Abstract
Gastric adenocarcinoma is the second leading cause of cancer death worldwide. Epstein-Barr virus (EBV) is present in the malignant cells of approximately 10% of cases. It is unclear whether EBV is being missed in some gastric adenocarcinomas due to insensitive test methods or partial EBV genome loss. In the current study, we screened 113 gastric adenocarcinomas from low and high incidence regions (United States and Central America) for the presence of EBV using a battery quantitative real-time PCR (Q-PCR) assays targeting disparate segments of the EBV genome (BamH1W, EBNA1, LMP1, LMP2, BZLF1, EBER1) and histochemical stains targeting EBV-encoded RNA (EBER), the latent proteins LMP1 and LMP2, and the lytic proteins BMRF1 and BZLF1. EBV DNA was detected by Q-PCR in 48/75 United States cancers (64%) and in 38/38 Central American cancers (100%), which was a significant differrence. EBER was localized to malignant epithelial cells in 8/48 (17%) United States and 3/38 (8%) Central American cancers. Viral loads were considerably higher for EBER-positive versus EBER-negative cancers (mean 162,986 versus 62 EBV DNA copies per 100,000 cells). A viral load of 2,000 copies per 100,000 cells is recommended as the threshold distinguishing EBER-positive from EBER-negative tumors. One infected cancer selectively failed to amplify the LMP2 gene because of a point mutation, while another cancer had an atypical pattern of Q-PCR positivity suggesting deletion of large segments of the EBV genome. Three different viral latency profiles were observed in the cancers based on constant expression of EBER and focal or variable expression of LMP1 or LMP2, without lytic protein expression. We conclude that EBV DNA levels generally reflect EBER status, and a panel of at least two Q-PCR assays is recommended for sensitive identification of infected cancers.
Keywords: Epstein-Barr virus, gastric adenocarcinoma, latency, replication, viral load
Gastric cancer represents the fourth most common type of cancer and the second leading cause of cancer death worldwide.(1) The incidence of gastric cancer varies up to ten fold by geographic region, suggesting that genetic or environmental factors influence carcinogenesis and that different carcinogenesis models may be operative.(2) Patients often present with advanced and incurable disease. Even with resection and chemotherapy, high rates of recurrence result in poor overall survival, especially in developing countries. For unclear reasons, the incidence of gastric cancer in the proximal one-third of the stomach, where Helicobacter pylori (H. pylori) infection is less frequent, has been increasing worldwide.(2, 3) Interestingly, Epstein-Barr virus (EBV) is more frequently identified in these proximal cancers.(4-8)
Patients who are at high risk of treatment failure are candidates for novel or aggressive treatment regimens. One promising therapeutic target is EBV, a ubiquitous gamma herpesvirus that has been implicated in the pathogenesis of a variety of benign and malignant diseases.(9, 10) EBV DNA is present in the malignant epithelial cells of about 10% of gastric adenocarcinomas across geographic regions. The gastric adenocarcinomas most likely to be EBV-related are those of the “diffuse” histologic subtype with abundant infiltrating lymphocytes, poor to moderate differentiation, and those involving the proximal stomach.(4, 5, 7, 11-13) EBV association is also noted in cancers of the gastric stump following surgery, especially with Billroth II reconstruction after gastrectomy for benign gastroduodenal disease.(7, 14-17)
The gold standard assay for EBV targets EBV-encoded RNA (EBER) by in situ hybridization in paraffin-embedded tissue sections.(10, 18) This assay detects the most abundant latent viral transcripts and also localizes the viral infection to malignant cells by microscopy. However, the sensitivity of EBER staining has been called into question by investigators who have shown, by molecular or immunohistochemical assays, that EBV is present in some EBER-negative tumors.(19-25) Therefore, it is unclear whether EBV is being missed in some gastric adenocarcinomas due to insensitive testing methods or partial EBV genome loss.
Reliable diagnosis of EBV-related malignancy requires not only detection of the viral genome or its gene products, but also localization of the virus to the malignant cell fraction. Viral LMP2 and EBNA1 are target analytes since they are often expressed in EBV-related gastric cancers and they can be localized to tumor cells by immunohistochemistry,(10, 26, 27) although EBNA1 histochemical stains are less reliable due to crossreactivity with a human protein.(28) LMP1 is rarely expressed, and the restricted pattern of viral gene expression categorizes gastric cancers as latency type I, although latency types II, III and lytic infection have also been described.(12, 29-33)
To detect EBV at the DNA level, semiquantitative PCR has been used to screen gastric adenocarcinoma tissues for presence of the EBV genome and to demonstrate that high viral load correlates with EBER localization to malignant cells.(34) Modern real-time PCR methods now permit even more precise measurement of EBV viral load in DNA extracted from tissue specimens.(10)
Using a battery of tests for EBV DNA, RNA and protein, we examined 113 gastric adenocarcinomas from high and low incidence regions. This included 75 from the United States and 38 from Western Honduras. Western Honduras has been characterized as a region of high gastric cancer incidence, with standardized annual incidence rates of approximately 39 cases per 100,000 for males and 21 per 100,000 for females.(35) In addition, endemic H. pylori infection has been confirmed in the region (85%). By comparison, the United States has a standardized annual incidence of 11 per 100,000 males and 6 per 100,000 females, with a lifetime risk of nearly 1%. To detect EBV in cancer tissues, we used a battery of sensitive and specific quantitative real-time PCR (Q-PCR) assays targeting six disparate regions of the EBV genome (BamH1W, EBNA1, LMP1, LMP2, EZLF1, EBER1). The virus was localized using EBER in situ hybridization and immunohistochemical assays targeting selected viral proteins expressed during latent infection (LMP1, LMP2A) or during active viral replication (BZLF1, BMRF1).
Materials and Methods
Gastric Adenocarcinoma Samples
Formalin fixed, paraffin-embedded blocks representing 113 gastric adenocarcinomas were obtained from UNC Hospitals tissue archives in Chapel Hill, NC (n=55), Massachusetts General Hospital in Boston, MA (n=20) and Western Regional Hospital, Santa Rosa de Copan, Honduras, Central America (n=38). The gastric adenocarcinomas obtained from Massachusetts General Hospital were selectively enriched for gastric stump site, with 16/20 of these being stump cancers. The remaining cancers from all three sites were serial cases.
Clinical information included age, anatomic site of the cancer within the stomach, and, for the Central American cases, H. pylori serologic status. H. pylori serology was not performed on the gastric cancer patients from the United States, rather histologic evidence was sought for infection by bacteria consistent with H. pylori, but evidence was found in only one US case. Note that the sensitivity of microscopic detection of H.pylori is low given that surface mucosa was sometimes not even present in the cancer specimen used for this study. Furthermore, there is loss of H. pylori colonization as cancer develops.(1) For each cancer, histologic subtype was classified as intestinal versus diffuse using a modern version of Lauren’s criteria.(36) Paraffin sections were placed on coated glass slides for histochemical stains, or placed in a microfuge tube for manual DNA extraction as previously described in Ryan et al.(37)
Quantitative real-time PCR
A battery of Q-PCR assays targeting six disparate regions of the EBV genome was used to measure EBV DNA. We previously validated five of these Q-PCR assays targeting BamH1W, EBNA1, LMP1, LMP2, and BZLF1 regions of the EBV genome(37), while the sixth assay targeting EBER1 was developed by Ling et al.(38) A Q-PCR assay targeting the human APOB gene was used, as previously described, to control for efficacy of DNA extraction and to normalize for the number of cells amplified per reaction.(37)
PCR was performed and products were detected on ABI Prism 7900 and 7500 Real-Time PCR instruments with Sequence Detection System software (Applied Biosystems, Foster City, CA). Thermocycling conditions were: 50°C for 2 minutes, 95°C for 10 minutes, and then 95°C for 15 seconds and 60°C for 1 minute for 40 cycles. Each 25μl reaction contained: 1X TaqMan Universal Master Mix and TaqMan probe (10ρmol). Forward and reverse primers (15ρmol each) were used for all assays except those targeting LMP1 and BZLF1 in which 30ρmol was used. DNA template volume was 1μl, and failed APOB reactions were repeated at higher or lower template volumes. A standard curve was generated using serial 10-fold dilutions of Namalwa Burkitt lymphoma cell line DNA (at two copies of EBV genome per cell) varying from 50,000 copies to 0.5 copies of EBV DNA per reaction. This curve was acceptable if sensitivity was at least 50 copies of EBV DNA per PCR, a difference of 3.3 +/- 0.3 cycles was demonstrated between each of the 10-fold dilutions, and if the correlation coefficient was at least 0.99. To check for amplicon contamination, every run contained at least two “no template” controls in which nuclease-free H2O was substituted for template.
All experimental samples were run in duplicate and a mean viral load was calculated based on the ratio of the copies of EBV to APOB in a given volume of extracted DNA, with APOB quantity representing the number of cells in the reaction. The resulting ratio was adjusted to provide the number of copies of EBV DNA per 100,000 cells. Samples with no measurable EBV DNA were reported as having a viral load of zero.
EBV-encoded RNA (EBER) in situ hybridization
EBER in situ hybridization was performed on paraffin sections of cases having detectable EBV DNA by at least one Q-PCR assay (n=86). This was accomplished using either a manual method with fluorescein-labeled oligonulceotide EBER and oligo(d)T control probes and the Super-Sensitive Poly-HRP ISH Non-Biotin Detection Kit (Biogenex, San Ramon, CA) with methyl green counterstain, or by an automated method using fluorescein-labeled EBER and oligo(d)T control probes on the Ventana Benchmark in situ hybridization system (Ventana Medical Systems, Tuscon AZ). Validation work on 56 samples showed no discrepancies between the two staining procedures. The oligo(d)T probe served as a control for RNA preservation in histological sections. A tumor was considered EBER-negative if EBER staining was undetected or was only expressed in benign-appearing lymphoid cells, and EBER-positive if the signal was localized to malignant epithelial cells.
Immunohistochemistry for viral LMP1, LMP2, BMRF1, and BZLF1
Immunohistochemical stains for viral protein were performed on the 86 gastric cancer samples having detectable EBV DNA by Q-PCR. Stains for EBV LMP1 and LMP2 proteins were done as previously described(37) using citrate retrieval and the CS1-4 cocktail of mouse monoclonal antibodies against LMP1 (1:100, Dako, Capinteria, CA) and the E411 rat monoclonal antibody against LMP2A (1mg/ml, Asencion, Munich, Germany). Paraffin sections of EBV-related Hodgkin lymphoma served as positive controls.
Immunohistochemical analysis of the EBV replicative proteins BMRF1 and BZLF1 was performed using anti-BMRF1 clone G3-E31 (1:200 dilution, Research Diagnostics, Inc., Flanders, NJ) and anti-BZLF1 clone BZ.1 (1:25 dilution, Dako, Carpinteria, CA) on paraffin sections pretreated with citrate for antigen retrieval. Sections were incubated with primary antibody for 30 minutes at 37°C using the manufacturer’s blocking and detection protocols in the Super-Sensitive Non Biotin HRP Detection Kit (Biogenex). Bound antibody was detected by diaminobenzidine chromogen (Biogenex) and tissues were counter-stained with hematoxylin (Dako). Oral hairy leukoplakia paraffin sections served as a positive control.
Qualitative LMP2 PCR and DNA Sequencing
To further investigate a case in which the EBV LMP2 segment selectively failed to amplify by Q-PCR, a primer set was designed to amplify a 130bp region of the LMP2 gene encompassing the 69bp segment that had been targeted by the standard LMP2 Q-PCR assay. The following PCR primers were used: LMP2Ext forward 5′-CTGTTTTGCAGCTGAGTCC-3′ and LMP2Ext reverse 5′-CAATGTTAAAAGGGCTGCACC-3′. The 50μl reaction consisted of: 1X PCR Buffer, 2mM MgCl2 2.5 units Platinum Taq DNA Polymerase (Invitrogen, Carlsbad, CA), 0.2mM dNTPs (Applied Biosystems), 50ρmol of each LMP2Ext primer, and nuclease-free water. Reaction conditions were: 95°C for 2 minutes; 35 cycles of 95°C for 30 seconds, 60°C for 30 seconds, and 72°C for 1 minute; and then 72°C for 10 minutes. The product was confirmed by electrophoresis in a 2% agarose gel containing 0.5mg/ml ethidium bromide. The product was sequenced in both directions by first removing unincorporated primers and dNTPs, and then incubating with shrimp alkaline phosphatase and exonuclease I (Amersham, Piscataway, NJ) at 37°C for 30 minutes, then 80°C for 15 minutes. The 12.5μl sequencing reaction consisted of: 1.1μl of template, 1μl LMP2Ext forward primer (5ρmol/ml), 5μl BD Terminator Dye (Applied Biosystems), and water. The reaction was performed on the ABI 9700 thermocycler under the following conditions: 96°C for 5 seconds; 25 cycles of 96°C for 10 seconds, 50°C for 5 seconds, 60°C for 4 minutes; 60°C for 10 minutes followed by a hold at 4°C. The product was purified using the Qiagen DyeEx 2.0 Spin Kit, denatured at 95°C for 3 minutes, and separated by size in POP6 polymer by capillary electrophoresis on an ABI 3100 Gene Analyzer instrument. The sequence was compared to consensus EBV reference sequence in GenBank (NC_007605) and to wild type viral DNA from an EBER-positive AIDS lymphoma and another gastric carcinoma that were tested in parallel.
Results
Histopathologic findings
Gastric adenocarcinomas from the United States (n=75) and Central America (n=38) were classified by histopathology into intestinal pattern (n=50), diffuse pattern (n=55), or mixed pattern of tumor growth (n=8). The anatomic site of origin was classified as distal (n=93) for tumors occurring in the antrum or body, proximal (n=14) for tumors occurring in the cardia, or whole stomach (n=6). H. pylori infection was detected in 33/38 (87%) Central American gastric cancer patients by standard serological testing.
EBV DNA Measurement by Quantitative Real-Time PCR
Gastric adenocarcinoma tissues were screened for EBV genome using six separate Q-PCR assays targeting six different regions of the viral genome (BamH1W, EBNA1, LMP1, LMP2, BZLF1, and EBER1). Out of the 75 United States gastric cancers, 48 (64%) had detectable EBV DNA by at least one Q-PCR assay, and 11 (15%) were positive by all six Q-PCR assays (Table 1), while 27 (36%) were negative by all six assays. The Central American cases more frequently contained EBV DNA: 38/38 (100%) by at least one Q-PCR assay, and 11 (29%) by all six Q-PCR assays (Table 2).
Table 1.
Clinicopathologic Findings in EBV Q-PCR Positive United States Gastric Cancers
| EBV DNA copies / 100 000 cells | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Case # |
Histological Classification |
Site | Hp Status* |
APOB (# of cells) | BamH1W | EBNA1 | LMP1 | LMP2 | BZLF1 | EBER1 |
EBER in situ
hybridization |
| 1 | Intestinal | Stump | Unk | 2 974 | 44 956 | 57 969 | 10 558 | 12 441 | 58 238 | 85 945 | Tumor + |
| 2 | Diffuse | Stump | Neg | 271 | 200 000 | 569 004 | 338 007 | 217 712 | 318 450 | 453 506 | Tumor + |
| 3 | Mixed | Whole Stomach | Neg | 2 896 | 90 919 | 117 818 | 38 640 | 18 750 | 178 280 | 255 939 | Tumor + |
| 4 | Intestinal | Distal | Unk | 1 198 | 193 823 | 370 534 | 318 865 | 104 257 | 230 134 | 336 394 | Tumor + |
| 5 | Intestinal | Distal | Unk | 2 734 | 144 221 | 390 856 | 402 341 | 149 268 | 287 564 | 307 206 | Tumor + |
| 6 | Diffuse/Signet | Distal | Neg | 80 033 | 117 755 | 219 321 | 213 938 | 20 183 | 26 240 | 24 526 | Tumor + |
| 7 | Diffuse | Distal | Neg | 8 770 | 130 935 | 263 592 | 292 873 | 65 211 | 98 141 | 32 132 | Tumor + |
| 8 | Diffuse | Distal | Neg | 23 209 | 70 757 | 141 260 | 183 722 | 31 509 | 38 912 | 27 429 | Tumor + |
| 9 | Intestinal | Distal | Neg | 11 655 | 17 | 60 | 43 | 0 | 34 | 43 | Tumor -; Lymph + |
| 10 | Diffuse/Signet | Stump | Unk | 2 148 | 93 | 116 | 186 | 1 629 | 93 | 186 | Tumor -; Lymph + |
| 11 | Intestinal | Distal | Neg | 1 106 | 0 | 0 | 0 | 0 | 90 | 0 | Tumor -; Lymph + |
| 12 | Diffuse | Distal | Neg | 9 101 | 66 | 132 | 99 | 0 | 66 | 22 | Tumor -; Lymph + |
| 13 | Diffuse/Signet | Whole Stomach | Neg | 45 494 | 4 | 4 | 0 | 4 | 0 | 2 | Tumor -; Lymph + |
| 14 | Intestinal | Distal | Unk | 14 047 | 5 | 14 | 0 | 14 | 14 | 21 | Tumor -; Lymph + |
| 15 | Intestinal | Distal | Neg | 5 338 | 880 | 918 | 1 068 | 262 | 225 | 262 | Tumor -; Lymph + |
| 16 | Diffuse | Proximal | Neg | 8 866 | 10 | 34 | 0 | 23 | 0 | 11 | Tumor -; Lymph + |
| 17 | Diffuse | Distal | Neg | 10 507 | 10 | 10 | 0 | 0 | 0 | 0 | Tumor -; Lymph + |
| 18 | Diffuse/Signet | Distal | Neg | 5 007 | 20 | 40 | 0 | 0 | 0 | 50 | Tumor -; Lymph + |
| 19 | Diffuse | Distal | Neg | 8 406 | 5 | 0 | 0 | 0 | 0 | 0 | Tumor -; Lymph + |
| 20 | Diffuse/Signet | Distal | Positive | 17 630 | 1 | 0 | 0 | 0 | 0 | 0 | Tumor -; Lymph + |
| 21 | Intestinal | Distal | Neg | 2 183 | 137 | 32 | 0 | 137 | 0 | 64 | Tumor -; Lymph + |
| 22 | Diffuse | Distal | Neg | 22 500 | 116 | 84 | 182 | 18 | 18 | 22 | Tumor -; Lymph + |
| 23 | Mixed | Distal | Neg | 242 | 8 | 165 | 0 | 0 | 0 | 0 | Tumor -; Lymph + |
| 24 | Diffuse/Signet | Proximal | Neg | 423 | 0 | 0 | 18 913 | 0 | 0 | 0 | Tumor -; Lymph + |
| 25 | Diffuse/Signet | Proximal | Neg | 9 507 | 0 | 1 | 0 | 0 | 1 | 0 | Tumor -; Lymph + |
| 26 | Intestinal | Stump | Neg | 3 289 | 9 | 15 | 0 | 0 | 15 | 0 | Tumor - |
| 27 | Mixed | Distal | Neg | 562 | 36 | 0 | 178 | 0 | 36 | 0 | Tumor - |
| 28 | Diffuse | Distal | Neg | 30 257 | 2 | 0 | 0 | 0 | 0 | 4 | Tumor - |
| 29 | Diffuse | Proximal | Neg | 798 | 50 | 0 | 0 | 0 | 0 | 63 | Tumor - |
| 30 | Mixed/Signet | Distal | Neg | 16 198 | 6 | 12 | 0 | 6 | 0 | 6 | Tumor - |
| 31 | Diffuse/Signet | Proximal | Neg | 709 | 14 | 0 | 0 | 0 | 0 | 0 | Tumor - |
| 32 | Diffuse | Distal | Neg | 9 268 | 11 | 0 | 0 | 0 | 0 | 6 | Tumor - |
| 33 | Intestinal | Distal | Neg | 218 | 92 | 0 | 0 | 0 | 0 | 275 | Tumor - |
| 34 | Intestinal | Distal | Neg | 14 907 | 1 | 0 | 0 | 0 | 0 | 0 | Tumor - |
| 35 | Diffuse/Signet | Distal | Neg | 9 105 | 22 | 33 | 0 | 0 | 0 | 0 | Tumor - |
| 36 | Intestinal | Distal | Neg | 7 466 | 4 | 5 | 0 | 0 | 0 | 0 | Tumor - |
| 37 | Diffuse/Signet | Distal | Neg | 3 723 | 5 | 27 | 27 | 1 | 5 | 0 | Tumor - |
| 38 | Diffuse | Distal | Neg | 11 976 | 0 | 3 | 0 | 0 | 0 | 0 | Tumor - |
| 39 | Intestinal | Stump | Unk | 1 784 | 11 | 17 | 28 | 0 | 56 | 28 | Tumor - |
| 40 | Diffuse | Stump | Neg | 431 | 232 | 0 | 0 | 0 | 0 | 0 | Tumor - |
| 41 | Diffuse/Signet | Stump | Unk | 614 | 0 | 65 | 0 | 0 | 0 | 0 | Tumor - |
| 42 | Intestinal | Stump | Neg | 943 | 0 | 0 | 0 | 0 | 21 | 0 | Tumor - |
| 43 | Intestinal | Stump | Unk | 108 | 93 | 0 | 0 | 0 | 0 | 0 | Tumor - |
| 44 | Diffuse/Signet | Stump | Unk | 774 | 0 | 0 | 0 | 0 | 129 | 0 | Tumor - |
| 45 | Intestinal | Stump | Unk | 6 597 | 8 | 15 | 0 | 0 | 6 | 0 | Tumor - |
| 46 | Intestinal | Stump | Neg | 5 594 | 0 | 0 | 89 | 0 | 0 | 0 | Tumor - |
| 47 | Mixed/Signet | Proximal | Unk | 2 782 | 0 | 0 | 0 | 0 | 11 | 0 | Tumor - |
| 48 | Intestinal | Proximal | Unk | 5 152 | 0 | 0 | 0 | 0 | 2 | 0 | Tumor - |
| Totals (N=48) | 25 Diffuse 18 Intestinal 5 Mixed 14 Signet | 27 Distal 7 Proximal 12 Stump 2 Other | 1/36 Positive | 48/48 Positive | 38/48 Positive | 31/48 Positive | 17/48 Positive | 17/48 Positive | 25/48 Positive | 24/48 Posituve | 23 Negative 17 Lymphocytes + 8 Tumor + |
Hp status = Helicobacter pylori as determined by histopathology.
Signet = Signet ring cells; Stump = Cancer arose in gastric remnant after prior surgery; Neg = negative; Unk = unknown.
Table 2.
Clinicopathologic Findings in Central American Gastric Cancers
| EBV DNA copies / 100 000 cells | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Case # | Histological Classification |
Site | Hp Status* |
APOB (# of cells) | BamH1W | EBNA1 | LMP1 | LMP2 | BZLF1 | EBER1 |
EBER in situ
hybridization |
| 49 | Diffuse | Distal | S+ | 38 727 | 112 875 | 186 573 | 413 866 | 2 | 215 594 | 141 963 | Tumor + |
| 50 | Diffuse | Proximal | Neg | 5 623 | 44 496 | 295 554 | 282 447 | 18 140 | 175 280 | 249 742 | Tumor + |
| 51 | Intestinal | Distal | Neg | 2 812 | 64 047 | 145 306 | 30 121 | 12 660 | 37 162 | 68 457 | Tumor + |
| 52 | Diffuse/Signet | Distal | S+ | 225 | 222 | 489 | 1.333 | 533 | 489 | 222 | Tumor -; Lymph + |
| 53 | Intestinal | Distal | S+ | 19 857 | 29 | 50 | 101 | 0 | 20 | 0 | Tumor -; Lymph + |
| 54 | Diffuse/Signet | Distal | S+ | 15 568 | 135 | 347 | 225 | 8 | 180 | 276 | Tumor -; Lymph + |
| 55 | Intestinal | Distal | S+/P+ | 23 224 | 142 | 469 | 306 | 22 | 95 | 177 | Tumor -; Lymph + |
| 56 | Diffuse | Distal | S+ | 8 100 | 370 | 444 | 494 | 25 | 716 | 370 | Tumor -; Lymph + |
| 57 | Diffuse/Signet | Distal | S+/P+ | 2 721 | 110 | 221 | 368 | 74 | 74 | 110 | Tumor -; Lymph + |
| 58 | Mixed/Signet | Distal | S+ | 2 967 | 236 | 944 | 472 | 135 | 303 | 236 | Tumor -; Lymph + |
| 59 | Intestinal | Distal | S+ | 3 194 | 188 | 626 | 282 | 63 | 219 | 188 | Tumor -; Lymph + |
| 60 | Diffuse/Signet | Distal | P+ | 1 315 | 61 | 228 | 0 | 0 | 53 | 61 | Tumor -; Lymph + |
| 61 | Intestinal | Distal | Neg | 1 052 | 29 | 124 | 0 | 57 | 0 | 29 | Tumor -; Lymph + |
| 62 | Diffuse/Signet | Distal | S+ | 602 | 166 | 0 | 0 | 0 | 100 | 166 | Tumor -; Lymph + |
| 63 | Diffuse/Signet | Distal | S+ | 4 476 | 134 | 424 | 313 | 0 | 201 | 134 | Tumor -; Lymph + |
| 64 | Diffuse | Distal | S+ | 3 806 | 21 | 32 | 0 | 0 | 26 | 0 | Tumor - |
| 65 | Intestinal | Distal | Neg | 8 504 | 176 | 353 | 294 | 0 | 41 | 0 | Tumor - |
| 66 | Intestinal | Distal | S+ | 10 035 | 0 | 0 | 0 | 0 | 4 | 0 | Tumor - |
| 67 | Intestinal | Distal | Neg | 10 933 | 1 | 0 | 0 | 0 | 0 | 1 | Tumor - |
| 68 | Diffuse/Signet | Proximal | S+/P+ | 7 103 | 42 | 99 | 56 | 0 | 42 | 42 | Tumor - |
| 69 | Diffuse/Signet | Distal | S+ | 1 744 | 0 | 0 | 0 | 0 | 17 | 0 | Tumor - |
| 70 | Intestinal | Distal | S+ | 7 943 | 5 | 25 | 0 | 0 | 15 | 13 | Tumor - |
| 71 | Diffuse | Distal | Neg | 2 869 | 0 | 0 | 0 | 0 | 35 | 0 | Tumor - |
| 72 | Diffuse | Distal | S+ | 2 151 | 14 | 0 | 0 | 0 | 23 | 0 | Tumor - |
| 73 | Intestinal | Distal | S+ | 18 386 | 131 | 239 | 261 | 3 | 109 | 136 | Tumor - |
| 74 | Intestinal | Distal | S+ | 1 726 | 0 | 0 | 0 | 0 | 93 | 116 | Tumor - |
| 75 | Intestinal | Proximal | S+ | 8 757 | 11 | 16 | 0 | 0 | 6 | 0 | Tumor - |
| 76 | Intestinal | Distal | S+ | 847 | 80 | 320 | 0 | 0 | 533 | 0 | Tumor - |
| 77 | Intestinal | Distal | Neg | 10 060 | 1 | 0 | 0 | 0 | 7 | 0 | Tumor - |
| 78 | Diffuse | Distal | S+ | 6 356 | 55 | 173 | 94 | 0 | 24 | 47 | Tumor - |
| 79 | Intestinal | Distal | S+ | 11 728 | 6 | 10 | 12 | 0 | 3 | 6 | Tumor - |
| 80 | Intestinal | Distal | S+/P+ | 809 | 12 | 371 | 0 | 0 | 0 | 12 | Tumor - |
| 81 | Intestinal | Distal | S+ | 288 | 104 | 0 | 0 | 0 | 69 | 104 | Tumor - |
| 82 | Intestinal | Distal | S+ | 738 | 68 | 136 | 0 | 0 | 203 | 68 | Tumor - |
| 83 | Diffuse/Signet | Distal | S+ | 4 454 | 7 | 0 | 0 | 0 | 0 | 7 | Tumor - |
| 84 | Diffuse/Signet | Distal | Neg | 1 255 | 8 | 0 | 0 | 0 | 0 | 8 | Tumor - |
| 85 | Diffuse/Signet | Distal | S+ | 2 009 | 10 | 199 | 0 | 0 | 20 | 10 | Tumor - |
| 86 | Diffuse | Distal | S+ | 3 281 | 49 | 183 | 0 | 0 | 18 | 49 | Tumor - |
| Totals (N=38) | 19 Diffuse 18 Intestinal 1 Mixed 12 Signet | 35 Distal 3 Proximal | 30/38 Positive | 38/38 Positive | 36/38 Positive | 27/38 Positive | 17/38 Positive | 12/38 Positive | 33/38 Positive | 28/38 Positive | 23 Negative 12 Lymphocytes + 3 Tumor + |
Hp status = Helicobacter pylori infection as determined by serology positive (S+) or histopathology positive (P+).
Signet = Signet ring cells; Stump = Cancer arose in gastric stump after prior surgery; Neg = negative; Unk = unknown.
Virologic correlates in gastric adenocarcinomas
EBER in situ hybridization is considered the gold standard assay for localizing latent EBV infection in lesional cells. EBER in situ hybridization revealed EBV within the malignant epithelial cells in 8/48 (17%) of the cancers from the United States and in 3/38 (8%) of the Central American cancers that had detectable EBV DNA by at least one Q-PCR. The EBER-negative cancer tissues often harbored scattered EBER-positive lymphocytes, including 17/40 (42%) United States and 12/35 (34%) Central American cancers. No EBER expression was seen in benign epithelial cells or stromal elements. In the malignant cells, EBER was either uniformly positive or uniformly negative, supporting that EBV infection may have occurred before malignant transformation and was transmitted to all daughter cells in the neoplastic clone.
Interestingly, the viral loads for gastric cancers with detectable EBV DNA could be split into two general categories: 1) viral load greater than 10,000 copies per 100,000 cells, or 2) viral load less than 2,000 copies per 100,000 cells. With two exceptions (discussed below), those viral loads consistently greater than 10,000 EBV DNA copies/100,000 cells (8 from the United States and 3 Central American) were all EBV-associated tumors as defined by EBER localization to the malignant epithelial cells, whereas those with low viral loads lacked EBER in the malignant cells. The viral loads in the EBER-positive gastric cancers were nearly 3,000 fold higher than in the EBER-negative cancers (mean = 162,986 versus 62 EBV DNA copies/100,000 cells, respectively; p=0.003). A cutoff value of 2,000 EBV copies per 100,000 cells is a reasonable and conservative level beyond which EBV was always localized to the malignant cells. A cost-effective screening strategy would be to reserve EBER staining for cases having viral loads over 2000 EBV copies per 100,000 cells. Among the EBER-negative malignancies, the 29 cases having visible EBER-positive lymphocytes by in situ hybridization had significantly higher EBV loads than those lacking visible EBER in lymphocytes (mean = 110 versus 30 EBV DNA copies/100,000 cells, respectively, by the BamH1W assay; p=0.02).
Association between EBV status and clinicopathologic findings
Surprisingly, EBV was more commonly detectable by Q-PCR in Central American than in United States gastric cancer tissues (p=0.0001). But EBV was not more commonly localized to the malignant epithelial cells in Central American compared with United States cancers (p=0.61). Statistical power was insufficient to examine subcategories, but trends showed that EBER-positivity was similarly frequent in the proximal tumors (1/10, 10%) compared to distal tumors (7/62, 11%), and it was slightly more common in stumps (2/12, 17%). In support of previous reports, EBER-positivity was significantly more common in diffuse compared to intestinal histology (p=0.01). However, when Q-PCR was used to identify the virus, detectable EBV DNA was not preferentially found in cancers with diffuse rather than intestinal histology (p=0.83).
All the Central American gastric cancers were located in non-cardia regions. Among US gastric cancers, EBER-positive cases were not preferentially localized in the cardia region of the stomach including cardia comprising a stump after prior surgery. Although our numbers were small: 3/8 EBER-positive cancers occurred in the antrum, 2/8 in the body, 1/8 were widespread, and 2/8 were stump cancers. A similar distribution of anatomical sites was observed for the 17 United States gastric cancers that expressed EBER only in lymphocytes, where 8/17 occurred in the body, 4/17 in the antrum, 3/17 in the cardia, 1/17 were widespread, and 1/17 was a stump cancer.
A negative correlation between EBER-positive cancer and H. pylori serologic status was observed in the Central American cases (p<0.0001). H. pylori status for the United States cases could not be evaluated because serological testing was not performed, and histologic visualization of H. pylori is not reliable in the setting of gastric cancer.
EBV latently infects gastric adenocarcinomas
Immunohistochemistry was performed on all gastric cancers having detectable EBV DNA by any of the Q-PCR assays to localize the viral infection and to further characterize it as latent or lytic based on the spectrum of expressed viral proteins. No lytic EBV infection was observed as signified by the absence of BMRF1 or BZLF1 replicative protein expression in the malignant cells or in reactive stromal tissue. The pattern of latent viral gene expression varied among the gastric cancers. Viral LMP2A staining was observed in 4/11 EBER-positive cancers and all four were among the eight United States cases. When present, LMP2A was restricted to the cytoplasm of malignant epithelial cells and was expressed diffusely in 1 case and focally in up to 10% of malignant cells in 3 cases (Figure 1A). Viral LMP1 expression was seen in only 1/11 EBER-positive gastric cancers, Central American case #51. In that case, membranous/cytoplasmic LMP1 was diffusely expressed and restricted to the malignant epithelial cells (Figure 1B), and the cells did not coexpress LMP2. No staining for LMP1 or LMP2 was observed in EBER-negative gastric cancers, nor was either protein expressed in infiltrating lymphocytes or benign epithelial cells.
Figure 1. Latent viral protein expression in gastric adenocarcinomas.
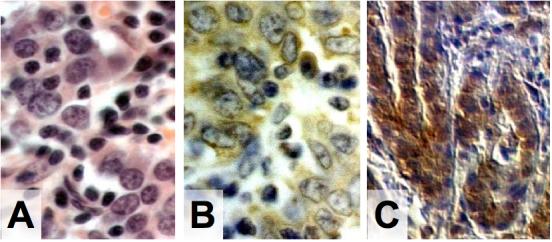
A) Hematoxylin and eosin stain reveals gastric adenocarcinoma in United States Case #6. B) Immunohistochemistry shows LMP2A expression localized to the malignant cells of United States Case #6. C) Immunohistochemistry reveals LMP1 is expressed in the malignant cells of Central American Case #51. (A, B 400×; C 100×)
Taken together, our results show three different viral latency profiles for EBV-associated gastric cancer: The majority of the EBER-positive cancers (4/8 United States and 2/3 Central American) were EBER+/LMP1-/LMP2A-. Less frequently, the latency profiles EBER+/LMP1-/LMP2A+ (4/8 United States) and EBER+/LMP1+/LMP2A-(1/3 Central American) were observed.
Viral genomic variation in an EBV-associated Central American Gastric Cancer
EBER-positive Central American gastric cancer #49 had selective dropout of amplifiable EBV DNA for the LMP2 locus whereas the viral loads were greater than 112,000 EBV DNA copies per 100,000 cells for the other five viral load assays (Table 2). Immunohistochemical analysis of this cancer revealed no LMP2A protein expression, however this is not necessarily a result of a viral genomic defect since most cancers in this series did not express LMP2A. Although the primers and probes were designed to target a highly conserved segment of the LMP2 gene, the selective non-amplification suggests either mutation or deletion interfering with primer/probe binding. To resolve this dilemma, amplification and sequencing was performed on the relevant LMP2 gene segment. A point mutation was found near the 5′ end of the reverse primer, only three bases in from where the polymerase initiates strand extension (Figure 2). This mutation involved a conversion from a cytosine to a thymine at position number 733 in the LMP2 gene sequence compared to the wild type sequence found in the positive control and in GenBank Accession No. NC_007605. Our findings suggest that the LMP2 733C>T mutation hinders annealing of the reverse primer, resulting in reduced amplicon production.
Figure 2. EBV LMP2 DNA sequencing revealed a mutation interfering with primer binding.
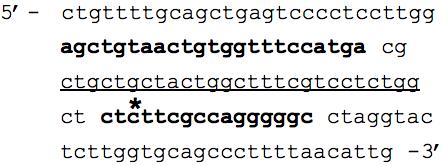
DNA sequencing was performed on a 130bp region of EBV LMP2 encompassing the 69bp region targeted by the LMP2 Q-PCR assay. The forward and reverse primer binding sites for the LMP2 Q-PCR assay are in bold font, while the TaqMan probe site is underlined. The star represents the position of a point mutation (C>T base substitution at position 733) in LMP2 exon 4 of Central American Case #49, which is predicted to result in substitution of serine by phenylalanine; this substitution of a large nonpolar for a small polar amino acid may have functional significance in addition to its apparent interference with laboratory testing.
Viral genomic variation in an EBER-negative US Gastric Cancer
The other exceptional case was an EBER-negative US gastric cancer case #24 with a single high LMP1 Q-PCR result and no other evidence of the virus beyond rare EBER-positive lymphocytes by in situ hybridization. The high level of LMP1 DNA is consistent with the levels of seen in EBV-associated gastric cancer rather than the levels seen in background infection of bystander lymphocytes. Repeat testing on four occasions verified the unusual results.
Discussion
This study is the first to demonstrate that quantitative real-time DNA amplification technology is equivalent to EBER in situ hybridization for identifying cases of gastric cancer that are EBV-related. We further showed that targeting viral DNA or EBER RNA is superior to immunohistochemical detection of selected viral proteins since these proteins are not consistently expressed at visible levels. Furthermore, expression patterns of viral proteins differed among cases, but always revealed latent infection. Use of multiple Q-PCR assays revealed that polymorphisms (mutation or deletion) of the viral genome may be more frequent than previously recognized.
When a battery of six different Q-PCR assays was applied to determine if one or a combination of these assays could be used to screen for EBV-related cancer, most of the Q-PCR assays were equally informative. A threshold could be set that clearly distinguished EBER-positive from EBER-negative cancers regardless of whether EBER-positive infiltrating lymphocytes were seen. With the exception of two unusual cases discussed below, no EBER-negative cancer had a viral load over 1,629, and no EBER-positive cancer had a level below 10,558 EBV copies per 100,000 cells. A cutoff value of 2,000 copies per 100,000 cells is suggested as a threshold above which an EBER stain should be done to insure that the EBV signal is localized to the malignant cells, and below which EBER is likely to be restricted to benign lymphocytes, assuming that the tumor is adequately represented in the sample. Note that this cutoff is substantially higher than the threshold of 100 EBV copies per 100,000 cells that had been set in a prior study of non-gastric malignancies, including non-Hodgkin lymphomas.(37) This difference suggests that EBER-negative gastric cancer tissues tend to have higher EBV loads than do EBER-negative lymphoma tissues, implicating higher levels of EBV in the non-malignant cells of the stomach.
Prior work localized EBV to benign lymphocytes in gastritis lesions and other preneoplastic gastric lesions.(14, 29, 39-43) In the current study, scattered EBER-positive lymphocytes were found in some EBER-negative cancer tissues where they tended to correlate with measurable viral loads. No other benign cell types were found to contain either latent or lytic viral infection by histochemical stains. Although we did not study preneoplastic tissue or lesions, it will be important to determine if patients who eventually develop cancer have higher levels of EBV beforehand. In this regard, it is worth considering if the high incidence of gastric cancer in Central America is related to the relatively high EBV loads seen in every gastric cancer from that region regardless of EBER status. Further studies are needed to explore whether host genetic, immunologic, infectious, or nutritional factors caused the levels of EBV to be higher in Central American compared to United States gastric cancer tissue.
The laboratory assays described in the current study were clearly useful for identifying infected cancers, and further work is needed to explore their utility for measuring low level EBV in preneoplastic tissue or in blood. Among the six assays that were used, the BamH1W assay had the lowest limit of detection, while the LMP1 and LMP2 assays were more likely to miss low-level infection as judged by the frequency of amplification in our EBER-negative cancer tissues. For applications in which analytic sensitivity is critical, the BamH1W Q-PCR is recommended.
Our results build on prior work by others demonstrating that EBV is found in distinct clinicopathologic subtypes of gastric cancer, suggesting that the virus is not just an innocent bystander but rather it tracks with fundamental biologic and immunologic characteristics that are likely to influence tumor development and maintenance. Although the incidence of gastric cancer is much higher in Central America than in the United States, our work shows that the proportion of EBV-related cases (as defined by EBER expression in malignant cells) is not radically different between the two countries (8% versus 17%, respectively; p=0.23) and is comparable to rates described by others worldwide.(4, 5, 7, 11, 29, 32, 44, 45) The geographic variation is not as dramatic as with other EBV-related diseases such as Burkitt lymphoma in which EBV-positivity rates vary from over 95% in tropical Africa to 25% in the United States, and nasopharyngeal carcinoma where rates vary from over 95% in Southern China to about 75% in the United States.(18)
A known contributing oncogenic factor is H. pylori infection. In fact, both EBV and H. pylori are considered class 1 oncogenic pathogens by the World Health Organization (WHO) and are associated with overlapping subsets of gastric carcinoma.(6, 46, 47) H. pylori is endemic among adults in Honduras, while the United States carrier rate is about 30% among adults. Unless treated with medications to eradicate the bacterium, the majority of individuals maintain chronic infection. In our Central American cohort on whom serologic testing was performed, a significant negative association was found between EBER-positive cancer and H. pylori infection. Further studies are warranted to determine if different mechanisms of carcinogenesis are operative.
Because mutation or partial genomic deletion could interfere with detection of viral DNA or gene products, a battery of six Q-PCR assays was used to assay multiple segments of the viral genome. In any given tumor specimen, the viral loads were fairly consistent across the six Q-PCR assays and were clearly informative with regard to EBER in situ hybridization status, suggesting that one or more of these Q-PCR assays could be used as a screening tool for EBV-related cancer in paraffin-embedded tissue. It is important to note that one of the gastric cancers in this series would have yielded a false negative interpretation of EBV status if only the LMP2 Q-PCR assay had been used. In a prior study, false negative Q-PCR assays targeting either LMP2, LMP1, or BZLF1 were seen in AIDS lymphomas.(37) These findings demonstrate the importance of targeting more than one segment of the viral genome to ensure sensitive detection of the virus.
US gastric cancer case #24 was exceptional in that a high LMP1 Q-PCR value was found despite undetectable virus by the other five Q-PCRs. The high level of LMP1 DNA is consistent with the levels seen in EBV-associated gastric cancers, however the gold standard EBER in situ assay showed no EBER localization to malignant cells, and LMP1 immunohistochemistry was likewise negative. While it is feasible that this LMP1 Q-PCR result is false positive (perhaps crossreacting with a rare pathogen), one must consider the possibility that the result legitimately reflects a segment of EBV DNA. Interestingly, the LMP1 gene is considered to be an oncogene in that it has many qualities that promote tumorigenesis. First, deletion of the LMP1 gene renders EBV incapable of immortalizing B cells.(48) Second, LMP1 functions as a TNF receptor that constitutively signals through multiple growth-promoting and anti-apoptotic pathways including EGFR, NFKB, PI3K and AKT.(49, 50) Its effect on the inhibitors of differentiation (ID1 and ID3) could help explain why infected carcinomas tend to appear so undifferentiated.(51) Transgenic mice expressing LMP1 behind a keratin promoter develop epithelial hyperplasia while LMP1 expressed behind an immunoglobulin promoter induces B cell lymphoma.(50) In this patient, it is feasible that a defective recombinant viral genome containing LMP1 could have been produced naturally by atypical rearrangement and partial deletion of the EBV genome. Integration of the defective LMP1-containing segment into host chromosomal DNA is a possible mechanism of persistence. The intact EBV genome may also persist in this patient as evidenced by rare EBER-expressing lymphocytes. If future work confirms that remnants of the EBV genome are indeed present within the malignant cells in cases such as this one, it would lend support to the hit-and-run hypothesis by which EBV might have contributed early during tumorigenesis but then was largely eliminated from the epithelial cells prior to clonal expansion.
The current study demonstrates that latent, not lytic, EBV infection characterizes gastric adenocarcinomas. This confirms previous reports showing that virally encoded EBER and LMP2 are often expressed, but LMP1 is very rarely expressed.(29, 30, 32, 52-56) EBER, a non-polyadenylated transcript, is thought to promote growth of gastric epithelial cells at least in part through induction of insulin-like growth factor (IGF1).(57) Only one case in the current series expressed LMP1 by histochemical analysis, while 4/11 infected cases expressed LMP2A. It is important to characterize viral gene expression not only because viral gene products serve as targets to assist in laboratory diagnosis, but also because they may serve as targets for therapy. The immune system seems to tolerate latent viral infection in this and other infected malignancies.(58) Some progress has been made in managing EBV-related cancers by infusing cytotoxic T cells which have been expanded in vitro through stimulation by viral proteins.(59) Another promising strategy is to use differentiating agents that induce lytic viral replication, which incites a strong antiviral immune reaction.(60-62)
Prior research showed lytic EBV infection in some gastric adenocarcinomas using either histochemistry or rtPCR.(29, 32) In the current study, lytic viral protein expression was not detected by either BZLF1 or BMRF1 immunohistochemistry, suggesting that cells undergoing replicative infection are rare. These findings are relevant to patient care since one would predict that well-tolerated oral antiviral agents like gancyclovir, which target replicative infection, are unlikely to eliminate malignant cells. Nevertheless, even rare cells expressing lytic viral proteins may contribute to angiogenesis and tumor growth.(63) In fact, Kenney and colleagues succeeded in killing malignant cells using gancyclovir combined with standard chemotherapeutic or radiation therapies, presumably because gancyclovir is activated by phosphorylation in cells that the standard therapies induce to switch from latency to lytic viral replication.(64-66) Not only are the lytically infected cells killed, but adjacent cells die as phosphorylated gancyclovir is transferred to them and exerts its cytotoxic activity.(64) Once EBV-directed therapies are established to be useful in patient management, it will be all the more important to identify virally-infected cancers that would be predicted to respond to the therapy. Meantime, the utility of knowing that a particular patient’s cancer is EBV-infected may be limited to prognosis(7, 67) and to measurement of circulating EBV DNA as a marker of tumor burden.(68) The laboratory assays described herein should prove useful for identifying affected patients.
Acknowledgements
The authors thank Rachel Edwards for providing LMP2 gene sequence data.
This study was sponsored by the University of North Carolina Department of Pathology and Laboratory Medicine and the Environmental Pathology Training Grant (T32-ES07017) supporting the graduate studies of Julie L. Ryan, PhD, MPH. The study was also sponsored in part by the NIH Mentored Scholars Research Award (K12 RR1667) and the UNC Center for Gastrointestinal Biology and Disease (P30 DK 034987) supporting Douglas Morgan, MD MPH.
Abbreviations
- (EBV)
Epstein-Barr virus
- (US)
United States
- (Q-PCR)
Quantitative polymerase chain reaction
- (EBER)
Epstein Barr virus-encoded RNA
- (H. pylori)
Helicobactor pylori
Footnotes
Disclosure/Duality of Interest: The authors have no duality of interest to disclose.
References
- 1.Correa P, Piazuelo MB, Camargo MC. The future of gastric cancer prevention. Gastric Cancer. 2004;7(1):9–16. doi: 10.1007/s10120-003-0265-0. [DOI] [PubMed] [Google Scholar]
- 2.Boussioutas A, Taupin D. Towards a molecular approach to gastric cancer management. Intern Med J. 2001;31(5):296–303. doi: 10.1046/j.1445-5994.2001.00057.x. [DOI] [PubMed] [Google Scholar]
- 3.Henson DE, Dittus C, Younes M, Nguyen H, Albores-Saavedra J. Differential trends in the intestinal and diffuse types of gastric carcinoma in the United States, 1973-2000: increase in the signet ring cell type. Arch Pathol Lab Med. 2004;128(7):765–770. doi: 10.5858/2004-128-765-DTITIA. [DOI] [PubMed] [Google Scholar]
- 4.Carrascal E, Koriyama C, Akiba S, Tamayo O, Itoh T, Eizuru Y, et al. Epstein-Barr virus-associated gastric carcinoma in Cali, Colombia. Oncol Rep. 2003;10(4):1059–1062. [PubMed] [Google Scholar]
- 5.Corvalan A, Koriyama C, Akiba S, Eizuru Y, Backhouse C, Palma M, et al. Epstein-Barr virus in gastric carcinoma is associated with location in the cardia and with a diffuse histology: a study in one area of Chile. Int J Cancer. 2001;94(4):527–530. doi: 10.1002/ijc.1510. [DOI] [PubMed] [Google Scholar]
- 6.Cho HJ, Kim JY, Yoo J, Lee SS. Gastric carcinoma with lymphoid stroma: incidence of EBV and Helicobacter pylori infection. Appl Immunohistochem Mol Morphol. 2003;11(2):149–152. doi: 10.1097/00129039-200306000-00010. [DOI] [PubMed] [Google Scholar]
- 7.van Beek J, zur Hausen A, Klein Kranenbarg E, van de Velde CJ, Middeldorp JM, van den Brule AJ, et al. EBV-positive gastric adenocarcinomas: a distinct clinicopathologic entity with a low frequency of lymph node involvement. J Clin Oncol. 2004;22(4):664–670. doi: 10.1200/JCO.2004.08.061. [DOI] [PubMed] [Google Scholar]
- 8.Oda K, Koda K, Takiguchi N, Nunomura M, Seike K, Miyazaki M. Detection of Epstein-Barr virus in gastric carcinoma cells and surrounding lymphocytes. Gastric Cancer. 2003;6(3):173–178. doi: 10.1007/s10120-003-0247-2. [DOI] [PubMed] [Google Scholar]
- 9.Pattle SB, Farrell PJ. The role of Epstein-Barr virus in cancer. Expert Opin Biol Ther. 2006;6(11):1193–1205. doi: 10.1517/14712598.6.11.1193. [DOI] [PubMed] [Google Scholar]
- 10.Gulley ML, Tang W. Laboratory assays for epstein-barr virus-related disease. J Mol Diagn. 2008;10(4):279–292. doi: 10.2353/jmoldx.2008.080023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koriyama C, Akiba S, Corvalan A, Carrascal E, Itoh T, Herrera-Goepfert R, et al. Histology-specific gender, age and tumor-location distributions of Epstein-Barr virus-associated gastric carcinoma in Japan. Oncol Rep. 2004;12(3):543–547. [PubMed] [Google Scholar]
- 12.Fukayama M, Chong JM, Kaizaki Y. Epstein-Barr virus and gastric carcinoma. Gastric Cancer. 1998;1(2):104–114. doi: 10.1007/s101200050003. [DOI] [PubMed] [Google Scholar]
- 13.Lee HS, Chang MS, Yang HK, Lee BL, Kim WH. Epstein-barr virus-positive gastric carcinoma has a distinct protein expression profile in comparison with epstein-barr virus-negative carcinoma. Clin Cancer Res. 2004;10(5):1698–1705. doi: 10.1158/1078-0432.ccr-1122-3. [DOI] [PubMed] [Google Scholar]
- 14.Schneider BG, Gulley ML, Eagan P, Bravo JC, Mera R, Geradts J. Loss of p16/CDKN2A tumor suppressor protein in gastric adenocarcinoma is associated with Epstein-Barr virus and anatomic location in the body of the stomach. Hum Pathol. 2000;31(1):45–50. doi: 10.1016/s0046-8177(00)80197-5. [DOI] [PubMed] [Google Scholar]
- 15.Zur Hausen A, van Rees BP, van Beek J, Craanen ME, Bloemena E, Offerhaus GJ, et al. Epstein-Barr virus in gastric carcinomas and gastric stump carcinomas: a late event in gastric carcinogenesis. J Clin Pathol. 2004;57(5):487–491. doi: 10.1136/jcp.2003.014068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nishikawa J, Yanai H, Hirano A, Okamoto T, Nakamura H, Matsusaki K, et al. High prevalence of Epstein-Barr virus in gastric remnant carcinoma after Billroth-II reconstruction. Scand J Gastroenterol. 2002;37(7):825–829. [PubMed] [Google Scholar]
- 17.Yamamoto N, Tokunaga M, Uemura Y, Tanaka S, Shirahama H, Nakamura T, et al. Epstein-Barr virus and gastric remnant cancer. Cancer. 1994;74(3):805–809. doi: 10.1002/1097-0142(19940801)74:3<805::aid-cncr2820740304>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 18.Gulley ML. Molecular diagnosis of Epstein-Barr virus-related diseases. J Mol Diagn. 2001;3(1):1–10. doi: 10.1016/S1525-1578(10)60642-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grinstein S, Preciado MV, Gattuso P, Chabay PA, Warren WH, De Matteo E, et al. Demonstration of Epstein-Barr virus in carcinomas of various sites. Cancer Res. 2002;62(17):4876–4878. [PubMed] [Google Scholar]
- 20.Sixbey J. Epstein-Barr virus DNA loss from tumor cells and the geography of Burkitt’s lymphoma. Epstein-Barr Virus Report. 2000;7:37–40. [Google Scholar]
- 21.Gan YJ, Razzouk BI, Su T, Sixbey JW. A defective, rearranged Epstein-Barr virus genome in EBER-negative and EBER-positive Hodgkin’s disease. Am J Pathol. 2002;160(3):781–786. doi: 10.1016/S0002-9440(10)64900-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takeuchi H, Kobayashi R, Hasegawa M, Hirai K. Detection of latent Epstein-Barr virus (EBV) DNA in paraffin sections of nasopharyngeal carcinomas expressing no EBV-encoded small RNAs using in situ PCR. Arch Virol. 1997;142(9):1743–1756. doi: 10.1007/s007050050194. [DOI] [PubMed] [Google Scholar]
- 23.Chen PC, Pan CC, Yang AH, Wang LS, Chiang H. Detection of Epstein-Barr virus genome within thymic epithelial tumours in Taiwanese patients by nested PCR, PCR in situ hybridization, and RNA in situ hybridization. J Pathol. 2002;197(5):684–688. doi: 10.1002/path.1141. [DOI] [PubMed] [Google Scholar]
- 24.Korabecna M, Ludvikova M, Skalova A. Molecular diagnosis of Epstein-Barr virus in paraffin-embedded tissues of tumors with abundant lymphoid infiltration. Neoplasma. 2003;50(1):8–12. [PubMed] [Google Scholar]
- 25.Lauritzen AF, Hording U, Nielsen HW. Epstein-Barr virus and Hodgkin’s disease: a comparative immunological, in situ hybridization, and polymerase chain reaction study. Apmis. 1994;102(7):495–500. doi: 10.1111/j.1699-0463.1994.tb05196.x. [DOI] [PubMed] [Google Scholar]
- 26.Niedobitek G, Herbst H. In situ detection of Epstein-Barr virus and phenotype determination of EBV-infected cells. Methods Mol Biol. 2006;326:115–137. doi: 10.1385/1-59745-007-3:115. [DOI] [PubMed] [Google Scholar]
- 27.Delecluse HJ, Feederle R, O’Sullivan B, Taniere P. Epstein Barr virus-associated tumours: an update for the attention of the working pathologist. J Clin Pathol. 2007;60(12):1358–1364. doi: 10.1136/jcp.2006.044586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hennard C, Pfuhl T, Buettner M, Becker KF, Knofel T, Middeldorp J, et al. The antibody 2B4 directed against the Epstein-Barr virus (EBV)-encoded nuclear antigen 1 (EBNA1) detects MAGE-4: implications for studies on the EBV association of human cancers. J Pathol. 2006;209(4):430–435. doi: 10.1002/path.1996. [DOI] [PubMed] [Google Scholar]
- 29.Gulley ML, Pulitzer DR, Eagan PA, Schneider BG. Epstein-Barr virus infection is an early event in gastric carcinogenesis and is independent of bcl-2 expression and p53 accumulation. Hum Pathol. 1996;27(1):20–27. doi: 10.1016/s0046-8177(96)90133-1. [DOI] [PubMed] [Google Scholar]
- 30.Hoshikawa Y, Satoh Y, Murakami M, Maeta M, Kaibara N, Ito H, et al. Evidence of lytic infection of Epstein-Barr virus (EBV) in EBV-positive gastric carcinoma. J Med Virol. 2002;66(3):351–359. doi: 10.1002/jmv.2152. [DOI] [PubMed] [Google Scholar]
- 31.Wang Y, Luo B, Yan LP, Huang BH, Zhao P. Relationship between Epstein-Barr virus-encoded proteins with cell proliferation, apoptosis, and apoptosis-related proteins in gastric carcinoma. World J Gastroenterol. 2005;11(21):3234–3239. doi: 10.3748/wjg.v11.i21.3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luo B, Wang Y, Wang XF, Liang H, Yan LP, Huang BH, et al. Expression of Epstein-Barr virus genes in EBV-associated gastric carcinomas. World J Gastroenterol. 2005;11(5):629–633. doi: 10.3748/wjg.v11.i5.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Andal N, Shanthi P, Krishnan KB, Taralaxmi V. The Epstein Barr virus and gastric carcinoma. Indian J Pathol Microbiol. 2003;46(1):34–36. [PubMed] [Google Scholar]
- 34.van Beek J, zur Hausen A, Kranenbarg EK, Warring RJ, Bloemena E, Craanen ME, et al. A rapid and reliable enzyme immunoassay PCR-based screening method to identify EBV-carrying gastric carcinomas. Mod Pathol. 2002;15(8):870–877. doi: 10.1097/01.MP.0000024147.43288.B1. [DOI] [PubMed] [Google Scholar]
- 35.Morgan DR, Dominguez RL, Keku TO, Heidt PE, Martin CF, Galanko JA, et al. Gastric cancer and the high combination prevalence of host cytokine genotypes and Helicobacter pylori in Honduras. Clin Gastroenterol Hepatol. 2006;4(9):1103–1111. doi: 10.1016/j.cgh.2006.05.025. [DOI] [PubMed] [Google Scholar]
- 36.Fenoglio-Presier C, Carneiro F, Correa P. Tumours of the Stomach. In: SR IH, editor. World Health Organization Classification of Tumours: Pathology and Genetics of Tumours of the Digestive System. IARC Press; Lyon, France: 2000. pp. 37–52. [Google Scholar]
- 37.Ryan JL, Fan H, Glaser SL, Schichman SA, Raab-Traub N, Gulley ML. Epstein-Barr virus quantitation by real-time PCR targeting multiple gene segments: a novel approach to screen for the virus in paraffin-embedded tissue and plasma. J Mol Diagn. 2004;6(4):378–385. doi: 10.1016/S1525-1578(10)60535-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ling PD, Vilchez RA, Keitel WA, Poston DG, Peng RS, White ZS, et al. Epstein-Barr virus DNA loads in adult human immunodeficiency virus type 1-infected patients receiving highly active antiretroviral therapy. Clin Infect Dis. 2003;37(9):1244–1249. doi: 10.1086/378808. [DOI] [PubMed] [Google Scholar]
- 39.Yanai H, Takada K, Shimizu N, Mizugaki Y, Tada M, Okita K. Epstein-Barr virus infection in non-carcinomatous gastric epithelium. J Pathol. 1997;183(3):293–298. doi: 10.1002/(SICI)1096-9896(199711)183:3<293::AID-PATH937>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 40.Arikawa J, Tokunaga M, Tashiro Y, Tanaka S, Sato E, Haraguchi K, et al. Epstein-Barr virus-positive multiple early gastric cancers and dysplastic lesions: a case report. Pathol Int. 1997;47(10):730–734. doi: 10.1111/j.1440-1827.1997.tb04450.x. [DOI] [PubMed] [Google Scholar]
- 41.Hungermann D, Muller S, Spieker T, Lisner R, Niedobitek G, Herbst H. Low prevalence of latently Epstein-Barr virus-infected cells in chronic gastritis. Microsc Res Tech. 2001;53(6):409–413. doi: 10.1002/jemt.1109. [DOI] [PubMed] [Google Scholar]
- 42.Hirano A, Yanai H, Shimizu N, Okamoto T, Matsubara Y, Yamamoto K, et al. Evaluation of epstein-barr virus DNA load in gastric mucosa with chronic atrophic gastritis using a real-time quantitative PCR assay. Int J Gastrointest Cancer. 2003;34(2-3):87–94. doi: 10.1385/IJGC:34:2-3:087. [DOI] [PubMed] [Google Scholar]
- 43.Imai S, Koizumi S, Sugiura M, Tokunaga M, Uemura Y, Yamamoto N, et al. Gastric carcinoma: monoclonal epithelial malignant cells expressing Epstein-Barr virus latent infection protein. Proc Natl Acad Sci U S A. 1994;91(19):9131–9135. doi: 10.1073/pnas.91.19.9131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Alipov G, Nakayama T, Nakashima M, Wen CY, Niino D, Kondo H, et al. Epstein-Barr virus-associated gastric carcinoma in Kazakhstan. World J Gastroenterol. 2005;11(1):27–30. doi: 10.3748/wjg.v11.i1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vo QN, Geradts J, Gulley ML, Boudreau DA, Bravo JC, Schneider BG. Epstein-Barr virus in gastric adenocarcinomas: association with ethnicity and CDKN2A promoter methylation. J Clin Pathol. 2002;55(9):669–675. doi: 10.1136/jcp.55.9.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chan WY, Liu Y, Li CY, Ng EK, Chow JH, Li KK, et al. Recurrent genomic aberrations in gastric carcinomas associated with Helicobacter pylori and Epstein-Barr virus. Diagn Mol Pathol. 2002;11(3):127–134. doi: 10.1097/00019606-200209000-00001. [DOI] [PubMed] [Google Scholar]
- 47.Saxena A, Nath Prasad K, Chand Ghoshal U, Krishnani N, Roshan Bhagat M, Husain N. Association of Helicobacter pylori and Epstein-Barr virus with gastric cancer and peptic ulcer disease. Scand J Gastroenterol. 2008;43(6):669–674. doi: 10.1080/00365520801909660. [DOI] [PubMed] [Google Scholar]
- 48.Kaye KM, Izumi KM, Li H, Johannsen E, Davidson D, Longnecker R, et al. An Epstein-Barr virus that expresses only the first 231 LMP1 amino acids efficiently initiates primary B-lymphocyte growth transformation. J Virol. 1999;73(12):10525–10530. doi: 10.1128/jvi.73.12.10525-10530.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Song YJ, Jen KY, Soni V, Kieff E, Cahir-McFarland E. IL-1 receptor-associated kinase 1 is critical for latent membrane protein 1-induced p65/RelA serine 536 phosphorylation and NF-kappaB activation. Proc Natl Acad Sci U S A. 2006;103(8):2689–2694. doi: 10.1073/pnas.0511096103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thornburg NJ, Kulwichit W, Edwards RH, Shair KH, Bendt KM, Raab-Traub N. LMP1 signaling and activation of NF-kappaB in LMP1 transgenic mice. Oncogene. 2006;25(2):288–297. doi: 10.1038/sj.onc.1209023. [DOI] [PubMed] [Google Scholar]
- 51.Everly DN, Jr., Mainou BA, Raab-Traub N. Induction of Id1 and Id3 by latent membrane protein 1 of Epstein-Barr virus and regulation of p27/Kip and cyclin-dependent kinase 2 in rodent fibroblast transformation. J Virol. 2004;78(24):13470–13478. doi: 10.1128/JVI.78.24.13470-13478.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Qiu K, Tomita Y, Hashimoto M, Ohsawa M, Kawano K, Wu DM, et al. Epstein-Barr virus in gastric carcinoma in Suzhou, China and Osaka, Japan: association with clinico-pathologic factors and HLA-subtype. Int J Cancer. 1997;71(2):155–158. doi: 10.1002/(sici)1097-0215(19970410)71:2<155::aid-ijc5>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 53.Chu PG, Chang KL, Chen WG, Chen YY, Shibata D, Hayashi K, et al. Epstein-Barr virus (EBV) nuclear antigen (EBNA)-4 mutation in EBV-associated malignancies in three different populations. Am J Pathol. 1999;155(3):941–947. doi: 10.1016/S0002-9440(10)65193-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Koriyama C, Shinkura R, Hamasaki Y, Fujiyoshi T, Eizuru Y, Tokunaga M. Human leukocyte antigens related to Epstein-Barr virus-associated gastric carcinoma in Japanese patients. Eur J Cancer Prev. 2001;10(1):69–75. doi: 10.1097/00008469-200102000-00008. [DOI] [PubMed] [Google Scholar]
- 55.Lee MA, Hong YS, Kang JH, Lee KS, You JY, Lee KY, et al. Detection of Epstein-Barr virus by PCR and expression of LMP1, p53, CD44 in gastric cancer. Korean J Intern Med. 2004;19(1):43–47. doi: 10.3904/kjim.2004.19.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sugiura M, Imai S, Tokunaga M, Koizumi S, Uchizawa M, Okamoto K, et al. Transcriptional analysis of Epstein-Barr virus gene expression in EBV-positive gastric carcinoma: unique viral latency in the tumour cells. Br J Cancer. 1996;74(4):625–631. doi: 10.1038/bjc.1996.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Iwakiri D, Eizuru Y, Tokunaga M, Takada K. Autocrine growth of Epstein-Barr virus-positive gastric carcinoma cells mediated by an Epstein-Barr virus-encoded small RNA. Cancer Res. 2003;63(21):7062–7067. [PubMed] [Google Scholar]
- 58.Lopes V, Young LS, Murray PG. Epstein-Barr virus-associated cancers: aetiology and treatment. Herpes. 2003;10(3):78–82. [PubMed] [Google Scholar]
- 59.Savoldo B, Goss JA, Hammer MM, Zhang L, Lopez T, Gee AP, et al. Treatment of solid organ transplant recipients with autologous Epstein Barr virus-specific cytotoxic T lymphocytes (CTLs) Blood. 2006;108(9):2942–2949. doi: 10.1182/blood-2006-05-021782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kenney S, Theodore E. Woodward Award: Development of Novel, EBV-Targeted Therapies for EBV-Positive Tumors. Trans Am Clin Climatol Assoc. 2006;117:55–74. [PMC free article] [PubMed] [Google Scholar]
- 61.Schwarzmann F, Jager M, Prang N, Wolf H. The control of lytic replication of Epstein-Barr virus in B lymphocytes (Review) Int J Mol Med. 1998;1(1):137–142. doi: 10.3892/ijmm.1.1.137. [DOI] [PubMed] [Google Scholar]
- 62.Laichalk LL, Thorley-Lawson DA. Terminal differentiation into plasma cells initiates the replicative cycle of Epstein-Barr virus in vivo. J Virol. 2005;79(2):1296–1307. doi: 10.1128/JVI.79.2.1296-1307.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hong GK, Kumar P, Wang L, Damania B, Gulley ML, Delecluse HJ, et al. Epstein-Barr virus lytic infection is required for efficient production of the angiogenesis factor vascular endothelial growth factor in lymphoblastoid cell lines. J Virol. 2005;79(22):13984–13992. doi: 10.1128/JVI.79.22.13984-13992.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Feng WH, Israel B, Raab-Traub N, Busson P, Kenney SC. Chemotherapy induces lytic EBV replication and confers ganciclovir susceptibility to EBV-positive epithelial cell tumors. Cancer Res. 2002;62(6):1920–1926. [PubMed] [Google Scholar]
- 65.Feng WH, Westphal E, Mauser A, Raab-Traub N, Gulley ML, Busson P, et al. Use of adenovirus vectors expressing Epstein-Barr virus (EBV) immediate-early protein BZLF1 or BRLF1 to treat EBV-positive tumors. J Virol. 2002;76(21):10951–10959. doi: 10.1128/JVI.76.21.10951-10959.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Feng WH, Hong G, Delecluse HJ, Kenney SC. Lytic induction therapy for Epstein-Barr virus-positive B-cell lymphomas. J Virol. 2004;78(4):1893–1902. doi: 10.1128/JVI.78.4.1893-1902.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Koriyama C, Akiba S, Itoh T, Kijima Y, Sueyoshi K, Corvalan A, et al. Prognostic significance of Epstein-Barr virus involvement in gastric carcinoma in Japan. Int J Mol Med. 2002;10(5):635–639. [PubMed] [Google Scholar]
- 68.Lo YM, Chan WY, Ng EK, Chan LY, Lai PB, Tam JS, et al. Circulating Epstein-Barr virus DNA in the serum of patients with gastric carcinoma. Clin Cancer Res. 2001;7(7):1856–1859. [PubMed] [Google Scholar]


