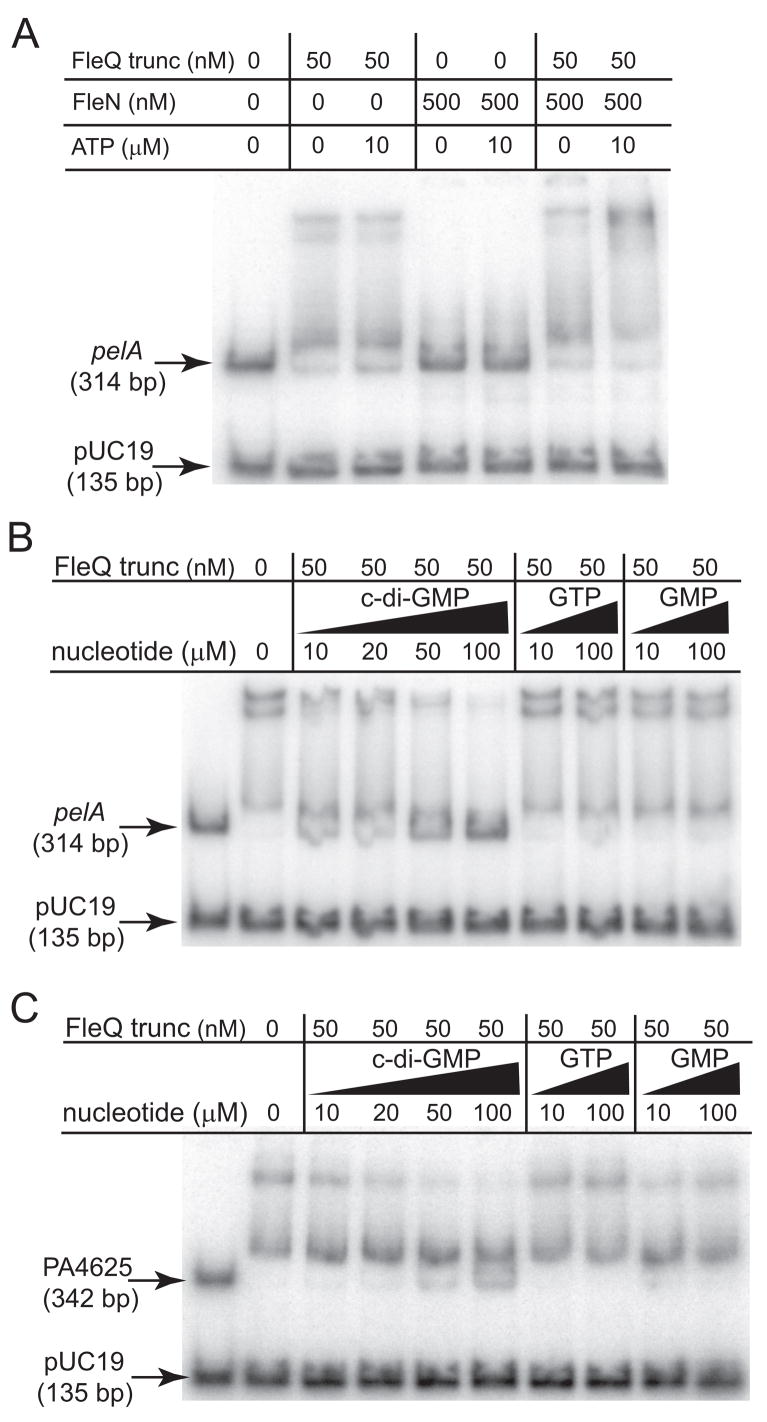
Figure 4. FleQ binds to the pel promoter and binding is inhibited by c-di-GMP.
A) Binding of FleQ or FleN to the pelA promoter in the absence and presence of ATP (10 μM). A fragment from the pUC19 vector is included as a negative control. The migration of unbound pelA promoter or pUC19 DNA though the gel is indicated by arrows. The concentrations of FleQ and FleN are indicated. B) The effect of FleN and ATP on FleQ-DNA complex formation. The FleQ and FleN proteins were provided in equimolar amounts at the concentrations indicated. ATP was added at a concentration of 10 μM where indicated. C) The effect of different nucleotides on FleQ-FleN-DNA complex formation. The nucleotide added and its concentration is indicated above each lane. D) Binding of FleQ and FleN to the pel promoter in the absence and presence of c-di-GMP. FleQ and FleN were provided in equimolar amounts at the concentrations indicated. c-di-GMP was added where indicated. All reactions contained 10 μM ATP.
FleN binds with FleQ to pel promoter DNA in an ATP-dependant manner
Previous work showed that FleN and FleQ physically interact with each other and that addition of FleN, together with FleQ, to EMSA reactions retarded the migration of fhlA promoter DNA through acrylamide gels more than did FleQ alone (Dasgupta and Ramphal, 2001). This was interpreted to mean that FleN binds to FleQ at the fhlA promoter to form a higher molecular weight complex. To test possible effects of FleN on pel promoter DNA migration though gels, we purified FleN and added it to our EMSA reactions either alone, or in equimolar amounts with full length FleQ. FleN alone had a barely detectable effect on pelA promoter migration in the EMSA assays (Figure 4A). FleN has a predicted nucleotide-binding site and is a predicted ATPase. When we added 10 μM ATP to FleN alone or to FleQ alone in gel shift reactions, we saw no effect of ATP on the ability of either of these proteins to bind the pelA promoter (Figure 4A). In contrast, the inclusion of FleN in reaction mixtures together with FleQ resulted in the formation of a complex with a slower mobility than that observed with FleQ alone when 10 μM ATP was present (Figure 4B). ADP also enhanced the ability of FleQ-FleN to shift pelA promoter DNA (Figure 4C). However the inclusion of AMP, GTP, or GMP in reactions had no effect on the ability of FleQ-FleN to shift the pelA promoter (Figure 4C).
C-di-GMP abrogates binding of the FleQ-FleN complex to pel promoter DNA
If c-di-GMP and FleQ regulate the transcription of the pel operon through a common system, then one would predict that the addition of c-di-GMP to EMSA reaction mixtures might influence the binding of FleQ and FleN to pel promoter DNA. We found that the addition of c-di-GMP at concentrations as low as 10 μM resulted in a decrease in the ability of FleQ-FleN to bind pel promoter DNA as assayed by retarded migration of the protein-DNA complex though gels (Figure 4D). These results indicate that FleQ-FleN constitute a c-di-GMP-responsive transcription regulatory complex. C-di-GMP addition did not influence the small shift in pelA mobility observed with 500 nM FleQ alone (data not shown).
We also tested if c-di-GMP addition affected the ability of FleQ-FleN to shift the promoter of the flagella genes fleSR. While FleQ-FleN shifted fleSR promoter DNA in the presence of ATP, we saw little to no effect of c-di-GMP on the ability of FleQ-FleN to shift fleSR promoter DNA (Supplementary Figure 1A, B). This result is consistent with our observation that elevated c-di-GMP resulted in a very small decrease of fleR transcript levels in vivo (Figure 2E).
N-terminally truncated FleQ has enhanced DNA-binding properties
Enhancer binding proteins like FleQ often have an N-terminal domain that controls the DNA binding properties of the protein in response to a variety of stimuli including small molecules or other proteins (Chen et al., 2005; Keener and Kustu, 1988; Little and Dixon, 2003; Martinez-Argudo et al., 2004). We constructed and purified a truncated version of FleQ that lacked its 126 amino acid N-terminal domain and tested it in EMSA reactions. We found that truncated FleQ bound to the pel promoter region with much higher affinity than the full-length protein (Figure 5A). A concentration of 10-fold less truncated FleQ (50 nM) gave a greater mobility shift than that observed with full length FleQ. As with full length FleQ, addition of ATP to truncated FleQ alone did not affect the ability of this protein to bind the pelA promoter (Figure 5A). The addition of FleN and ATP along with truncated FleQ in reaction mixtures resulted in a small increase in the shift of pelA promoter (Figure 5A), but the effect was much less than that seen with full length FleQ. In contrast to the situation with pelA, truncated FleQ did not bind with increased affinity to the fleSR promoter (Supplementary Figure 1C).
Figure 5. FleQ lacking its N-terminal domain has increased DNA binding and its binding is inhibited by c-di-GMP.
A) Binding of truncated FleQ (FleQ-trunc) or FleN to the pelA promoter in the absence and presence of ATP (10 μM). The amount of protein added is indicated above the lane. B) Binding of truncated FleQ to the pelA promoter in the presence of c-di-GMP, GTP, and GMP. The nucleotide and amount of each added is indicated above each lane. All reactions contained 50 nM truncated FleQ and 1 mM ATP. C) Binding of truncated FleQ to the PA4625 promoter in the presence of c-di-GMP, GTP, and GMP. The nucleotide and amount added is indicated above each lane. All reactions contained 50 nM truncated FleQ and 1 mM ATP.
C-di-GMP inhibits DNA binding by N-terminally truncated FleQ
We found that the addition of c-di-GMP to truncated FleQ abrogated the ability of this form of the protein to retard the migration of pelA promoter DNA in EMSA reactions. C-di-GMP concentrations as low as 10 μM affected the shift, with an almost complete inhibition of DNA binding at 100 μM in both the presence and absence of 1 mM ATP (Figure 5B and data not shown). GTP or GMP at concentrations of 10 μM had no effect on the gel mobility shift of pelA promoter DNA by truncated FleQ, and 100 μM of either of these nucleotides had a very small effect on DNA binding by truncated FleQ (Figure 5B). We also tested the ability of truncated FleQ to bind to the promoter of PA4625, another gene whose expression is co-regulated by FleQ and c-di-GMP. Truncated FleQ also specifically retarded the migration of this promoter fragment through gels and addition of c-di-GMP inhibited the binding of FleQ to PA4625 promoter DNA (Figure 5C). These results demonstrate that c-di-GMP acts directly on FleQ to control DNA binding at multiple promoters, FleN is not required for this effect, and that the N-terminal domain is dispensable for c-di-GMP-mediated inhibition of DNA binding by FleQ.
FleQ binds c-di-GMP in vitro
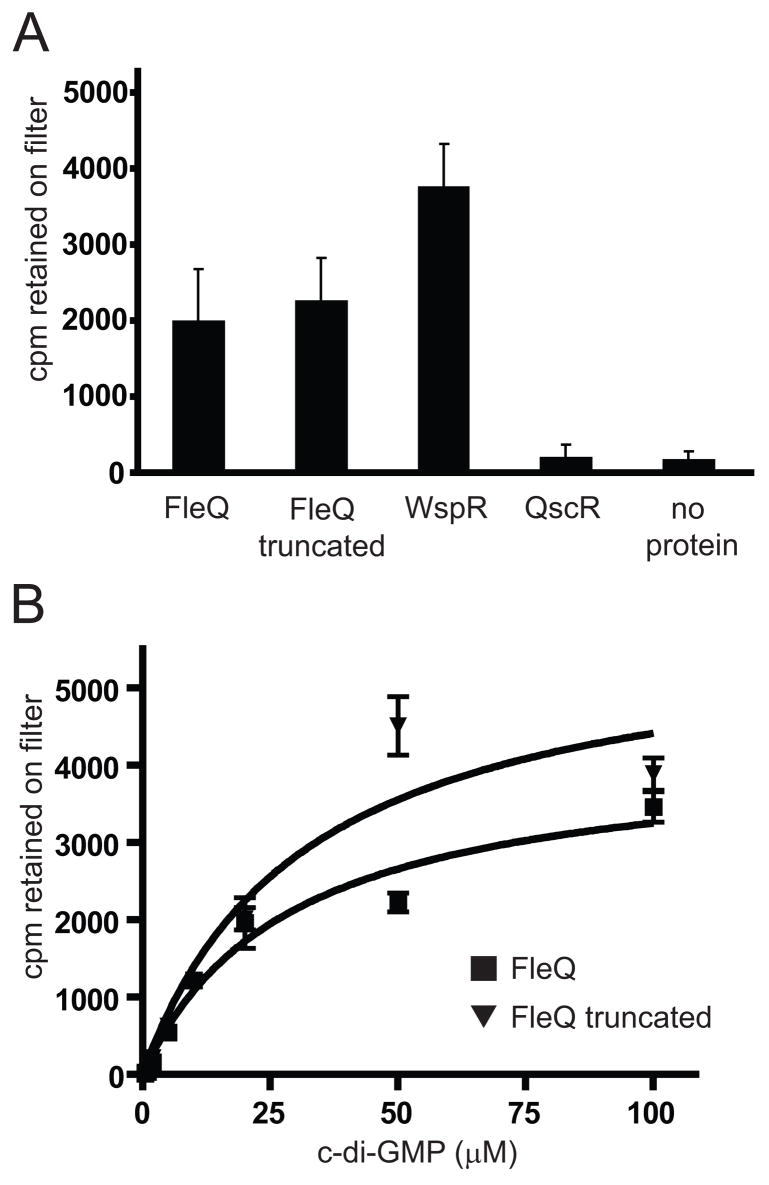
We carried out direct binding assays to confirm and expand our results indicating that FleQ binds c-di-GMP. FleQ (2 μM) incubated with 20 μM [32P]-c-di-GMP in filter binding assays bound approximately 20 times more c-di-GMP than a no protein control or reactions containing the P. aeruginosa transcriptional regulator QscR (Figure 6A). QscR is not known to bind c-di-GMP or any other nucleotide compounds, and did not bind more c-di-GMP than the no-protein control (Chugani et al., 2001; Ledgham et al., 2003; Lee et al., 2006; Lequette et al., 2006). The diguanylate cyclase WspR, bound approximately 1.9 fold more c-di-GMP than FleQ under the same conditions (Figure 6A). WspR has a c-di-GMP binding site, the I-site, that is involved in product inhibition of enzymatic activity by c-di-GMP (De et al., 2008). Truncated FleQ bound as much c-di-GMP as full length FleQ under these conditions, consistent with the model that the N-terminal domain has no role in c-di-GMP binding by FleQ (Figure 6A). Both FleQ and truncated FleQ bound c-di-GMP in a concentration dependant manner over a range of 0.5 to 100 μM c-di-GMP (Figure 6B). Both FleQ and truncated FleQ displayed binding characteristics consistent with first order kinetics. Half-maximal binding of c-di-GMP by FleQ occurred at a concentration of approximately 15–25 μM (Figure 6B).
Figure 6. FleQ binds c-di-GMP in vitro.
A) Binding of radiolabeled c-di-GMP by FleQ truncated FleQ, WspR (positive control), QscR (negative control), and no protein control. All reactions contained 20 μM [32P]–c-di-GMP. Data are presented as cpm retained on the nitrocellulose filter. B) Concentration dependent binding of [32P]–c-di-GMP by FleQ (squares) and truncated FleQ (triangles). All data are the average of at least three independent binding reactions. Error bars represent the standard deviations between replicates.
The addition of unlabeled c-di-GMP at ten-fold excess to reaction mixtures inhibited the binding of [32P]-c-di-GMP to FleQ or truncated FleQ, indicating that FleQ interacts specifically with this dinucleotide (Table 1). By contrast, addition of ATP, GMP, or GTP at ten fold excess concentration had little effect on the binding of [32P]-c-di-GMP to FleQ or truncated FleQ (Table 1). ATP or GTP partially competed with c-di-GMP for binding to full length FleQ when provided at 100-fold excess relative to radiolabled c-di-GMP, however these two nucleotides had a much smaller effect on c-di-GMP binding by truncated FleQ (Table 1). GMP at 100-fold excess had no effect on c-di-GMP binding by full length FleQ, and a relatively small effect on binding by truncated FleQ (Table 1).
Table 1.
Relative c-di-GMP binding by FleQ in the presence of competing nucleotides
| Competitor a | FleQ full length b | FleQ truncated c |
|---|---|---|
| None | 100 (35) | 100 (26) |
| 10 × c-di-GMP | 23 (9) | 20 (4) |
| 10 × ATP | 97 (12) | 83 (9) |
| 10 × GMP | 109 (14) | 112 (33) |
| 10 × GTP | 87 (17) | 111 (27) |
| 100 × ATP | 47 (17) | 77 (20) |
| 100 × GMP | 104 (3) | 84 (15) |
| 100 × GTP | 42 (10) | 76 (23) |
Nonradioactive nucleotides added to binding reactions at 10-fold (10X) and 100-fold (100X) excess concentrations over [32P]-c-di-GMP (20 μM). Results are the average of at least three independent binding reactions
Relative binding of [32P]-c-di-GMP by full length FleQ in the absence or presence of competing nucleotides. Numbers indicate percent binding relative to the no competitor control, which is set to 100 percent. Standard deviations between replicates are indicated in parentheses.
Relative binding of [32P]-c-di-GMP by truncated FleQ in the absence or presence of competing nucleotides. Numbers indicate percent binding relative to the no competitor control, which is set to 100 percent. Standard deviations between replicates are indicated in parentheses.
Discussion
Here we have presented three lines of evidence that c-di-GMP binds to the transcriptional regulator FleQ to cause it to derepress the expression of pel and other EPS genes necessary for biofilm formation. First, a fleQ mutation and high c-di-GMP each caused an increase in the relative levels of pelA, pslA, PA2441, and PA4625 transcripts and the effects of these two conditions were not additive (Figure 2). This suggests that FleQ and c-di-GMP act through a common mechanism. Second, electromobility shifts of pelA promoter DNA in the presence of FleQ and FleN, or FleQ lacking its N-terminal domain, were abrogated by addition of c-di-GMP. This suggests that c-di-GMP prevents binding of FleQ to pelA promoter DNA (Figures 4 and 5). The effect of c-di-GMP on the binding of truncated FleQ to the promoter region of PA4625 was similar to that seen at the pel promoter. Finally, radiolabeled c-di-GMP bound specifically to purified FleQ protein (Figure 6 and Table 1). Also, truncated FleQ bound c-di-GMP as well as full length protein, indicating that the N-terminal domain is dispensible for c-di-GMP binding (Figure 6 and Table 1).
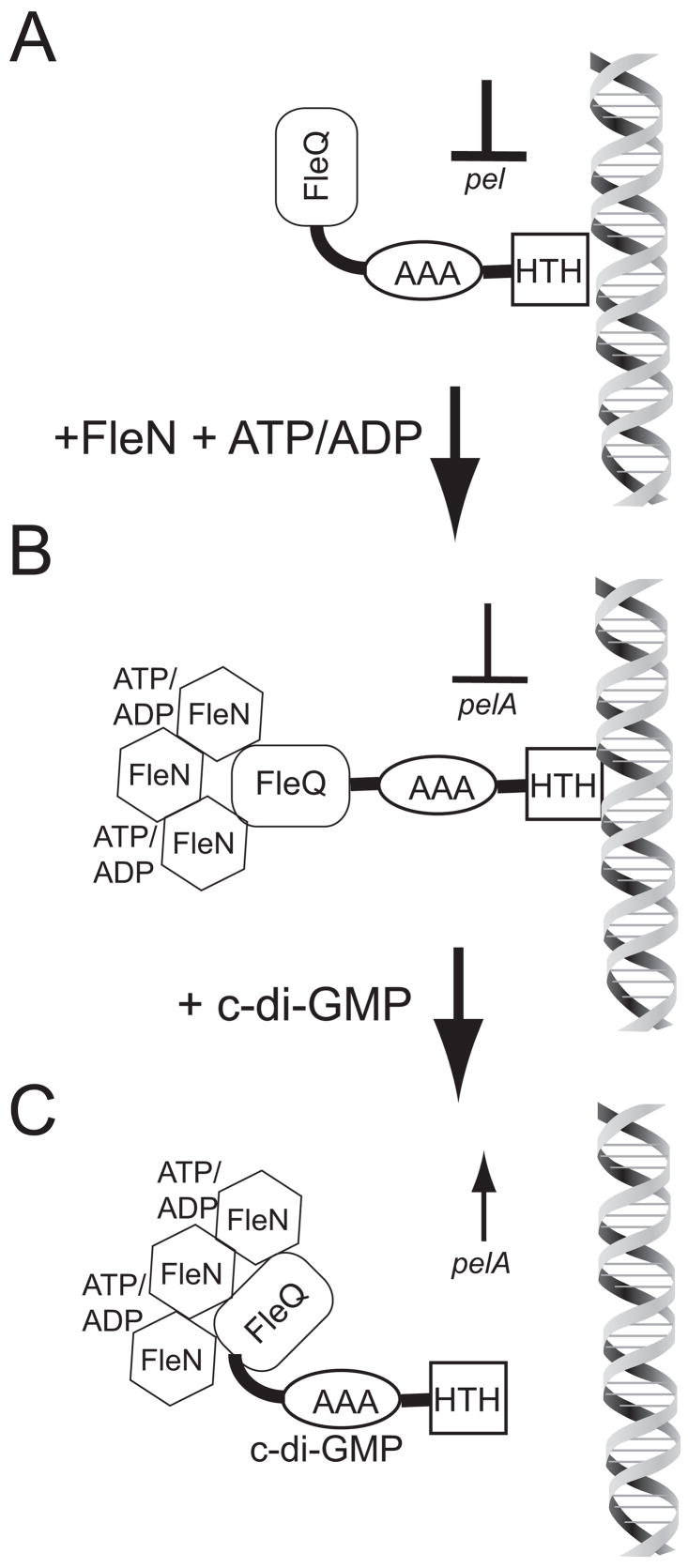
Our data on FleQ-mediated regulation of pelA expression are consistent with the model shown in Figure 7. We propose that in the absence of FleN or ATP, FleQ maximally represses pelA expression (Figure 7A). When FleN and ATP (or ADP) are present they partially inhibit the ability of FleQ to repress transcription of pelA (Figure 7B). FleN is a predicted ATPase with 35% sequence identity to the cell division inhibitor MinD and 25% identify to nitrogenase reductase NifH. By analogy with the known modes of action of these proteins (Lutkenhaus, 2007; Schindelin et al., 1997), we hypothesize that ATP/ADP promotes multimerization of FleN and binding of FleN to FleQ, leading to inhibition of FleQ activity. C-di-GMP stimulates the derepression of pel transcription. One possible mechanism for this is that binding of c-di-GMP to FleQ stimulates a change in protein complex conformation that results in the dissociation of FleQ from promoter DNA (Figure 7C).
Figure 7. Model for the regulation of gene expression by FleQ, FleN, and c-di-GMP.
A) FleQ in the absence of FleN or c-di-GMP maximally represses pel transcription. B) The situation in wild-type cells; FleQ binding at the pelA promoter is reduced by FleN and ATP/ADP, resulting in less pel repression than the situation in panel A. C) C-di-GMP binds to FleQ to cause it to dissociate from DNA thereby causing derepression of transcription from the pel promoter.
Our data indicate that FleQ has different effects at promoters it represses (like pel) as compared to promoters it activates (like fleSR). While removal of the N-terminal domain of FleQ enhanced DNA binding at pel and PA4625, it had little effect on binding to the fleSR promoter. Also, elevated c-di-GMP had a much greater effect on promoters that FleQ represses (pel) compared to those that it activates (fleSR). The mechanism of transcriptional regulation by FleQ at activated and repressed promoters is likely to differ in significant ways because transcriptional activation by FleQ is dependant on the alternative RNA polymerase sigma factor σ54, whereas published array data indicate that transcriptional repression of pel, psl, PA4625, and other genes by FleQ does not depend on RNA polymerase σ54 (Dasgupta et al., 2003). These differences in sigma factor participation may account for why c-di-GMP has such a small effect on activation of σ54-dependent genes including flagella genes and much larger effects, as much as 20-fold effects, on derepressing expression of genes that are transcriptionally repressed by FleQ in a σ54- independent manner. In addition, it seems plausible that the number and position of FleQ binding sites may be different between repressed and activated promoters. Only a few FleQ binding sites have been identified to date, all of which are upstream of flagella genes that FleQ activates (Jyot et al., 2002). No consensus DNA sequence for FleQ binding was discernable from these previous studies. Therefore it is difficult at this point to use bioinformatics to predict possible FleQ binding sites in the promoter regions of repressed genes.
FleQ homologues transcriptionally regulate flagella gene expression in other polarly flagellated γ-proteobacteria including other Pseudomonas species, Vibrio, and Legionella species (McCarter, 2006) and it is possible these homolgous proteins also regulate exopolysaccharide gene transcription in response to c-di-GMP. Recently, P. fluorescens FleQ was reported to negatively regulate expression of the wss operon necessary for cellulose production. Interestingly, expression of these genes is also activated by high c-di-GMP levels (Giddens et al., 2007).
FleQ represents the first member of a new class of c-di-GMP binding proteins that are transcriptional regulators. The fact that FleQ lacking its N-terminal domain binds c-di-GMP as well as the full-length protein indicates that c-di-GMP binds somewhere outside the N-terminal domain. The in vitro system that we have established for examining binding of FleQ to pelA promoter DNA provides an opportunity to examine in detail the mechanism by which c-di-GMP controls the biochemical activity of a transcriptional regulator. The FleQ protein does not contain any regions that have a predicted secondary structure that resembles those of known c-di-GMP binding regions, including the PelD protein from P. aeruginosa, PilZ domains, or the I-sites of diguanylate cyclases. (Amikam and Galperin, 2006; Chan et al., 2004; De et al., 2008; Wassmann et al., 2007). A detailed characterization of FleQ is expected to reveal alternative determinants of c-di-GMP binding.
Experimental procedures
Strains and growth conditions
Strains and plasmids used in this study are listed in Table 2. Primer sequences are available upon request. P. aeruginosa and E. coli strains were routinely cultivated on LB medium at 37°C unless otherwise indicated. Colony morphology was visualized on tryptone-congo red agar plates (10 g tryptone, 40mg Congo red, 10 mg comassie brilliant blue R-250 per liter) after 5 days growth at 25°C (Friedman and Kolter, 2004b). Antibiotics for E. coli and P. aeruginosa were added where appropriate. These were 100 μg/ml ampicillin for E. coli and 50 μg/ml gentamycin for P. aeruginosa.
Table 2.
Strains and Plasmids
| Strain or plasmid | Relevant phenotype or genotype | Source or reference |
|---|---|---|
| P. aeruginosa | ||
| PAO1 | Wild-type strain; twitching motility+ | (Jacobs et al., 2003) |
| PAO1100 | PAO1 derivative, in-frame deletion of wspF | (Hickman et al., 2005) |
| fleQ::Tn5 | PAO1 derivative, fleQ::Tn5; TcR | (Jacobs et al., 2003) |
| fleN::Tn5 | PAO1 derivative, fleN::Tn5; TcR | (Jacobs et al., 2003) |
| PAO1110 | fleQ::Tn5 with in-frame deletion of wspF | This study |
| PAO1111 | fleN::Tn5 with in-frame deletion of wspF | This study |
| PAO1114 | fleQ::Tn5 with deletion of pelA and pslBCD | This study |
| E. coli | ||
| DH5α | supE44 ΔlacU169 (φ80 lacZΔM15) hsdR178 recA1 endA1 gyrA96 thi-1 relA1 | Gibco-BRL |
| S17-1 | C600::RP-4 2-(Tc::Mu) (Kn::Tn7) thi pro hsdR hsdM+ recA | (Simon et al., 1983) |
| ER2566 | F− lamda− fhuA2 [lon] ompT lacZ:: T7 gene1 gal sulA11Δ(mcrC-mrr)114::IS10 R(mcr-73::miniTn 10--TetS)2 R(zgb-210::Tn10) (TetS) endA1 [dcm] | NEB |
| Plasmids | ||
| pTYB12 | Apr; vector used for n-terminal Intein-chitin-binding domain fusions | NEB |
| pFleQ-Int1 | Apr; NdeI-EcoRI fragment containing fleQ cloned into NdeI-EcoRI digested pTYB12 | This study |
| pFleN-Int1 | Apr; NdeI-EcoRI fragment containing fleN cloned into NdeI-EcoRI digested pTYB12 | This study |
| pFleQ-trunc-Int1 | Apr; NdeI-EcoRI fragment containing a truncated fleQ cloned into NdeI-EcoRI digested pTYB12 | This study |
| pJN105 | Gmr; araC-PBAD cassette cloned in pBBR1MCS-5 | (Newman and Fuqua, 1999) |
| pJNFleQ | Gmr; fleQ cloned as a 1.7 Kb EcoRI-SmaI fragment into EcoRI-SmaI digested pJN105 | This study |
| pJN1120 | Gmr; PA1120 cloned as a 1.3 Kb NheI-XbaI fragment into NheI-XbaI digested pJN105 | This study |
| pMPSL-KO1 | Apr, Gmr; Pseudomonas suicide vector containing deletion construct for pslBCD genes | M. Starkey, unpublished |
| pMPELA | Apr, Gmr; Pseudomonas suicide vector containing deletion construct for pelA gene | M. Starkey, unpublished |
Plasmid and strain construction
All primer sequences are available upon request. Plasmid pJNFleQ was constructed by ligating an EcoRI-SmaI fragment from plasmid pAT6 containing the FleQ gene into EcoRI-SmaI digested pJN105 (Tart et al., 2005). Plasmid pJN1120 was constructed by amplification of PA1120 with primers homologous to the 5′ and 3′ ends of PA1120 containing restriction sites for NheI and XbaI respectively. The PCR product was cloned into NheI-XbaI digested pJN105 and the resulting plasmid, pJN1120, was verified by sequencing. Plasmids were electroporated into wild-type PAO1 and the fleQ mutant using established protocols (Choi et al., 2006).
Deletions of pelA and pslBCD in a fleQ mutant were constructed using standard procedures for allelic exchange in P. aeruginosa. Two plasmids, pMPSL-KO1 and pMPELA, containing the deletion constructs (M. Starkey and M. Parsek, unpublished) were mated into a fleQ mutant and strains were selected on Pseudomonas isolation agar containing 50 gg/ml gentamycin. Double recombinant mutants were selected on LB plates containing 5% sucrose. Mutants were confirmed by PCR.
RNA isolation and real-time quantitative PCR
P. aeruginosa strains were grown in 5 ml cultures in test tubes overnight then subcultured twice into 125 ml baffled flasks to an OD600 of 0.02. After the second subculture and subsequent growth, cells were harvested at an OD600 of 0.4–0.6 for RNA isolation. RNA was isolated and cDNA synthesized using previously published procedures (Schuster et al., 2003). Primers for RT-PCR were designed using the program PrimerExpress (Applied Biosystems). Reactions were performed using SybrGreen master mix (Applied Biosystems) with 1 ng cDNA as template and 200 nM of each primer in a final reaction volume of 25 μl. Cycling parameters were 95°C for 10 min, followed by 40 cycles of 95°C for 15 sec and 60°C for 1 min. A final dissociation curve of all amplified products was performed as a quality control check. Standard curves for quantitation of transcripts were performed using dilutions of P. aeruginosa chromosomal DNA from 1 ×10−4 ng to 10 ng (Lequette et al., 2006). Transcript levels of all genes tested were normalized to transcript levels of the ampR gene (Lequette et al., 2006). Data presented are averages of three independent cultures grown on different days. Error bars represent the standard deviation between samples.
Quantitation of c-di-GMP levels by LC-MS
Cells were grown overnight in VBMM (Vogel-Bonner minimal medium) and subcultured the next day to an OD600 of 0.02 in 125 ml baffled shake flasks containing 15 ml of VBMM. VBMM, a defined medium, was used for c-di-GMP quantitation because an unknown component of LB medium co-eluted with c-di-GMP and masked the c-di-GMP signal during LC-MS analysis. When cells reached an OD600 of 0.4–0.6, 1 ml was removed, centrifuged for 1 min at 15,000 × g, and the supernatant was removed. The cell pellet was resuspended in 100 μl 0.6 M perchloric acid. Samples were incubated on ice for 30 min, and cell debris was removed by microcentrifugation at 4°C for 5 min at 15000 × g. Supernatants (100 μl) were removed and samples were neutralized by addition of 20 μl 2.5 M KHCO3. The resulting precipitate was removed by centrifugation at 4°C for 5 min at 15,000 × g. These neutralized supernatants were stored at −80°C until analyzed by LC-MS. For protein determinations, three 1-ml samples of each culture were removed and centrifuged to pellet cells. Cell pellets were resuspended in 100 μl 1M NaOH and boiled in a water bath for 10 minutes. Samples were then stored on ice until protein was assayed. Standard Bradford protein assays were carried on all samples in duplicate using the Bio-Rad Protein assay reagent (Bio-Rad, Hercules, CA). Bovine serum albumin (BSA) containing 1 M NaOH was used as a standard.
Samples were analyzed at the University of Washington Mass Spectrometry Center using an HPLC/MS/MS system. Protocols for c-di-GMP separations and analysis by mass spectrometry were based on a previously published method (Thormann et al., 2006). Separation was achieved with an Acuity UPLC (Waters; Milford, MA) using a 2.1 × 50 mm Synergi Hydro RP column (Phenomenex; Torrance, CA). A gradient system was used starting from 98% aqueous (10 mM ammonium formate, pH 4.0) and 2% organic (acetonitrile). The aqueous went to 10% at 6.0 minutes, 95% at 6.5 minutes, 10% at 7.0 minutes and 98% at 7.5 minutes. Temperature was uncontrolled, flow was 0.3 ml/min and cycle time was 10 minutes. The extra step from high to low aqueous and back again between 6.5 and 7.5 minutes served to eliminate carry-over between injections. The c-di-GMP was detected by MS/MS multiple reaction monitoring (MRM) using a Premiere XL triple quadrapole mass spectrometer (MicroMass; Milford, MA) in positive electrospray ionization (API-ES+). The m/z 691>152 transition was used for quantitation; 691>248 and 691>540 were monitored as confirmatory signals. The collision energies were 30, 24, and 24 eV respectively; the cone voltages were 40 V in all cases. For a standard curve, 50, 100, 250, 500, and 1000 fmol pure c-di-GMP (Axxora, San Diego, CA) were analyzed by the above method. C-di-GMP levels are normalized to total protein per ml of culture. Data represent averages of three independent cultures. Error bars are the standard deviation between replicate cultures. Intracellular concentrations of c-di-GMP were estimated from this data assuming that 1 mg of protein corresponds to 6.7 × 109 cells and that each P. aeruginosa cell has a volume of 1 × 10−15 liters.
Protein purification
FleQ, FleQ lacking its N-terminal domain (FleQ-trunc), and FleN were purified using the IMPACT system from NEB (Beverly, MA). The fleQ, fleQ-trunc, and fleN genes were each cloned into pTYB12 as NdeI-EcoRI fragments generating plasmids pFleQ-Int1, pFleQ-trunc-Int1, and pFleN-Int1. This generated N-terminal fusions of the Intein-Chitin binding domain (Intein-CBD) to FleQ, FleQ-trunc, and FleN, respectively. This resulted in the addition of Ala, Gly, and His residues to the N-terminus after cleavage of the Intein-CBD tag. Plasmids were transformed into E. coli ER2566 for expression of fusion proteins. For overexpression of the fusion proteins, E. coli carrying either pFleQ-Int1, pFleQ-trunc-Int1, or pFleN-Int1 were subcultured from overnights into 1 L of fresh LB in 3 L Fernbach flasks. Cultures were grown to an OD600 of 0.4 and then shifted to 15°C. After 30 minutes at 15°C, 0.3 mM IPTG was added to induce expression of fusion proteins, and cultures were grown for 16 h. Cells were harvested by centrifugation at 5000 × g, pellets resuspended in column buffer with Triton X-100 (20 mM Tris-Cl pH 8.0, 250 mM KCl, 0.1 mM EDTA, 0.1% (v/v) Triton X-100), and cells were lysed by sonication. Lysates were clarified by centrifugation at 15,000 × g for 30 minutes. The solubility of the expressed proteins was checked by analyzing soluble and insoluble fractions by SDS-PAGE. Expression at 15°C prevented the insolubility of overexpressed FleN that had been previously described (Dasgupta and Ramphal, 2001).
Intein-CBD fusions were purified as previously described with the following modifications (Hickman et al., 2005). Proteins were loaded in column buffer plus Triton X-100 onto a 5 ml chitin bead column. Columns were washed with 50 ml column buffer plus Triton, then washed with 50 ml column buffer lacking Triton X-100. Cleavage of the Intein-CBD was induced by addition of column buffer containing 50 mM DTT or β-mercaptoethanol and incubating overnight at room temperature. When necessary, protein was concentrated using Amicon ultrafiltration devices (Millipore, Billerica, MA.) Proteins were dialyzed into column buffer containing 50% glycerol and stored at −20°C. Both FleQ and FleN were stable under these storage conditions for up to 2 months.
In vitro DNA binding analysis
The pel promoter region spanning −284 to +30 base pairs relative to translational start of pelA was PCR amplified. As a negative control a 135 base pair fragment of pUC19 vector was also PCR amplified. These DNA fragments were end labeled with T4 polynucleotide kinase and γ[32P]-ATP. Equal amounts of each of these labeled DNA fragments (4 fmol, final total probe concentration 100 pM) were added to binding reactions with varying amounts of FleQ or FleQ lacking its N-terminal domain (FleQ-trunc) in binding buffer (10 mM Tris, pH 7.8, 8 mM magnesium acetate, 50 mM KCl, 5% glycerol, 250 ng/μl BSA, 20 μl total reaction volume). FleQ was incubated with DNA for 30 minutes on ice. Reactions carried out with FleN were performed as above with the addition of equimolar amounts of FleN and FleQ. Where indicated, 10 μM ATP or other nucleotides were also added to the reactions and incubated with the proteins for 30 minutes on ice prior to addition of DNA. Reactions containing c-di-GMP were performed as described above except c-di-GMP was incubated with proteins for 30 minutes before addition of DNA. All reactions mixtures were loaded onto a 5 % acrylamide gel containing 10 mM Tris-Cl pH 8.0, 400 mM glycine, 5 mM EDTA, and electrophoresed at 70 V at room temperature for 1–1.5 h. Gels were dried and exposed to a phosphorimaging screen overnight, visualized on a Storm phosphorimager (GE Healthcare, Pistcataway, NJ), and images analyzed using ImageQuant software (GE Healthcare, Pistcataway, NJ).
C-di-GMP binding assays
[32P]-c-di-GMP was generated using purified WspR protein using reaction conditions previously described (Hickman et al., 2005; Merighi et al., 2007) Reactions (100 μl total volume) contained 10 μM WspR protein, 250 μCi α[32P]-GTP and 2.5 mM acetyl phosphate in reaction buffer (75 mM Tris-Cl pH 7.8, 250 mM NaCl, 25 mM KCl, 10 mM MgCl2) and were incubated at 37°C for 2 h. Antarctic phosphatase (NEB, Beverly, MA) was added and reactions were incubated an additional 30 minutes to remove any residual α[32P]-GTP. Reactions were then heated to 95°C for 5 min and spun at 15000 × g for 5 min to precipitate protein and supernatants were spun through a Millipore Ultrafree filter device (5000 MWCO) at 5000 × g for 30 minutes. The filtrate was loaded onto a Sep-Pak Light C18 cartridge (Waters, Milford, MA), washed with 0.5 × reaction buffer containing 1% MeOH, and [32P]-c-di-GMP was eluted with 50% MeOH. Fractions from the Sep Pak cartridge were dried using a Speed-Vac and resuspended in 50 μl water. Samples were run on PEI-cellulose TLC plates using 1.5 M KH2PO4 pH 3.5 to determine that pure c-di-GMP was obtained (Hickman et al., 2005).
The binding of c-di-GMP by proteins was assayed using a filter binding technique. Protein (2 μM FleQ, FleQ truncated, WspR, or QscR) was incubated with 20 μM [32P]-c-di-GMP (0.2 Ci/mmol) in binding buffer (40 mM Tris pH 7.8, 10 mM magnesium acetate, 50 mM KCl.) Reactions were incubated on ice for 25 min then filtered through a 0.2 μM nitrocellulose membrane (Whatman, Dassel, Germany), using a slot blot apparatus (PR 600 SlotBlot, Hoeffer Scientific). Samples were washed with 1 ml cold reaction buffer, the membrane was removed, dried, and then individual wells were cut out and radioactivity was determined using a scintillation counter. Purified QscR from P. aeruginosa was used as a negative control and showed no binding of c-di-GMP above the no protein control. For competition experiments with ATP, GTP, GMP, or c-di-GMP, 2 μM FleQ or FleQ truncated was incubated with 20 μM [32P]-c-di-GMP for 12.5 minutes, then competing substrate was added and the reactions incubated another 12.5 minutes and processed as above. For binding curves, 2 μM FleQ or FleQ truncated was incubated with varying amounts of [32P]-c-di-GMP and reactions were processed as above.
Supplementary Material
Acknowledgments
Public Health Service Grant GM56665 from the National Institute of General Medical Sciences supported this work. JWH was supported by a Cystic Fibrosis Foundation Postdoctoral Fellowship (R565-CR02). We would like to thank Thomas F. Kalhorn and the University of Washington Mass Spectrometry Center for their invaluable assistance with c-di-GMP quantitation. Plasmids pMPSL-KO1 and pMPELA were generous gifts from Melissa Starkey and M. Parsek. Purified QscR protein was a generous gift from Ken-Ichi Oinuma and E. P. Greenberg. We would also like to thank Amy L. Schaefer and Breck A. Duerkop for reading the manuscript and for insightful discussions.
References
- Aldridge P, Paul R, Goymer P, Rainey P, Jenal U. Role of the GGDEF regulator PleD in polar development of Caulobacter crescentus. Mol Microbiol. 2003;47:1695–1708. doi: 10.1046/j.1365-2958.2003.03401.x. [DOI] [PubMed] [Google Scholar]
- Amikam D, Galperin MY. PilZ domain is part of the bacterial c-di-GMP binding protein. Bioinformatics. 2006;22:3–6. doi: 10.1093/bioinformatics/bti739. [DOI] [PubMed] [Google Scholar]
- Arora SK, Ritchings BW, Almira EC, Lory S, Ramphal R. A transcriptional activator, FleQ, regulates mucin adhesion and flagellar gene expression in Pseudomonas aeruginosa in a cascade manner. J Bacteriol. 1997;179:5574–5581. doi: 10.1128/jb.179.17.5574-5581.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyhan S, Tischler AD, Camilli A, Yildiz FH. Transcriptome and phenotypic responses of Vibrio cholerae to increased cyclic di-GMP level. J Bacteriol. 2006;188:3600–3613. doi: 10.1128/JB.188.10.3600-3613.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boles BR, McCarter LL. Vibrio parahaemolyticus scrABC, a novel operon affecting swarming and capsular polysaccharide regulation. J Bacteriol. 2002;184:5946–5954. doi: 10.1128/JB.184.21.5946-5954.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan C, Paul R, Samoray D, Amiot NC, Giese B, Jenal U, Schirmer T. Structural basis of activity and allosteric control of diguanylate cyclase. Proc Natl Acad Sci U S A. 2004;101:17084–17089. doi: 10.1073/pnas.0406134101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y, Tang T, Li JL. Isolation of a flagellar operon in Azospirillum brasilense and functional analysis of FlbD. Res Microbiol. 2007;158:521–528. doi: 10.1016/j.resmic.2007.04.005. [DOI] [PubMed] [Google Scholar]
- Chen S, Liu L, Zhou X, Elmerich C, Li JL. Functional analysis of the GAF domain of NifA in Azospirillum brasilense: effects of Tyr-->Phe mutations on NifA and its interaction with GlnB. Mol Genet Genomics. 2005;273:415–422. doi: 10.1007/s00438-005-1146-5. [DOI] [PubMed] [Google Scholar]
- Choi KH, Kumar A, Schweizer HP. A 10-min method for preparation of highly electrocompetent Pseudomonas aeruginosa cells: application for DNA fragment transfer between chromosomes and plasmid transformation. J Microbiol Methods. 2006;64:391–397. doi: 10.1016/j.mimet.2005.06.001. [DOI] [PubMed] [Google Scholar]
- Christen M, Christen B, Allan MG, Folcher M, Jeno P, Grzesiek S, Jenal U. DgrA is a member of a new family of cyclic diguanosine monophosphate receptors and controls flagellar motor function in Caulobacter crescentus. Proc Natl Acad Sci U S A. 2007;104:4112–4117. doi: 10.1073/pnas.0607738104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chugani SA, Whiteley M, Lee KM, D’Argenio D, Manoil C, Greenberg EP. QscR, a modulator of quorum-sensing signal synthesis and virulence in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A. 2001;98:2752–2757. doi: 10.1073/pnas.051624298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costerton JW. Introduction to biofilm. Int J Antimicrob Agents. 1999;11:217–221. doi: 10.1016/s0924-8579(99)00018-7. discussion 237–219. [DOI] [PubMed] [Google Scholar]
- Costerton JW, Stewart PS, Greenberg EP. Bacterial biofilms: a common cause of persistent infections. Science. 1999;284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- Costerton W, Veeh R, Shirtliff M, Pasmore M, Post C, Ehrlich G. The application of biofilm science to the study and control of chronic bacterial infections. J Clin Invest. 2003;112:1466–1477. doi: 10.1172/JCI20365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotter PA, Stibitz S. c-di-GMP-mediated regulation of virulence and biofilm formation. Curr Opin Microbiol. 2007;10:17–23. doi: 10.1016/j.mib.2006.12.006. [DOI] [PubMed] [Google Scholar]
- Dasgupta N, Arora SK, Ramphal R. fleN, a gene that regulates flagellar number in Pseudomonas aeruginosa. J Bacteriol. 2000;182:357–364. doi: 10.1128/jb.182.2.357-364.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasgupta N, Ramphal R. Interaction of the antiactivator FleN with the transcriptional activator FleQ regulates flagellar number in Pseudomonas aeruginosa. J Bacteriol. 2001;183:6636–6644. doi: 10.1128/JB.183.22.6636-6644.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasgupta N, Wolfgang MC, Goodman AL, Arora SK, Jyot J, Lory S, Ramphal R. A four-tiered transcriptional regulatory circuit controls flagellar biogenesis in Pseudomonas aeruginosa. Mol Microbiol. 2003;50:809–824. doi: 10.1046/j.1365-2958.2003.03740.x. [DOI] [PubMed] [Google Scholar]
- De N, Pirruccello M, Krasteva PV, Bae N, Raghavan RV, Sondermann H. Phosphorylation-independent regulation of the diguanylate cyclase WspR. PLoS Biol. 2008;6:e67. doi: 10.1371/journal.pbio.0060067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira RB, Antunes LC, Greenberg EP, McCarter LL. Vibrio parahaemolyticus ScrC modulates cyclic dimeric GMP regulation of gene expression relevant to growth on surfaces. J Bacteriol. 2008;190:851–860. doi: 10.1128/JB.01462-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman L, Kolter R. Two genetic loci produce distinct carbohydrate-rich structural components of the Pseudomonas aeruginosa biofilm matrix. J Bacteriol. 2004a;186:4457–4465. doi: 10.1128/JB.186.14.4457-4465.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman L, Kolter R. Genes involved in matrix formation in Pseudomonas aeruginosa PA14 biofilms. Mol Microbiol. 2004b;51:675–690. doi: 10.1046/j.1365-2958.2003.03877.x. [DOI] [PubMed] [Google Scholar]
- Garcia B, Latasa C, Solano C, Garcia-del Portillo F, Gamazo C, Lasa I. Role of the GGDEF protein family in Salmonella cellulose biosynthesis and biofilm formation. Mol Microbiol. 2004;54:264–277. doi: 10.1111/j.1365-2958.2004.04269.x. [DOI] [PubMed] [Google Scholar]
- Giddens SR, Jackson RW, Moon CD, Jacobs MA, Zhang XX, Gehrig SM, Rainey PB. Mutational activation of niche-specific genes provides insight into regulatory networks and bacterial function in a complex environment. Proc Natl Acad Sci U S A. 2007;104:18247–18252. doi: 10.1073/pnas.0706739104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goymer P, Kahn SG, Malone JG, Gehrig SM, Spiers AJ, Rainey PB. Adaptive divergence in experimental populations of Pseudomonas fluorescens. II. Role of the GGDEF regulator WspR in evolution and development of the wrinkly spreader phenotype. Genetics. 2006;173:515–526. doi: 10.1534/genetics.106.055863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickman JW, Tifrea DF, Harwood CS. A chemosensory system that regulates biofilm formation through modulation of cyclic diguanylate levels. Proc Natl Acad Sci U S A. 2005;102:14422–14427. doi: 10.1073/pnas.0507170102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoiby N. Understanding bacterial biofilms in patients with cystic fibrosis: current and innovative approaches to potential therapies. J Cyst Fibros. 2002;1:249–254. doi: 10.1016/s1569-1993(02)00104-2. [DOI] [PubMed] [Google Scholar]
- Jacobs MA, Alwood A, Thaipisuttikul I, Spencer D, Haugen E, Ernst S, Will O, Kaul R, Raymond C, Levy R, Chun-Rong L, Guenthner D, Bovee D, Olson MV, Manoil C. Comprehensive transposon mutant library of Pseudomonas aeruginosa. Proc Natl Acad Sci U S A. 2003;100:14339–14344. doi: 10.1073/pnas.2036282100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jyot J, Dasgupta N, Ramphal R. FleQ, the major flagellar gene regulator in Pseudomonas aeruginosa, binds to enhancer sites located either upstream or atypically downstream of the RpoN binding site. J Bacteriol. 2002;184:5251–5260. doi: 10.1128/JB.184.19.5251-5260.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kader A, Simm R, Gerstel U, Morr M, Romling U. Hierarchical involvement of various GGDEF domain proteins in rdar morphotype development of Salmonella enterica serovar Typhimurium. Mol Microbiol. 2006;60:602–616. doi: 10.1111/j.1365-2958.2006.05123.x. [DOI] [PubMed] [Google Scholar]
- Keener J, Kustu S. Protein kinase and phosphoprotein phosphatase activities of nitrogen regulatory proteins NTRB and NTRC of enteric bacteria: roles of the conserved amino-terminal domain of NTRC. Proc Natl Acad Sci U S A. 1988;85:4976–4980. doi: 10.1073/pnas.85.14.4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolter R, Greenberg EP. Microbial sciences: the superficial life of microbes. Nature. 2006;441:300–302. doi: 10.1038/441300a. [DOI] [PubMed] [Google Scholar]
- Kulasakara H, Lee V, Brencic A, Liberati N, Urbach J, Miyata S, Lee DG, Neely AN, Hyodo M, Hayakawa Y, Ausubel FM, Lory S. Analysis of Pseudomonas aeruginosa diguanylate cyclases and phosphodiesterases reveals a role for bis-(3′–5′)-cyclic-GMP in virulence. Proc Natl Acad Sci U S A. 2006;103:2839–2844. doi: 10.1073/pnas.0511090103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledgham F, Ventre I, Soscia C, Foglino M, Sturgis JN, Lazdunski A. Interactions of the quorum sensing regulator QscR: interaction with itself and the other regulators of Pseudomonas aeruginosa LasR and RhlR. Mol Microbiol. 2003;48:199–210. doi: 10.1046/j.1365-2958.2003.03423.x. [DOI] [PubMed] [Google Scholar]
- Lee JH, Lequette Y, Greenberg EP. Activity of purified QscR, a Pseudomonas aeruginosa orphan quorum-sensing transcription factor. Mol Microbiol. 2006;59:602–609. doi: 10.1111/j.1365-2958.2005.04960.x. [DOI] [PubMed] [Google Scholar]
- Lee VT, Matewish JM, Kessler JL, Hyodo M, Hayakawa Y, Lory S. A cyclic-di-GMP receptor required for bacterial exopolysaccharide production. Mol Microbiol. 2007;65:1474–1484. doi: 10.1111/j.1365-2958.2007.05879.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lequette Y, Lee JH, Ledgham F, Lazdunski A, Greenberg EP. A distinct QscR regulon in the Pseudomonas aeruginosa quorum-sensing circuit. J Bacteriol. 2006;188:3365–3370. doi: 10.1128/JB.188.9.3365-3370.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim B, Beyhan S, Yildiz FH. Regulation of Vibrio polysaccharide synthesis and virulence factor production by CdgC, a GGDEF-EAL Domain protein, in Vibrio cholerae. J Bacteriol. 2007;189:717–729. doi: 10.1128/JB.00834-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little R, Dixon R. The amino-terminal GAF domain of Azotobacter vinelandii NifA binds 2-oxoglutarate to resist inhibition by NifL under nitrogen-limiting conditions. J Biol Chem. 2003;278:28711–28718. doi: 10.1074/jbc.M301992200. [DOI] [PubMed] [Google Scholar]
- Lutkenhaus J. Assembly dynamics of the bacterial MinCDE system and spatial regulation of the Z ring. Annu Rev Biochem. 2007;76:539–562. doi: 10.1146/annurev.biochem.75.103004.142652. [DOI] [PubMed] [Google Scholar]
- Martinez-Argudo I, Little R, Dixon R. A crucial arginine residue is required for a conformational switch in NifL to regulate nitrogen fixation in Azotobacter vinelandii. Proc Natl Acad Sci U S A. 2004;101:16316–16321. doi: 10.1073/pnas.0405312101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarter LL. Regulation of flagella. Curr Opin Microbiol. 2006;9:180–186. doi: 10.1016/j.mib.2006.02.001. [DOI] [PubMed] [Google Scholar]
- Mendez-Ortiz MM, Hyodo M, Hayakawa Y, Membrillo-Hernandez J. Genome-wide transcriptional profile of Escherichia coli in response to high levels of the second messenger 3′,5′-cyclic diguanylic acid. J Biol Chem. 2006;281:8090–8099. doi: 10.1074/jbc.M510701200. [DOI] [PubMed] [Google Scholar]
- Merighi M, Lee VT, Hyodo M, Hayakawa Y, Lory S. The second messenger bis-(3′–5′)-cyclic-GMP and its PilZ domain-containing receptor Alg44 are required for alginate biosynthesis in Pseudomonas aeruginosa. Mol Microbiol. 2007;65:876–895. doi: 10.1111/j.1365-2958.2007.05817.x. [DOI] [PubMed] [Google Scholar]
- Newman JR, Fuqua C. Broad-host-range expression vectors that carry the L-arabinose-inducible Escherichia coli araBAD promoter and the araC regulator. Gene. 1999;227:197–203. doi: 10.1016/s0378-1119(98)00601-5. [DOI] [PubMed] [Google Scholar]
- North AK, Weiss DS, Suzuki H, Flashner Y, Kustu S. Repressor forms of the enhancer-binding protein NrtC: some fail in coupling ATP hydrolysis to open complex formation by sigma 54-holoenzyme. J Mol Biol. 1996;260:317–331. doi: 10.1006/jmbi.1996.0403. [DOI] [PubMed] [Google Scholar]
- Pratt JT, Tamayo R, Tischler AD, Camilli A. PilZ domain proteins bind cyclic diguanylate and regulate diverse processes in Vibrio cholerae. J Biol Chem. 2007 doi: 10.1074/jbc.M611593200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romling U, Gomelsky M, Galperin MY. C-di-GMP: the dawning of a novel bacterial signalling system. Mol Microbiol. 2005;57:629–639. doi: 10.1111/j.1365-2958.2005.04697.x. [DOI] [PubMed] [Google Scholar]
- Romling U, Amikam D. Cyclic di-GMP as a second messenger. Curr Opin Microbiol. 2006;9:218–228. doi: 10.1016/j.mib.2006.02.010. [DOI] [PubMed] [Google Scholar]
- Ross P, Weinhouse H, Aloni Y, Michaeli D, Weinberger-Ohana P, Mayer R, Braun S, de Vroom E, van der Marel GA, van Boom JH, Benziman M. Regulation of cellulose synthesis in Acetobacter xylinus by cyclic diguanylic acid. Nature. 1987;325:279–281. doi: 10.1038/325279a0. [DOI] [PubMed] [Google Scholar]
- Ross P, Mayer R, Benziman M. Cellulose biosynthesis and function in bacteria. Microbiol Rev. 1991;55:35–58. doi: 10.1128/mr.55.1.35-58.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryjenkov DA, Simm R, Romling U, Gomelsky M. The PilZ domain is a receptor for the second messenger c-di-GMP: the PilZ domain protein YcgR controls motility in enterobacteria. J Biol Chem. 2006;281:30310–30314. doi: 10.1074/jbc.C600179200. [DOI] [PubMed] [Google Scholar]
- Sakuragi Y, Kolter R. Quorum-sensing regulation of the biofilm matrix genes (pel) of Pseudomonas aeruginosa. J Bacteriol. 2007;189:5383–5386. doi: 10.1128/JB.00137-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin H, Kisker C, Schlessman JL, Howard JB, Rees DC. Structure of ADP x AIF4(−)-stabilized nitrogenase complex and its implications for signal transduction. Nature. 1997;387:370–376. doi: 10.1038/387370a0. [DOI] [PubMed] [Google Scholar]
- Schuster M, Lostroh CP, Ogi T, Greenberg EP. Identification, timing, and signal specificity of Pseudomonas aeruginosa quorum-controlled genes: a transcriptome analysis. J Bacteriol. 2003;185:2066–2079. doi: 10.1128/JB.185.7.2066-2079.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simm R, Morr M, Kader A, Nimtz M, Romling U. GGDEF and EAL domains inversely regulate cyclic di-GMP levels and transition from sessility to motility. Mol Microbiol. 2004;53:1123–1134. doi: 10.1111/j.1365-2958.2004.04206.x. [DOI] [PubMed] [Google Scholar]
- Simon R, Priefer U, Pühler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology. 1983;1:784–789. [Google Scholar]
- Singh PK, Schaefer AL, Parsek MR, Moninger TO, Welsh MJ, Greenberg EP. Quorum-sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature. 2000;407:762–764. doi: 10.1038/35037627. [DOI] [PubMed] [Google Scholar]
- Stewart PS, Costerton JW. Antibiotic resistance of bacteria in biofilms. Lancet. 2001;358:135–138. doi: 10.1016/s0140-6736(01)05321-1. [DOI] [PubMed] [Google Scholar]
- Tamayo R, Pratt JT, Camilli A. Roles of cyclic diguanylate in the regulation of bacterial pathogenesis. Annu Rev Microbiol. 2007;61:131–148. doi: 10.1146/annurev.micro.61.080706.093426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tart AH, Wolfgang MC, Wozniak DJ. The alternative sigma factor AlgT represses Pseudomonas aeruginosa flagellum biosynthesis by inhibiting expression of fleQ. J Bacteriol. 2005;187:7955–7962. doi: 10.1128/JB.187.23.7955-7962.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thormann KM, Duttler S, Saville RM, Hyodo M, Shukla S, Hayakawa Y, Spormann AM. Control of formation and cellular detachment from Shewanella oneidensis MR-1 biofilms by cyclic di-GMP. J Bacteriol. 2006;188:2681–2691. doi: 10.1128/JB.188.7.2681-2691.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tischler AD, Camilli A. Cyclic diguanylate (c-di-GMP) regulates Vibrio cholerae biofilm formation. Mol Microbiol. 2004;53:857–869. doi: 10.1111/j.1365-2958.2004.04155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassmann P, Chan C, Paul R, Beck A, Heerklotz H, Jenal U, Schirmer T. Structure of BeF3−-modified response regulator PleD: implications for diguanylate cyclase activation, catalysis, and feedback inhibition. Structure. 2007;15:915–927. doi: 10.1016/j.str.2007.06.016. [DOI] [PubMed] [Google Scholar]
- Weiss DS, Batut J, Klose KE, Keener J, Kustu S. The phosphorylated form of the enhancer-binding protein NTRC has an ATPase activity that is essential for activation of transcription. Cell. 1991;67:155–167. doi: 10.1016/0092-8674(91)90579-n. [DOI] [PubMed] [Google Scholar]
- Wolfe AJ, Visick KL. Get the message out: cyclic-di-GMP regulates multiple levels of flagellum-based motility. J Bacteriol. 2008;190:463–475. doi: 10.1128/JB.01418-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yahr TL, Greenberg EP. The genetic basis for the commitment to chronic versus acute infection in Pseudomonas aeruginosa. Mol Cell. 2004;16:497–498. doi: 10.1016/j.molcel.2004.11.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.