Abstract
BACKGROUND
Formalin fixation and paraffin embedding (FFPE) is the standard practice to process surgical specimens in preparation for pathological evaluation. The FFPE procedure is known to introduce adverse effects on RNA quality and the ensuing RNA based expression analysis. However, the overall impact of the FFPE procedure alone on the reliability and accuracy of expression data, without influences from additional compromising factors introduced during long-term storage, has not been vigorously assessed.
METHODS
The quality of RNA extracted from recently processed FFPE prostate tissues was examined. FFPE and frozen blocks were prepared from matched surgical specimens and processed in parallel for RNA extraction, two rounds of linear RNA amplification, and genome wide expression analysis. Expression ratios of prostate cancer versus benign prostatic hyperplasia (BPH) were compared between data derived from the paired FFPE tissues and frozen tissues.
RESULTS
RNA extracted from recently processed FFPE prostate tissues was of high quality and suitable for RNA expression analysis. An unbiased analysis of expression data revealed nearly 80% concordance between FFPE tissues and frozen tissues, in genome-wide gene expression differences of cancer vs. BPH, and across a wide spectrum of fold expression differences.
CONCLUSIONS
FFPE procedure itself does not lead to adverse effects on genome wide expression analysis. Prospective molecular archiving or modified long-term storage conditions should be implemented to expand the utility of FFPE tissues without compromising standard surgical pathology routines.
Keywords: FFPE, microarray, prostate
Formalin fixation and paraffin embedding (FFPE) is a standard practice to process surgical specimens in preparation for pathological evaluation. The FFPE procedure introduces chemical modification and cross-linking of RNA molecules through the addition of mono-methylol (-CH2OH) to the amino groups of bases and the formation of methylene bridges between amino groups, compromising the yield and quality of RNA extracted from such tissues and their utility for RNA expression analysis [1]. A number of recent studies have demonstrated and validated the general feasibility of obtaining high quality total RNA from a subset of FFPE tissues for a variety of RNA-based expression studies [2-6]. However, data derived from these studies were also consistent with the general consensus that global gene expression data derived from FFPE specimens suffered from poor quality and low detection sensitivity when compared to frozen specimens, and the adverse effects are more pronounced and more frequently observed in aged archival FFPE specimens. Another recent study revealed that extended storage of FFPE blocks in currently practiced conditions (i.e., at room temperature), rather than the FFPE procedure itself, may have accounted for the poor preservation of RNA in archived FFPE specimens [7,8]. Given these recent developments, it is imperative to determine whether and to what extent the FFPE procedure alone impacts on genome wide expression analysis, without further compromises on preservation of RNA due to long-term storage of FFPE specimens. A rigorous interrogation of this critical question mandates an unbiased comparison of FFPE versus frozen specimens derived from the same original specimen and processed in parallel, and an assessment of multiple intermediate quality control measures and genome wide expression array data endpoints.
We first focused on the quality assessment of total RNA extracted from recently processed (within 3 months from the time of fixation to the time of RNA extraction) FFPE tissues, and determined whether current tissue fixation methodologies employed in our institution result in differential RNA quality. We used the Paradise™ Reagent System (PRS) (Arcturus Bioscience, Mountain View, CA) for RNA extraction from FFPE specimens processed using three different formalin fixation procedures as shown in Figure 1. High quality total RNA can be consistently extracted from FFPE tissues processed using any of the 3 formalin fixation methods, as evidenced by the presence of 28S and 18S ribosomal RNA bands following analysis using the Agilent 2100 Bioanalyzer (Agilent Technologies, Wilmington, DE) (Figure 1), as well as the RNA integrity numbers that ranged from 2.0 to 5.8 for the majority (16/17) of the samples (Figure 1).
Figure 1.
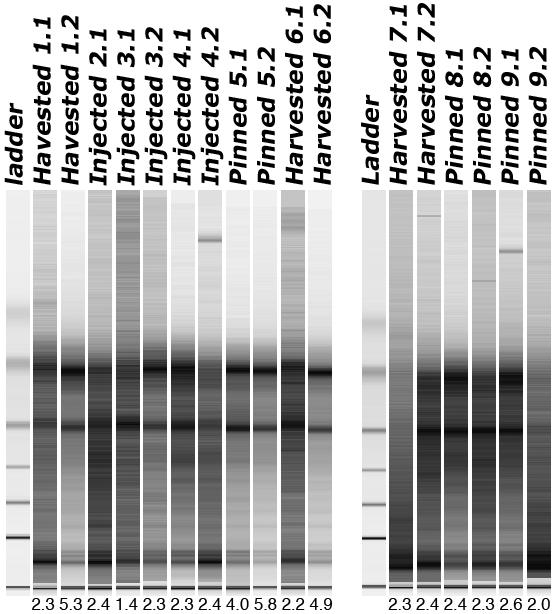
Gel image of 17 samples dissected from FFPE specimens prepared using 3 different formalin fixation methods employed in Surgical Pathology lab at Johns Hopkins University. Harvested: tissue slices were processed for FFPE after standard tissue harvesting procedure [9], during which areas of cancer were cut in duplicate for fresh-freezing and FFPE processing in the surgical pathology lab; Pinned: tissue slices were processed for FFPE after punch cores of cancer tissue were taken while pinned down; Injected: whole prostate fixation by formalin injection and microwaving. A total of 9 cases, 8 of which with two (denoted by “.1” and “.2” following the case number) sections taken from different parts of the specimen, were subjected to RNA extraction. RNA integrity numbers associated with each sample were provided below the corresponding electropherogram. One section for samples no. 2 peeled off during processing.
We then focused on analyzing the impact of the FFPE procedure on the detection of global gene expression differences between prostate cancer and benign prostatic hyperplasia (BPH). We designed the experiments to ensure an accurate assessment of the data output as a function of the two tissue processing methods. A pair of surgical specimens, prostate cancer and BPH, was each divided in two, one processed for formalin fixation and paraffin embedding (in the Surgical Pathology lab) and the other snap frozen, according to established tissue harvesting procedures [9]. Tissue lesions of interest (i.e., prostate cancer and BPH) were subsequently isolated following the sectioning of FFPE and frozen tissue blocks and processed for RNA extraction, according to the manufacturer’s instructions in the Paradise™ Reagent System (PRS) (Arcturus Biosciences) for FFPE sections, or as described previously for frozen sections [10]. All 4 RNA samples (FFPE cancer, FFPE BPH, frozen cancer, frozen BPH) were subjected to T7 based linear RNA amplification using the PRS reagents (Arcturus Biosciences) for 1.5 rounds according to the manufacturer’s instructions (Arcturus Bioscience). During the second round of amplification, a T7 based linear RNA amplification reaction incorporated either cyanine 3-CTP or cyanine 5-CTP using the Low RNA Input Linear Amplification Kit (Agilent Technologies). The labeled products were purified and then quantified using the NanoDrop® Spectrophotometer (NanoDrop Technologies, Wilmington, DE), followed by microarray analysis according to instructions in the Agilent Whole Genome Expression Microarray system (Agilent Technologies).
We analyzed multiple intermediate quality control measures including total RNA yield, RNA quality, amplified RNA (aRNA) yield, aRNA quality, and fluorescent dye labeling efficiency, by parallel comparison of relevant data derived from FFPE and frozen prostate tissues. As anticipated, the FFPE procedure resulted in a generally decreased RNA yield, approximately one fourth of those extracted from equivalent amount of frozen tissues (data not shown). Also as anticipated, RNA samples extracted from the FFPE samples were associated with decreased RNA integrity numbers (RIN at 2.2 and 2.4 for tumor and BPH, respectively) when compared to those from frozen tissues (RIN at 7.1 and 8.0 for tumor and BPH, respectively). However, RNA extracted from both tissue processing methods yielded comparable amounts of amplified RNA in both the first and second rounds of amplification when using equal amount of input RNA (not shown). The size of first round amplified RNA derived from FFPE tissues was slightly decreased (peak at ∼500bp) when compared to amplified RNA from frozen tissues (peak at ∼800bp), likely contributing to a lower labeling efficiency for RNA from FFPE tissues, which was roughly half of those from frozen tissues (not shown).
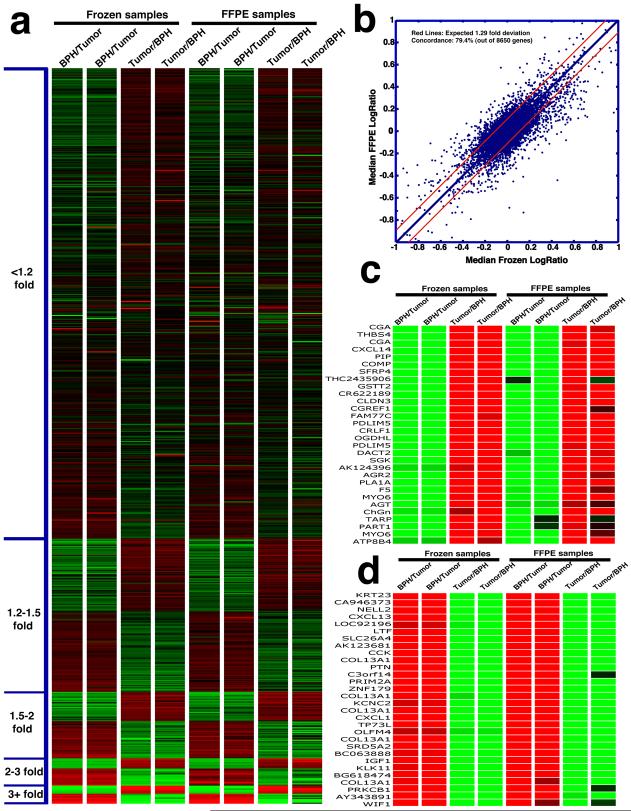
Finally we analyzed the end-point expression data derived from tissues prepared by the two processing methods (i.e., FFPE and frozen). Using a 2-color design in which BPH and prostate tumor were labeled using different dyes (Cye 3 or Cye 5) and directly compared in each array experiment, we generated 8 sets of ratios (from 8 arrays), 4 replicates each for the frozen BPH/tumor (B/T) pair and the FFPE BPH/tumor (B/T) pair. In 2 of the 4 replicates, we performed a dye-swap to assess potential dye labeling bias. Expression ratios were extracted from 8,650 genes meeting the single quality filtration criterion of average signal intensity greater than 1000. We then ordered these detected genes according to median fold expression changes between prostate cancer and BPH in the 8 arrays. As shown in Figure 2a, gene expression data derived from the FFPE B/T pair showed remarkable concordance, both within the replicates and when compared with data derived the frozen B/T pair. As visually evident in Figure 2a, reliable detection of expression difference in FFPE tissues can be achieved across a wide range of fold expression changes, when compared with the frozen tissue standard. Figure 2b summarizes the general concordance between the median (of the 4 replicates) ratio of the frozen pair and the FFPE pair. Based on 4 replicated sets of ratios derived from the frozen pair, we estimated the variance of the median ratios for each gene at 0.043. This translates to an estimation of 1.29-fold deviation from the median ratio at the 99% confidence interval. In other words, if another 4 sets of ratios were to be derived from the same frozen pair, we expect 99% of the genes would deviate from the median ratio by less than 1.29-fold (i.e., within the red lines, Figure 2b). By plotting the median ratios from the FFPE pair against the median ratios from the frozen pair (Figure 2b), 20.6% of the 8650 genes showed significant deviation (greater than 1.29-fold) from what would be expected if the ratios from FFPE pair were true replicates of the frozen pair. Therefore, expression profiling using FFPE tissues resulted in a 20.6% drop-out, and thus a 79.4% concordance rate. The correlation coefficient between expression ratios derived from FFPE and frozen tissue was 0.813. It is worth noting that due to tissue heterogeneity, the tissue specimens for freezing and FFPE processing were not strictly identical, and therefore biological differences as opposed to FFPE processing could account for an unknown percentage of the drop-outs. This high concordance between expression data derived from the two tissue processing methods was further illustrated by parallel comparison of top ranked genes (by fold expression change), as shown in Figure 2c (genes overexpressed in prostate cancer relative to BPH), and Figure 2d (genes underexpressed in prostate cancer relative to BPH). Moreover, the majority of these top genes that were ranked by averaged values of 8 sets of ratios (Supplemental Figure 1), as well as genes ranked by averaged values of 4 sets of ratios from frozen tissues (Supplemental Figure 2), or by averaged values of 4 sets of ratios from FFPE tissues (Supplemental Figure 3), were validated in an independent data set we previously described (11).
Figure 2.
Reliability of whole genome expression differences detected using FFPE tissues. a. Heatmap of the 8650 genes ranked (in ascending order) based on median fold expression change between BPH and prostate cancer. Each column represents one of the 8 ratios derived from the annotated comparisons. b. Scatter plot of median expression ratios from FFPE tissues against median ratios from frozen tissues. Red lines denote the expected 1.29-fold deviation at p<0.01. c. Parallel comparison (FFPE vs. frozen) of top ranked genes over-expressed in prostate cancer vs. BPH. d. Parallel comparison (FFPE vs. frozen) of top 25 genes over-expressed in BPH vs. prostate cancer.
In this study, we assessed the overall impact of the FFPE procedure on RNA quality and genome-wide mRNA expression analysis, by using recently processed FFPE tissues and through comparison with frozen tissues processed in parallel. Our results demonstrate that when additional compromising factors introduced during long-term storage of the FFPE specimens were minimized, FFPE specimens should be compatible with routine RNA based microarary analysis using T7 based linear RNA amplification. These results have important implications regarding the utility of FFPE specimens for high quality molecular analysis. First, surgical specimens not amenable to harvesting by snap freezing procedures, such as small volume prostate cancer or biopsies, can be analyzed using expression microarrays following extraction of RNA from standard FFPE sections. Second, tissue lesions that require the FFPE procedure for reliable diagnosis, such as high-grade prostatic intraepithelial neoplasia, can be utilized for subsequent expression analysis. Lastly, since revised processing and storage conditions for FFPE specimens may reduce the negative impact of long-term storage on RNA quality [7, 8, 12, 13], we propose prospective archiving, which would involve storage of punched FFPE tissues in reduced temperature, or storage of RNA, extracted shortly after the FFPE procedure, to allow for future molecular profiling without compromising current surgical pathology routines.
Supplementary Material
Acknowledgments
Financial Support: NIH/NCI R21CA106425 (to J.L.), NIH/NCI Specialized Program in Research Excellence in Prostate Cancer P50CA58236 (Johns Hopkins University).
References
- 1.Masuda M, Ohnishi T, Kawamoto S, Monden M, Okubo K. Analysis of chemical modification of RNA from formalin-fixed samples and optimization of molecular biology applications for such samples. Nucleic Acid Res. 1999;27(22):4436–4443. doi: 10.1093/nar/27.22.4436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coudry RA, Meireles SI, Stoyanova R, Coopers HS, Carpino A, Wang X, Engstrom PF, Clapper ML. Successful application of microarray technology to microdissected formalin-fixed, paraffin-embedded tissue. J Mol Diagn. 2007;9(1):70–79. doi: 10.2353/jmoldx.2007.060004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bibikova M, Chudin E, Arsanjani A, Zhou L, Garcia EW, Modder J, Kostelec M, Barker D, Downs T, Fan JB, Wang-Rodriguez J. Expression signatures that correlated with Gleason score and relapse in prostate cancer. Genomics. 2007;89(6):666–72. doi: 10.1016/j.ygeno.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 4.Setlur SR, Mertz KD, Hoshida Y, Demichelis F, Lupien M, Perner S, Sboner A, Pawitan Y, Andrén O, Johnson LA, Tang J, Adami HO, Calza S, Chinnaiyan AM, Rhodes D, Tomlins S, Fall K, Mucci LA, Kantoff PW, Stampfer MJ, Andersson SO, Varenhorst E, Johansson JE, Brown M, Golub TR, Rubin MA. Estrogen-dependent signaling in a molecularly distinct subclass of aggressive prostate cancer. J Natl Cancer Inst. 2008;100(11):815–25. doi: 10.1093/jnci/djn150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xi Y, Nakajima G, Gavin E, Morris CG, Kudo K, Hayashi K, Ju J. Systematic analysis of microRNA expression of RNA extracted from fresh frozen and formalin-fixed paraffin-embedded samples. RNA. 2007;13(10):1668–74. doi: 10.1261/rna.642907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li J, Smyth P, Flavin R, Cahill S, Denning K, Aherne S, Guenther SM, O’Leary JJ, Sheils O. Comparison of miRNA expression patterns using total RNA extracted from matched samples of formalin-fixed paraffin-embedded (FFPE) cells and snap frozen cells. BMC Biotechnol. 2007;29(7):36. doi: 10.1186/1472-6750-7-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.von Ahlfen S, Missel A, Bendrat K, Schlumpberger M. Determinants of RNA quality from FFPE samples. PLoS Online. 2007;12:1–7. doi: 10.1371/journal.pone.0001261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cronin M, Pho M, Dutta D, Stephens JC, Shak S, Kiefer MC, Esteban JM, Baker JB. Measurement of gene expression in archival paraffin-embedded tissues: Development and performance of a 92-gene reverse transcriptase-polymerase chain reaction. Am J Pathology. 2004;164:35–42. doi: 10.1016/S0002-9440(10)63093-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bova GS, Fox WM, Epstein JI. Methods of radical prostatectomy specimen processing: a novel technique for harvesting fresh prostate cancer tissue and review of processing techniques. Mod Pathol. 1993;6(2):201–7. [PubMed] [Google Scholar]
- 10.Luo J, Duggan DJ, Chen Y, Sauvageot J, Ewing CM, Bittner ML, Trent JM, Isaacs WB. Human prostate cancer and benign prostatic hyperplasia: Molecular dissection by gene expression profiling. Cancer Research. 2001;61(12):4683–4688. [PubMed] [Google Scholar]
- 11.Dunn T, Chen S, Faith D, Hicks J, Platz E, Chen Y, Ewing C, Sauvageot J, Isaacs W, De Marzo A, Luo J. A Novel Role of Myosin VI in Human Prostate Cancer. American Journal of Pathology. 2006;169(5):1843–54. doi: 10.2353/ajpath.2006.060316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Srinivassan M, Sedmick D, Jewell S. Effects of fixatives and tissue processing on the content and integrity of nucleic acids. Am J Pathology. 2006;161:1961–71. doi: 10.1016/S0002-9440(10)64472-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Maldegem F, de Wit M, Morsink F, Musler A, Weegenaar J, van Noesel CJM. Effects of processing delay, formalin fixation, and immunohistochemistry on RNA recovery from formalin-fixed paraffin-embedded tissue sections. Diagn Mol Pathol. 2008;17(1):51–58. doi: 10.1097/PDM.0b013e31814b8866. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.