Abstract
Two partial mRNA sequences predicted to encode anti-lipopolysaccharide factors (ALFs) were identified among expressed sequence tags generated from the American lobster Homarus americanus and complete cDNA sequences were obtained from library clones. Comparison of the translated amino acid sequences to those publicly available confirmed similarity to arthropod anti-lipopolysaccharide factors. Both protein sequences, designated ALFHa-1 and ALFHa-2, contained an N-terminal signal peptide and two half-cysteines participating in a disulfide bridge, features conserved in other ALFs. Predicted secondary structures were similar to that described for the ALF from the horseshoe crab Limulus polyphemus. As part of an exploratory study of immunity in H. americanus, lobsters were injected with the bacterium Vibrio fluvialis and gill, hematopoietic, and hepatopancreas tissues were sampled for analysis of gene expression of ALFHa-1 and ALFHa-2 by quantitative PCR. The relative abundance of ALFHa-2 mRNA was not significantly affected by Vibrio injection in any of the three tissues tested. In contrast, ALFHa-1 mRNA levels in gills were increased by the treatment some 17-fold. Our results support a molecularly specific regulation of antimicrobial proteins in response to bacterial infection in H. americanus.
Keywords: crustacean, antimicrobial peptides, real-time quantitative PCR, DNA sequencing, gill tissue, hepatopancreas tissue, hematopoetic tissue
1. Introduction
Species and strains of Vibrio bacteria have become an increasing problem in the crustacean mariculture industry, affecting lobster, crab, and shrimp. In the American lobster Homarus americanus, the subgroup Vibrio fluvialis has been implicated in limp lobster disease (Tall et al. 2003) and as a member of the bacterial community associated with epizootic shell disease (Smolowitz et al. 2005). This gram-negative bacterium has caused significant economic detriment to the lobster industries of the eastern coasts of the US and Canada, with infections resulting in losses exceeding $2.5 million annually (Tall et al. 2003). Although antibiotics could be a possible means of treatment, they are only feasible in holding facilities where antibiotic resistance could become an issue. The industry prides itself on being sustainable and environmentally friendly, making the use of broad spectrum antibiotics a particularly unattractive option.
The immune system of arthropods does not exhibit adaptive immunity in response to potential pathogens, but uses instead a system of innate immunity that includes pathogen recognition, phagocytosis, encapsulation, cytotoxicity, and melanization (Söderhäll et al. 1992; Muta et al. 1996). Much of this activity takes place in hemocytes, where antimicrobial peptides and proteins are synthesized, stored, and released into the hemolymph upon exposure to infective agents (Iwanaga et al. 1998; Johansson et al. 2000).
Although a number of antimicrobial proteins have been identified in crustaceans, primarily in shrimp species, little research has been conducted to characterize these factors in the American lobster Homarus americanus. One antimicrobial protein from lobster, crustin, has been characterized in our laboratories (Christie et al. 2007). The present study identified two additional antimicrobial proteins related to known crustacean anti-lipopolysaccharide factors and examined their expression in lobster tissues in response to challenge by Vibrio fluvialis.
2. Materials and Methods
2.1 Identification and sequencing of cDNA clones encoding anti-lipopolysaccharide factors
Expressed sequence tags predicted to encode two different anti-lipopolysaccharide factors were identified in a normalized multiple tissue cDNA library prepared from H. americanus (Towle et al. 2006). The library clones selected for complete sequencing were HA_MX0_13f11 (Accession No. CN853488), designated as encoding anti-lipopolysaccharide factor Ha-1 (ALFHa-1), and HA_MX2_15b07 (FC556430), designated as encoding anti-lipopolysaccharide factor Ha-2 (ALFHa-2). Plasmids were isolated from overnight cultures of each clone and the inserts were sequenced using universal primers SP6 and M13F with a capillary-based sequencer (ABI 3100, Applied Biosystems, Foster City, CA, USA). Contiguous sequences were produced with the SeqMan component of Lasergene software (DNASTAR, Madison, WI, USA) and open reading frames were predicted and translated with DNASIS (Hitachi, S. San Francisco, CA, USA). Hydrophobicity indices (Kyte et al. 1982) of the resulting amino acid sequences were analyzed through the ExPaSy ProtScale server (Swiss Institute of Bioinformatics) and secondary structure predictions were made via PSIPRED (McGuffin et al. 2000). Alignment with arthropod anti-lipopolysaccharide protein sequences and a neighbor-joining tree of evolutionary relationships were generated via MEGA4 (Tamura et al. 2007).
2.2 Animals and tissue sampling
At the Virginia Institute of Marine Science, American lobsters (Homarus americanus) were obtained from a local supplier and acclimated overnight in individual aerated 9.5-liter tanks containing 35 psu seawater at 10–11°C. Three lobsters were injected at the base of a walking leg with 1 mL Vibrio fluvialis (108 cfu/mL) in tryptone soya broth (TSB) medium and 3 control lobsters were injected with 1:10 dilution of TSB. After 24 hours, all lobsters were euthanized. Gill, hematopoietic, and hepatopancreas tissues were aseptically removed and stored in RNAlater (Sigma-Aldrich, St. Louis, MO, USA) at −20°C for subsequent transport to the Mount Desert Island Biological Laboratory.
2.3 Total RNA isolation and cDNA synthesis
Total RNA was extracted from hepatopancreas, hematopoietic tissue, and gill of 3 control and 3 experimental lobsters under RNAse-free conditions (Chomczynski et al. 1987) using RNAgents Total RNA Isolation System (Promega, Madison, WI, USA). To determine quantity and quality of the RNA, samples were analyzed on a nanodrop spectrophotometer (Thermo Scientific, Wilmington, DE, USA) and by microfluidic electrophoresis (Agilent 2100 Bioanalyzer, Santa Clara, CA, USA), showing complete absence of degradation products or genomic DNA. Aliquots of 2 µg of each sample of total RNA were reverse transcribed to single-stranded cDNA using SuperScript III reverse transcriptase (Invitrogen, Carlsbad, CA, USA) and oligo(dT)20 as a primer according to the manufacturer’s protocol. cDNA was stored at −20°C between uses.
2.4 PCR amplification of cDNA templates, electrophoresis, and DNA sequencing
Two sets of non-degenerate primers were designed to target specifically ALFHa-1 or ALFHa-2 using Primer Premier software (Premier Biosoft, Palo Alto, CA, USA) and were synthesized by Integrated DNA Technologies (Coralville, IA, USA). Polymerase chain reactions were carried out using RedTaq Ready Mix PCR Reaction Mix (Sigma-Aldrich) under the following conditions: 30 cycles of 92°C for 1 min, 55°C for 1min, and 72°C for 2 min, then 72°C for 5 min and 4°C for storage. Following amplification, PCR products were analyzed by agarose gel electrophoresis (1% agarose in TBE buffer, 100 V, 30 min), stained with SYBR Green, and viewed over a UV light source. Amplification products of the correct size were excised from the agarose gel using a sterile scalpel blade and purified using a QiaQuick gel extraction kit (Qiagen, Valencia, CA, USA). Resulting nucleotide sequences were analyzed and edited using ChromasPro 1.3 (Technelysium, Tewantin, QLD, Australia). Forward and reverse sequences for each target were aligned using SeqMan (Lasergene, DNASTAR, Madison, WI, USA). BLASTN and BLASTX (Altschul et al. 1997) comparisons with non-redundant sequence databases (National Center for Biotechnology Information, Bethesda, MD, USA) were performed to confirm the presence of the intended target cDNAs.
2.5 Quantification of gene expression
Analysis of ALFHa-1 and ALFHa-2 mRNA expression was carried out via real-time quantitative PCR (QPCR) using Brilliant SYBR Green QPCR mix and MX4000 instrumentation (Stratagene, LaJolla, CA, USA). The primer combinations selected for ALFHa-1 were Ha-1-3F (5’-TAAAGTGGTTGGGTTATGGC-3’) and Ha-1-4R (5’-TGAGTAGAGGTAAAGGGTGA- 3’), producing an amplicon of 289 bp. For ALFHa-2, the primers were Ha-2-7F (5’-AGCGAACACTACAAGATGCG-3’) and Ha-2-6R (5’-CACCTTCTTATCTTGGGTTTCA-3’), producing an amplicon of 215 bp. The thermal profile for QPCR included an activation step at 95°C for 15 min and then 40 cycles of 94°C for 40 sec, 55°C for 40 sec, and 72°C for 60 sec. Following amplification, a thermal denaturation profile was generated to assess product homogeneity. For both targets, a single inflection point of the denaturation curve at 82.5° C for ALFHa-1 and 85.0° C for ALFHa-2 indicated a homogenous product. To enable calculation of relative expression values, a dilution series of cDNA prepared from control hematopoetic tissue was analyzed in parallel with the other samples, all of which were run in triplicate. Amplification efficiency was 89.8% for ALFHa-1 and 87.9% for ALFHa-2. Expression values were normalized to total RNA content (2.0 µg) in each sample employed in the reverse transcription step, a method well accepted in systems lacking a validated housekeeping gene {Bustin et al. 2005}. Means and SD were calculated for each triplicate and then pooled to obtain relative expression data for control and experimental gill, hepatopancreas and hematopoetic tissues (n=3). Statistical analysis using two-way ANOVA and Bonferroni posttests was accomplished with Prism GraphPad (LaJolla, CA, USA).
3. Results
Two partial nucleotide sequences encoding putative anti-lipopolysaccharide factors were identified in a normalized multiple-tissue cDNA library prepared from the American lobster Homarus americanus (Towle et al. 2006). Sequencing the inserts contained in the respective library clones yielded complete cDNA sequences with open reading frames encoding a 123-amino-acid protein for ALFHa-1 (Accession No. EU625516, Figure 1) and a 124-amino-acid protein for ALFHa-2 (EU625517, Figure 2). Calculated molecular weights for the two proteins are 13,683 for ALFHa-1 and 13,909 for ALFHa-2. The predicted ALFHa-1 protein is substantially more cationic than ALFHa-2, its calculated isoelectric point being 10.65 compared to 8.85 for ALFHa-2. Contributing to this difference, the ALFHa-1 protein contains 10 arginine residues and 6 lysine residues, compared with 7 arginine residues and 4 lysine residues for ALFHa-2.
Figure 1.
Nucleotide and amino acid sequence of anti-lipopolysaccharide factor cDNA ALFHa-1 from the lobster Homarus americanus, derived from the library clone identified by expressed sequence tag CN853488. The predicted amino acid sequence encoded by the open reading frame is indicated by single letter abbreviations, and the 25-amino-acid signal peptide predicted by SignalP 3.0 (Bendtsen et al. 2004) is underlined. Conserved cysteine residues are indicated by arrows, defining a predicted loop region containing the highly cationic KPXXRR motif (˜˜˜˜˜). (Accession No. EU625516.).
Figure 2.
Nucleotide and amino acid sequence of anti-lipopolysaccharide factor cDNA ALFHa-2 from the lobster Homarus americanus, derived from the library clone identified by expressed sequence tag FC556430. The predicted amino acid sequence encoded by the open reading frame is indicated by single letter abbreviations, and the 26-amino-acid signal peptide predicted by SignalP 3.0 is underlined. Conserved cysteine residues are indicated by arrows, defining a predicted loop region containing the highly cationic KPXXRR motif (˜˜˜˜˜) (Accession No. EU625517).
Both protein sequences contain a pair of conserved cysteine residues, at positions 55 and 76 in ALFHa-1 and positions 56 and 77 in ALFHa-2 (Figure 1 and Figure 2). Between these positions lies a cationic region containing a conserved KPXXRR motif, comprising the looped portion of the sequence that is thought to interact with target lipopolysaccharides in bacterial cell walls (Hoess et al. 1993). Hydrophobic signal peptides were predicted by SignalP 3.0 (Bendtsen et al. 2004) for both protein sequences, encompassing the N-terminal 25 amino acid residues in ALFHa-1 and 26 amino acid residues in ALFHa-2 (Figure 3). A second hydrophobic region lies adjacent to the signal peptide, but the remainder of the sequence is generally hydrophilic in both proteins.
Figure 3.
Hydrophobicity plots (Kyte et al. 1982) of amino acid sequences of anti-lipopolysaccharide factors ALFHa-1 and ALFHa-2 translated from cDNA sequences from Homarus americanus. The hydrophobicity index was calculated over a window of 9 residues by the ExPaSy ProtScale server (Swiss Institute of Bioinformatics). The signal peptide predicted by SignalP 3.0 (Bendtsen et al. 2004) is indicated by a solid bar.
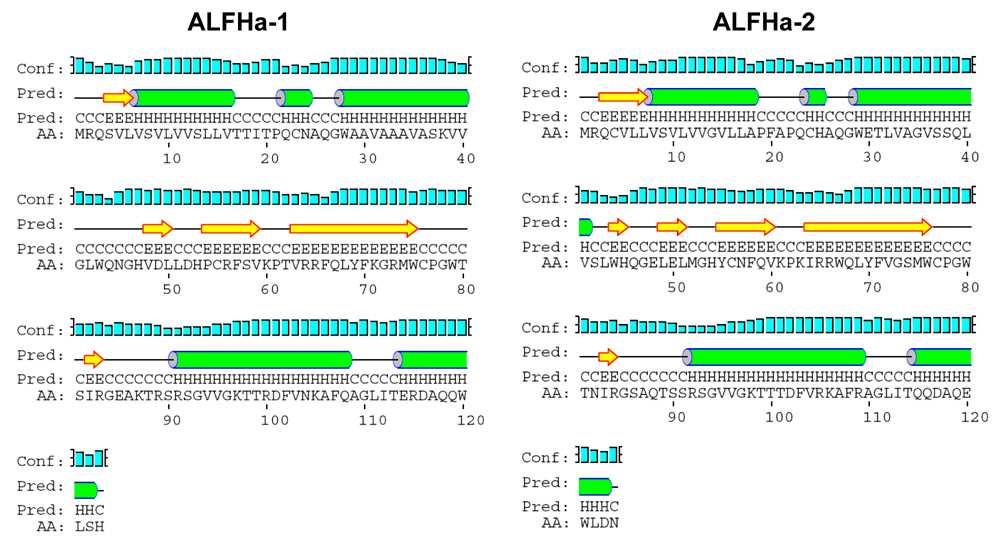
Secondary structure predictions using PSIPRED (McGuffin et al. 2000) identified α- helical regions in the signal peptide and the adjacent hydrophobic region, as well as two segments at the C-terminus (Figure 4). Between these regions are segments of β-strand and random coil that include the amphipathic loop defined by the conserved cysteine residues at positions 55 and 76 (ALFHa-1) or 56 and 77 (ALFHa-2). These predictions are surprisingly consistent with the known crystal structure of the mature anti-lipopolysaccharide factor of Limulus polyphemus in which the corresponding amphipathic loop includes amino acids 32–50, the position coinciding precisely with the cleavage of a 25-amino-acid signal peptide (Hoess et al. 1993).
Figure 4.
Secondary structure predictions via PSIPRED (McGuffin et al. 2000) for anti-lipopolysaccharide factors ALFHa-1 and ALFHa-2 from Homarus americanus. α-Helical regions (H) are indicated by cylinders, β-strand regions (E) by block arrows, and random coil (C) by straight lines. Confidence of the prediction is indicated by bars associated with the “Conf” line.
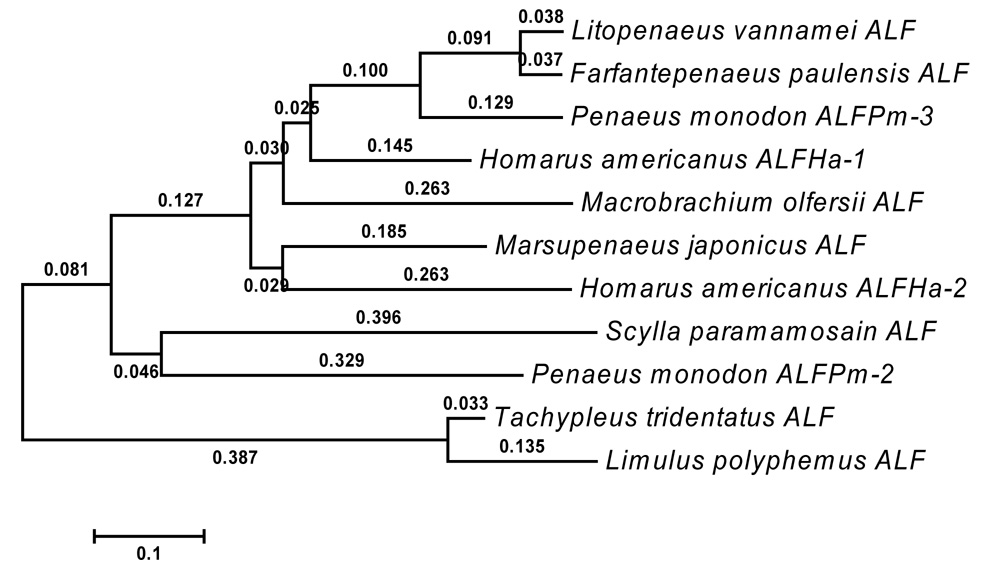
Alignment via MEGA4 (Tamura et al. 2007) of ALFHa-1 and ALFHa-2 amino acid sequences with representative arthropod anti-lipopolysaccharide factor amino acid sequences revealed several regions of sequence conservation, particularly around the conserved cysteine residues (Figure 5). Although the signal peptides of each sequence are consistently hydrophobic, amino acids within these regions are not highly conserved. A neighbor-joining tree based on this alignment sorted the chelicerates Limulus polyphemus and Tachypleus tridentatus into a group separate from the crustaceans, as expected (Figure 6). ALFHa-1 of Homarus americanus appears to be most similar to the ALFs of shrimps Litopenaeus vannamei and Farfantepenaeus paulensis and ALFPm3 of giant tiger prawn Penaeus monodon, while ALFHa-2 is grouped with an ALF from the kuruma prawn Marsupenaeus japonicus.
Figure 5.
Alignment of representative arthropod anti-lipopolysaccharide protein sequences, selected from chelicerate and decapod crustacean examples. Intensity of color reflects percentage identity calculated via MEGA4 (Tamura et al. 2007) and displayed via GeneDoc (Nicholas et al. 1997). ALFs presented include those from Pacific white shrimp Litopenaeus vannamei (DQ208703), pink shrimp Farfantepenaeus paulensis (EF601051), giant tiger prawn Penaeus monodon (ALFPm2 EF523561, ALFPm3 EF523559), American lobster Homarus americanus (ALFHa-1 EU625516, ALFHa-2 EU625517, this study), freshwater shrimp Macrobrachium olfersii (EU289220), kuruma prawn Marsupenaeus japonicus (AB210110), mud crab Scylla paramamosain (EF207786), and mature ALF peptides in the chelicerates Japanese horseshoe crab Tachypleus tridentatus (AF227150) and Atlantic horseshoe crab Limulus polyphemus (P07086). Arrows indicate conserved cysteine residues participating in a disulfide bridge that forms a cationic loop in the protein.
Figure 6.
Neighbor-joining tree of representative arthropod anti-lipopolysaccharide protein sequences based on the alignment in Figure 5, as calculated via MEGA4 (Tamura et al. 2007). Branch lengths were computed using the Poisson correction method and are expressed as number of amino acid substitutions per site. Accession numbers associated with the indicated species of origin are listed in the legend of Figure 5.
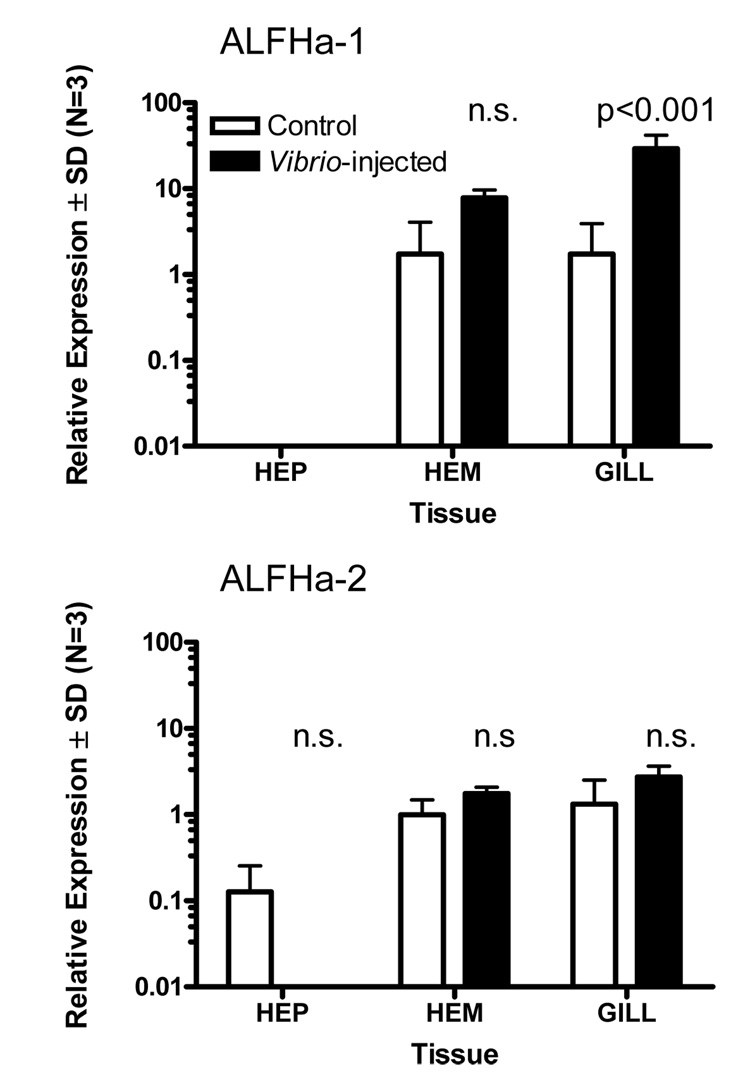
To investigate the transcriptional response of ALFHa-1 and ALFHa-2 to a pathogen, we injected adult lobsters with Vibrio fluvialis and sampled tissues 24 hours later for analysis of the respective RNAs by quantitative PCR. Employing sequence-specific primers that amplified the respective cDNAs, as affirmed by direct sequencing of PCR products, we found very low levels of the two mRNAs in hepatopancreas (Figure 7). However, hematopoietic tissue and gill showed substantial levels of both ALFHa-1 and ALFHa-2 mRNA. Injection of V. fluvialis elicited no response in the abundance of ALFHa-2, but induced a statistically significant 17-fold increase in ALFHa-1 mRNA in gill tissue (Figure 7). A high degree of specificity in the immune response to V. fluvialis was indicated since one of the genes encoding an anti-lipopolysaccharide factor (ALFHa-1) appeared to be transcriptionally upregulated, while the other (ALFHa-2) was not.
Figure 7.
Relative expression of two anti-lipopolysaccharide factor mRNAs, ALFHa-1 and ALFHa-2, in hepatopancreas (HEP), hematopoietic tissue (HEM), and gill of Homarus americanus 24 hours following injection with tryptone soya broth medium (TSB) (light bars) or Vibrio fluvialis in TSB (filled bars), determined by quantitative PCR. A statistically significant effect of Vibrio injection was observed only for ALFHa-1 in gill tissue (ANOVA with Bonferroni posttests, N=3).
4. Discussion
We report the finding of two putative anti-lipopolysaccharide factors in the tissues of the American lobster, Homarus americanus. Anti-lipopolysaccharide factors (ALFs) are small basic proteins of approximately 100 amino acids that were initially characterized from the hemolymph of distantly related arthropods, the horseshoe crabs T. tridentatus (Aketagawa et al. 1986) and L. polyphemus (Muta et al. 1987). ALFs bind to cell walls of gram-negative bacteria and inhibit the lipopolysaccharide-mediated coagulation cascade (Chaby 2004). The cDNA nucleotide sequence encoding ALFs includes a 5’ section encoding a hydrophobic signal peptide of approximately 25 amino acid residues, producing a nascent protein prior to signal cleavage of approximately 125 amino acids (Supungul et al. 2002). The mature protein consists of a single domain with three α-helices crowded against a four-strand β-sheet (Hoess et al. 1993). Two of the β-strands are linked by a disulfide bond to form an amphipathic loop rich in cationic amino acid side chains. Synthetic cyclic versions of this loop are functionally active against gram-negative bacteria including Vibrio harveyi (Imjongjirak et al. 2007) and Pseudomonas aeruginosa (Pan et al. 2007) and against gram-positive bacteria such as Micrococcus luteus (Imjongjirak et al. 2007). A recombinant ALF from the giant tiger prawn Penaeus monodon is highly effective against Vibrio species, including several that are pathogenic to shrimp (Somboonwiwat et al. 2005). Injection of double-stranded RNA corresponding to the mRNA encoding an anti-lipopolysaccharide factor from the Pacific white shrimp Litopenaeus vannamei into shrimp challenged with low doses of Vibrio penaeicida lead to knock-down of the ALF message and increased mortality (de la Vega et al. 2008). It is clear from this evidence that anti-lipopolysaccharide factors are potent antimicrobial agents in marine arthropods, with important implications in prevention of disease in aquacultured and farmed species.
A variety of ALF isoforms have been described in shrimp, including five (ALFPm1-5) in the giant tiger prawn P. monodon discovered during the course of expressed sequence tag analysis (Supungul et al. 2004; Tassanakajon et al. 2006). Two of these (ALFPm1 and 2) appear to be transcribed from a single gene locus and arise as a result of differential exon splicing (Tharntada et al. 2008). The remaining three ALF mRNAs are transcribed from a second gene locus, with ALFPm3 and 5 being essentially identical and ALFPm4 derived via differential exon splicing. In the present study, it is apparent that the two ALFs described in H. americanus are transcribed from separate gene loci and are not the result of differential splicing, since the nucleotide sequences, though closely related, show differences throughout their lengths.
Studies of the transcriptional response to pathogen exposure have shown increased abundance of anti-lipopolysaccharide factor mRNA in several crustacean species. In the Chinese shrimp Fenneropenaeus chinensis, infection with Vibrio anquillarum resulted in enhanced transcription of ALF mRNA in hemocytes within the first 24 hours (Liu et al. 2005). Anti-lipopolysaccharide factor mRNA was also abundant in gill tissue of F. chinensis, but not in hepatopancreas, in agreement with the present study in H. americanus. Injection of Vibrio harveyi into the giant tiger prawn P. monodon led to increased abundance of ALF mRNA in hemocytes, while mRNAs for other antimicrobial proteins including penaeidin and crustin decreased (Supungul et al. 2004; Somboonwiwat et al. 2006). Interestingly, treatment of Pacific white shrimp Litopenaeus vannamei with white spot syndrome virus resulted in upregulation of an ALF gene in hepatopancreas, suggesting that there may be some overlap between the antiviral response and the antibacterial response (Robalino et al. 2007).
We have shown in this study that one of the two anti-lipopolysaccharide factors identified in the American lobster, ALFHa-1, is transcriptionally upregulated in response to bacterial challenge with Vibrio fluvialis, but the other, ALFHa-2, is not (Figure 7). Messenger RNAs for both ALFHa-1 and ALFHa-2 are present in gill and hematopoietic tissue, but only ALFHa-1 responds to infection. Gills of bacteria-infected crustaceans are known to accumulate hemocytes (Burnett et al. 2006), and thus it is not surprising that gills would exhibit high levels of mRNAs that may be derived from hemocytes. However, the observation that ALFHa-2 transcription is not induced by Vibrio challenge, while ALFHa-1 is upregulated, indicates some degree of ALF specificity in the immune response to V. fluvialis.
Structural differences between the ALFHa-1 and ALFHa-2 proteins may suggest a mechanism by which they are capable of selecting a specific target. The amphipathic loop that is thought to interact with lipopolysaccharides in bacterial cell walls (Kloczewiak et al. 1994) contains one additional cationic residue in ALFHa-2 compared with ALFHa-1, possibly supporting interaction with more highly acidic regions of its target. On the other hand, ALFHa-1 contains a single pair of half-cysteines, compared to two pairs in ALFHa-2, likely producing a different 3-dimensional conformation of the protein that may further dictate specificity of binding.
Acknowledgements
This study was supported by a grant from the Atlantic Canada Opportunities Agency to the AVC Lobster Science Centre, University of Prince Edward Island, by the Maine IDeA Network of Biomedical Research Excellence (NIH P20-RR016463), and by the National Science Foundation (IOB-0543860).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aketagawa J, Miyata T, Ohtsubo S, Nakamura T, Morita T, Hayashida H, Miyata T, Iwanaga S, Takao T, Shimonishi Y. Primary structure of limulus anticoagulant anti-lipopolysaccharide factor. J. Biol. Chem. 1986;261:7357–7365. [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- Burnett LE, Holman JD, Jorgensen DD, Ikerd JL, Burnett KG. Immune defense reduces respiratory fitness in Callinectes sapidus, the Atlantic blue crab. Biol. Bull. 2006;211:50–57. doi: 10.2307/4134577. [DOI] [PubMed] [Google Scholar]
- Bustin SA, Benes V, Nolan T, Pfaffl MW. Quantitative real-time RT-PCR -- a perspective. J. Mol. Endocrinol. 2005;34:597–601. doi: 10.1677/jme.1.01755. [DOI] [PubMed] [Google Scholar]
- Chaby R. Lipopolysaccharide-binding molecules: transporters, blockers and sensors. Cell. Mol. Life Sci. 2004;61:1697–1713. doi: 10.1007/s00018-004-4020-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanatephenol-chloroform extraction. Anal. Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Christie AE, Rus S, Goiney CC, Smith CM, Towle DW, Dickinson PS. Identification and characterization of a cDNA encoding a crustin-like, putative antibacterial protein from the American lobster Homarus americanus. Mol. Immunol. 2007;44:3333–3337. doi: 10.1016/j.molimm.2007.02.028. [DOI] [PubMed] [Google Scholar]
- de la Vega E, O'Leary NA, Shockey JE, Robalino J, Payne C, Browdy CL, Warr GW, Gross PS. Anti-lipopolysaccharide factor in Litopenaeus vannamei (LvALF): a broad spectrum antimicrobial peptide essential for shrimp immunity against bacterial and fungal infection. Mol. Immunol. 2008;45:1916–1925. doi: 10.1016/j.molimm.2007.10.039. [DOI] [PubMed] [Google Scholar]
- Hoess A, Watson S, Siber GR, Liddington R. Crystal structure of an endotoxin-neutralizing protein from the horseshoe crab, Limulus anti-LPS factor, at 1.5 A resolution. EMBO J. 1993;12:3351–3356. doi: 10.1002/j.1460-2075.1993.tb06008.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imjongjirak C, Amparyup P, Tassanakajon A, Sittipraneed S. Antilipopolysaccharide factor (ALF) of mud crab Scylla paramamosain: molecular cloning, genomic organization and the antimicrobial activity of its synthetic LPS binding domain. Mol. Immunol. 2007;44:3195–3203. doi: 10.1016/j.molimm.2007.01.028. [DOI] [PubMed] [Google Scholar]
- Iwanaga S, Kawabata S, Muta T. New types of clotting factors and defense molecules found in horseshoe crab hemolymph: their structures and functions. J. Biochem. 1998;123:1–15. doi: 10.1093/oxfordjournals.jbchem.a021894. [DOI] [PubMed] [Google Scholar]
- Johansson MW, Keyser P, Sritunyalucksana K, Söderhäll K. Crustacean haemocytes and haematopoiesis. Aquaculture. 2000;191:45–52. [Google Scholar]
- Kloczewiak M, Black KM, Loiselle P, Cavaillon JM, Wainwright N, Warren HS. Synthetic peptides that mimic the binding site of horseshoe crab antilipopolysaccharide factor. J. Infect. Dis. 1994;170:1490–1497. doi: 10.1093/infdis/170.6.1490. [DOI] [PubMed] [Google Scholar]
- Kyte J, Doolittle RF. A simple method for displaying hydropathic character of a protein. J. Mol. Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Liu F, Liu Y, Li F, Dong B, Xiang J. Molecular cloning and expression profile of putative antilipopolysaccharide factor in Chinese shrimp (Fenneropenaeus chinensis) Mar. Biotechnol. 2005;7:600–608. doi: 10.1007/s10126-005-5006-4. [DOI] [PubMed] [Google Scholar]
- McGuffin LJ, Bryson K, Jones DT. The PSIPRED protein structure prediction server. Bioinformatics. 2000;16:404–405. doi: 10.1093/bioinformatics/16.4.404. [DOI] [PubMed] [Google Scholar]
- Muta T, Iwanaga S. The role of hemolymph coagulation in innate immunity. Curr. Opin. Immunol. 1996;8:41–47. doi: 10.1016/s0952-7915(96)80103-8. [DOI] [PubMed] [Google Scholar]
- Muta T, Miyata T, Tokunaga F, Nakamura T, Iwanaga S. Primary structure of anti-lipopolysaccharide factor from American horseshoe crab, Limulus polyphemus. J. Biochem. 1987;101:1321–1330. doi: 10.1093/oxfordjournals.jbchem.a121999. [DOI] [PubMed] [Google Scholar]
- Nicholas KB, Nicholas HB., Jr GeneDoc: a tool for editing and annotating multiple sequence alignments. 1997 www.psc.edu/biomed/genedoc.
- Pan CY, Chao TT, Chen JC, Chen JY, Liu WC, Lin CH, Kuo CM. Shrimp (Penaeus monodon) anti-lipopolysaccharide factor reduces the lethality of Pseudomonas aeruginosa sepsis in mice. Int. Immunopharmacol. 2007;7:687–700. doi: 10.1016/j.intimp.2007.01.006. [DOI] [PubMed] [Google Scholar]
- Robalino J, Almeida JS, McKillen D, Colglazier J, Trent HF, 3rd, Chen YA, Peck ME, Browdy CL, Chapman RW, Warr GW, Gross PS. Insights into the immune transcriptome of the shrimp Litopenaeus vannamei: tissue-specific expression profiles and transcriptomic responses to immune challenge. Physiol. Genomics. 2007;29:44–56. doi: 10.1152/physiolgenomics.00165.2006. [DOI] [PubMed] [Google Scholar]
- Smolowitz R, Christoserdov A, Hsu A. A description of the pathology of epizootic shell disease in the American lobster, Homarus americanus, H. Milne Edwards 1837. J. Shellfish Res. 2005;24:749–756. [Google Scholar]
- Söderhäll K, Cerenius L. Crustacean immunity. Ann. Rev. Fish Dis. 1992;2:3–23. [Google Scholar]
- Somboonwiwat K, Marcos M, Tassanakajon A, Klinbunga S, Aumelas A, Romestand B, Gueguen Y, Boze H, Moulin G, Bachere E. Recombinant expression and anti-microbial activity of anti-lipopolysaccharide factor (ALF) from the black tiger shrimp Penaeus monodon. Dev. Comp. Immunol. 2005;29:841–851. doi: 10.1016/j.dci.2005.02.004. [DOI] [PubMed] [Google Scholar]
- Somboonwiwat K, Supungul P, Rimphanitchayakit V, Aoki T, Hirono I, Tassanakajon A. Differentially expressed genes in hemocytes of Vibrio harveyi-challenged shrimp Penaeus monodon. J. Biochem. Mol. Biol. 2006;39:26–36. doi: 10.5483/bmbrep.2006.39.1.026. [DOI] [PubMed] [Google Scholar]
- Supungul P, Klinbunga S, Pichyangkura R, Hirono I, Aoki T, Tassanakajon A. Antimicrobial peptides discovered in the black tiger shrimp Penaeus monodon using the EST approach. Dis. Aquat. Organ. 2004;61:123–135. doi: 10.3354/dao061123. [DOI] [PubMed] [Google Scholar]
- Supungul P, Klinbunga S, Pichyangkura R, Jitrapakdee S, Hirono I, Aoki T, Tassanakajon A. Identification of immune-related genes in hemocytes of black tiger shrimp (Penaeus monodon) Mar. Biotechnol. 2002;4:487–494. doi: 10.1007/s10126-002-0043-8. [DOI] [PubMed] [Google Scholar]
- Tall BD, Fall S, Pereira MR, Ramos-Valle M, Curtis SK, Kothary MH, Chu DM, Monday SR, Kornegay L, Donkar T, Prince D, Thunberg RL, Shangraw KA, Hanes DE, Khambaty FM, Lampel KA, Bier JW, Bayer RC. Characterization of Vibrio fluvialis-like strains implicated in limp lobster disease. Appl. Environ. Microbiol. 2003;69:7435–7446. doi: 10.1128/AEM.69.12.7435-7446.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Tassanakajon A, Klinbunga S, Paunglarp N, Rimphanitchayakit V, Udomkit A, Jitrapakdee S, Sritunyalucksana K, Phongdara A, Pongsomboon S, Supungul P, Tang S, Kuphanumart K, Pichyangkura R, Lursinsap C. Penaeus monodon gene discovery project: the generation of an EST collection and establishment of a database. Gene. 2006;384:104–112. doi: 10.1016/j.gene.2006.07.012. [DOI] [PubMed] [Google Scholar]
- Tharntada S, Somboonwiwat K, Rimphanitchayakit V, Tassanakajon A. Anti-lipopolysaccharide factors from the black tiger shrimp, Penaeus monodon, are encoded by two genomic loci. Fish Shellfish Immunol. 2008;24:46–54. doi: 10.1016/j.fsi.2007.07.010. [DOI] [PubMed] [Google Scholar]
- Towle DW, Smith CM. Gene discovery in Carcinus maenas and Homarus americanus via expressed sequence tags. Integ. Comp. Biol. 2006;46:912–918. doi: 10.1093/icb/icl002. [DOI] [PubMed] [Google Scholar]