Abstract
Bacteriophages are bacterial viruses that infect bacterial cells and they have developed ingenious mechanisms to modify the bacterial RNA polymerase. Using a rapid, specific, single-step immunoisolation procedure to purify Escherichia coli RNA polymerase from bacteriophage T4 infected cells; we have identified bacteriophage T4-dependent modifications of the host RNA polymerase. We suggest that this methodology is applicable for the identification of bacteriophage-dependent alterations of the host synthesis machinery.
Keywords: Escherichia coli, bacteriophage, RNA polymerase, bacteriophage infection, immunoisolation, MALDI MS
Introduction
In the bacterium Escherichia coli (Ec), transcription is dependent upon a single ∼400 kDa multisubunit RNA polymerase (RNAP) that can exist in two forms: the catalytic core enzyme (α2ββ′ω) and the promoter-specific holoenzyme (α2ββ′ωσ). Transcription can be divided into three distinct stages: initiation, elongation, and termination. Regulation of RNAP can occur at each stage of transcription by protein, RNA, and small molecule trans-acting factors that associate with the enzyme to modulate its activity.
Bacteriophages (phages) are bacterial viruses dependent upon a host organism in order to propagate; therefore, phages are endowed with mechanisms that subvert the host's cellular processes to serve the needs of the virus. Consequently, bacteriophages offer an exciting and tractable system by which to study developmental processes. Recently, resurgence in the use of bacteriophages for the treatment or prophylaxis of bacterial infectious diseases – bacteriophage therapy – has increased the demands for understanding bacteriophage/host interactions1. In particular, identification of phage proteins that bind to and inhibit essential host enzymes should allow the identification of novel drug targets. Therefore, it is highly desirable to have efficient methods for the identification of phage-encoded proteins that associate with host proteins.
Many phages encode proteins that directly bind to and regulate the activity of the bacterial host RNAP, and some phages encode enzymes that covalently modify the host transcriptional apparatus. These phage-induced modifications ensure coordinated temporal transcription of the phage genome and inhibition of host transcription2.
Bacteriophage T4 (T4) does not encode its own RNAP and relies on Ec RNAP for expression of its genes. In order to appropriate the host RNAP, T4 employs several redundant mechanisms that target Ec RNAP to the T4 genome2 and promote the transcription of T4 DNA. One such mechanism involves the T4-encoded Alc protein that functions to increase the pool of host RNAP available for transcribing T4 DNA by selectively terminating RNAP transcription on Ec DNA without affecting transcription of T4 DNA3,4,5.
In this paper, we present a method for the rapid isolation of RNAP from phage-infected Ec cells, followed by matrix-assisted laser desorption ionization mass spectrometric (MALDI MS) analysis of proteins co-isolating with RNAP. This method allowed unequivocal identification of most of the known T4-encoded proteins that bind directly to host RNAP, identification of Alc whose binding to RNAP had not previously been reported, and also identified covalent modifications of the host RNAP by T4-encoded enzymes. This general procedure should be broadly applicable for the identification of phage-encoded proteins that interact with components of the host macromolecular synthesis machinery, and will become increasingly important in characterizing the many bacteriophage/host systems.
Experimental Section
Ec strains and growth conditions
A strain of Ec MG1655 encoding two Fc-binding repeats of the Protein A (2PrA) affinity tag6 appended to the 3′ end of the rpoC gene (which encodes the RNAP β′ subunit) was constructed as previously described7. Both the MG1655 Ec wild-type and MG1655 Ec rpoC::2PrA strains exhibited almost identical growth curves in Luria-Bertani broth (LB; data not shown) and doubling times (Ec wild-type, 25.3 +/− 1.6 minutes; Ec rpoC::2PrA, 28.5 +/− 2.1 minutes).
All Ec strains were grown in LB at 37 °C with shaking. To prepare T4-infected biomass, wild-type Ec and Ec rpoC::2PrA strains (4 L of each) at an A600 nm 0.5 – 0.6 were infected with T4 (alt-) at a multiplicity of infection of 10. Infection was halted 15 minutes post-infection by rapidly cooling the samples in icy water baths. Cells were harvested by centrifugation, washed once with ice-cold 10% (v/v) glycerol, and frozen as pellets in liquid nitrogen. Cells were cryogenically disrupted with a Retsch MM301 mixer mill (Retsch) that was maintained at liquid nitrogen temperature, and stored at minus 80 °C8.
Immunoisolation and mass spectrometric identification of protein complexes
Cryogenically lysed cells (1 g) were suspended in 5 ml of extraction buffer (20 mM Hepes (pH 7.4); 2 mM MgCl2; 150 mM NaCl) supplemented with 1 protease inhibitor cocktail tablet (Roche Diagnostics). Suspended lysate was treated with 0.002 % (w/v) DNase I (Sigma-Aldrich) with agitation. The soluble fraction was isolated by centrifugation for 10 minutes; the supernatant was decanted and subjected to another 10 minute centrifugation step. The supernatant was incubated with 25 mg of Dynabeads (Invitrogen) cross-linked to rabbit IgG (MP Biomedicals) with gentle agitation for 10 minutes. For β′-2PrA, and other affinity tagged proteins, rapid immunoisolations are optimal (data not shown;9). Dynabeads were collected with a magnet and washed with wash buffer (20 mM Hepes (pH 7.4); 150 mM NaCl). The β′-2PrA-tagged protein and co-purifying proteins were eluted from the IgG-Dynabeads with 0.5 M NH4OH; 0.5 mM EDTA. The eluted proteins were frozen in liquid nitrogen and evaporated to dryness in a SpeedVac (Thermo Savant). Dried protein samples were dissolved in SDS-PAGE loading buffer and heated at 95 °C. Samples were resolved by SDS-PAGEwith 4-12 % (w/v) Bis-Tris polyacrylamide gels (Invitrogen), and bands due to proteins were visualized by Coomassie blue staining. The entire gel lane was sliced into 1 mm slices and the proteins in each gel slice were destained with a mixture of 50 % (v/v) acetonitrile and 50 mM ammonium bicarbonate. After destaining, the proteins were reduced with 10 mM TCEP-HCl (Pierce) for 30 minutes and then alkylated with 50 mM iodoacetamide (Sigma-Aldrich) for 1 hour. Finally, proteins in each gel slice were digested with Trypsin (Roche Diagnostics) at 37 °C for 4 hours. Proteins were identified by MALDI-QqTOF single stage MS and MALDI-ion-trap MS2 as previously described10,11. Single-stage and multi-stage mass spectrometric data were used for protein identification with the programs Profound12 and Sonar MS/MS13, respectively.
Cloning and overexpression of Alc
Due to the toxicity of T4 Alc to Ec cells, a T7 RNAP-directed Alc overexpression plasmid was constructed in several steps. First, using purified T4 genomic DNA as a template, the DNA encoding Alc was amplified by the Polymerase Chain Reaction (PCR). The resultant PCR product was blunt-end cloned into pT7Blue (Novagen) in an anti-sense orientation relative to the T7 RNAP f10 and Ec RNAP lacUV5 promoters to reduce basal expression of Alc, creating pT7-alcrev. Second, pT7-alcrev was cleaved with AfIIII; treated with Klenow DNA polymerase to generate blunt ends, and digested with BamHI to obtain a fragment encoding Alc and the ϕ10 and lacUV5 promoters. The resultant fragment was cloned into pET28a (Novagen) cleaved with HindIII; treated with Klenow to generate blunt ends, and then cleaved with BamHI to create pET28-alc(T7-lacUV5). Finally, a DNA fragment encoding only the lacUV5 promoter was generated by digesting pUC19 with AflIII; followed by incubation with Klenow to generate blunt-ends, and then cleaved with HindIII. The resultant lacUV5 fragment was cloned between the NotI site, also treated with Klenow, and the HindIII sites of pET28-alc(T7-lacUV5), thus creating pET28-alc(lacUV5). Thus, Alc overexpression is under the control of the plasmid-encoded T7 RNAP ϕ10 promoter, while the Ec RNAP lacUV5 promoter drives transcription anti-sense to Alc reducing Alc toxicity.
The expression plasmid pET28-alc(lacUV5) was transformed into Ec cells harboring the paf rpoB allele (3; which partially nullifies the toxicity of Alc) and the lambda (DE3) lysogen encoding T7 RNAP, and transformants were selected in the presence of 50 μg/ml kanamycin. Cultures were grown at 37 °C to an A600 nm ∼0.6 and induced with 1 mM IPTG for 14 hours at 37 °C. Cells containing overexpressed proteins were harvested by centrifugation and stored at minus 80°C.
Protein Purification
Recombinant Alc formed inclusion bodies and was solubilised by adding urea to a final concentration of 8 M. A polyhistidine tag (derived from the vector) was appended to the amino-terminus of Alc, thus Alc was purified by HiTrap Ni2+-charged affinity chromatography (GE Healthcare) with 8 M urea present in all the column buffers. Purified (His)6-Alc was exchanged into renaturation buffer (40 mM Tris-HCl (pH 8.0), 0.1 M NaCl, 0.5 mM EDTA, 2 mM β-mercaptoethanol, 5 % (v/v) glycerol) by dialysis and concentrated using centrifugal concentration units (Pall Life Sciences). Finally, the protein was exchanged into storage buffer (20 mM Tris-HCl (pH 8.0), 0.1 M NaCl, 1 mM EDTA, 0.5 mM DTT, 50 % (v/v) glycerol) by dialysis and stored at minus 20 °C.
Wild-type Ec σ70 was purified as previously described14. Ec core enzyme and σ70-associated holoenzyme were reconstituted in vitro14 and fractionated by anion-exchange chromatography (Mono Q; GE Healthcare;15). Fractions containing core and σ70-associated enzyme were pooled separately and stored as described previously15.
Analysis of Alc/RNAP interactions
Reactions (8 μl) containing 0.75 μg Alc and either 0.125 mg core RNAP, or 0.07 μg σ70-associated holoenzyme in transcription buffer (30 mM Tris-HCl (pH 7.9), 40 mM KCl, 10 mM MgCl2, 1 mM β-mercaptoethanol) were incubated at room temperature for 10 minutes.
Samples (4 μl of each reaction mixture) were resolved by non-denaturing PAGE on 4-15 % (w/v) gradient polyacrylamide gel and the bands due to proteins stained with Commassie blue. To determine the composition of the bands due to proteins separated by gel electrophoresis under non-denaturing conditions, the bands were excised from the non-denaturing gel, equilibrated in SDS-PAGE loading buffer, and their composition determined by electrophoresis on a 10 % (w/v) SDS-PAGE denaturing gel.
Results and Discussion
Noncovalent modifications of Ec RNAP: Immunoisolation and mass spectrometric identification of T4-encoded proteins that co-isolate with Ec RNAP
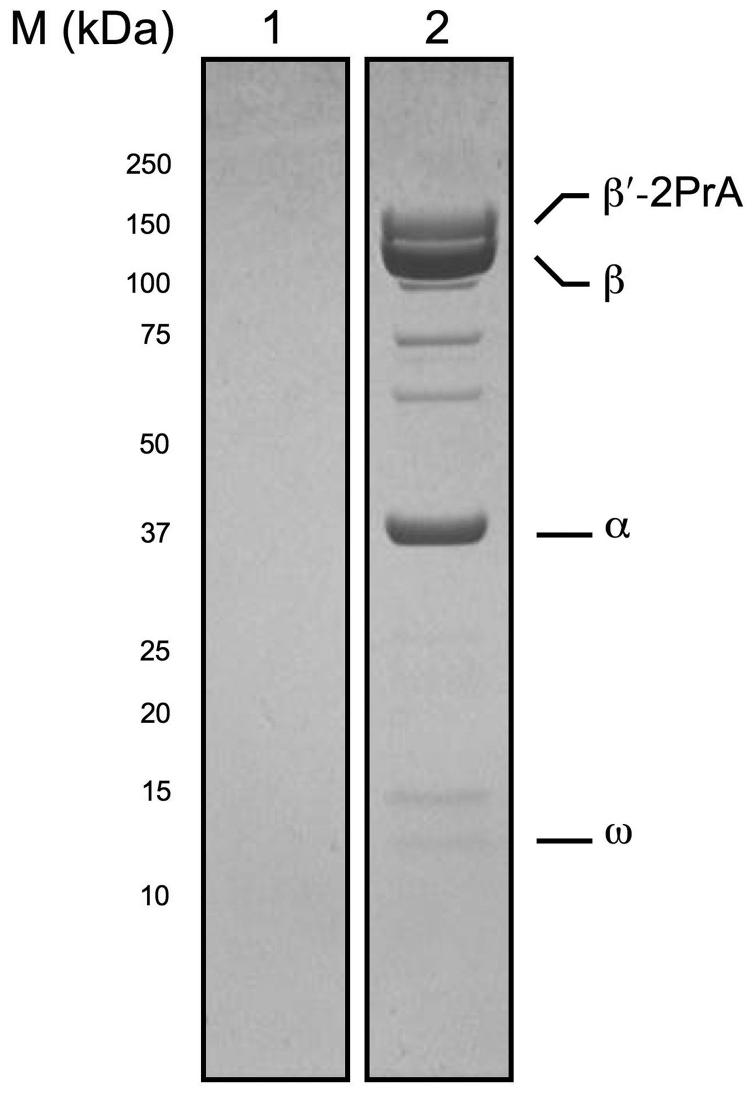
To identify T4-encoded Ec RNAP co-isolating proteins, we immunoisolated RNAP from T4-infected Ec cells (Figure 1, lane 2); isolation was performed using the genomically 2PrA-tagged β′ subunit of RNAP. The conditions of immunoisolation were sufficiently gentle and rapid to maintain interactions with other cellular macromolecules8,9,16-19. As a control, parallel isolation of proteins from T4-infected wild-type cells (untagged β′) was performed. Little or no protein bands are present in the control, indicating that our conditions are stringent enough to prevent non-specific association of most Ec and T4 proteins with the immunoaffinity matrix (Figure 1, lane 1). The material from the tagged strain contained major bands corresponding to the RNAP subunits (Figure 1, lane 2), indicating that the procedure allowed specific single-step purification of RNAP from whole-cell lysates.
Figure 1.
Immunoisolation of β′-2PrA and co-isolating proteins from T4-infected Ec cells. Complexes were isolated via a PrA tag under conditions that co-isolated interacting proteins. Proteins were resolved by denaturing SDS-PAGE, visualized by Coomassie blue staining, and analyzed by MALDI MS. Lanes are loaded as follows: lane 1, Immunoisolated proteins from Ec wild-type cells infected with T4; lane 2, immunoisolated proteins from EcrpoC::2PrA cells infected with T4. RNAP subunits in the 2PrA-tagged sample (lane 2) are labeled.
Immunoisolated T4-encoded proteins were identified by mass spectrometry (Table 1). Traces of contaminating proteins that were present in the untagged control sample that non-specifically associated with the immunoaffinity matrix (lane 1) were computationally filtered out of the dataset obtained from the tagged strain (lane 2). As is clearly seen in Figure 1, and as confirmed by MS analysis, all RNAP subunits were present in the immunoisolated (lane 2) but were absent from the untagged control (lane 1).
Table 1.
| Tagged protein |
T4-encoded RNAP binding proteins |
Peptide coverage (%) |
Mr (kDa) |
Profound score |
Sonar score |
|---|---|---|---|---|---|
| MotA | 58 | 23.6 | 2.5×10−4 | 1.0×10−11 | |
| gp55 | 70 | 21.6 | 6.2×10−8 | 5.0×10−11 | |
| β′-PrA | Alc | 57 | 19.2 | 4.2×10−4 | 3.7×10−12 |
| RpbA | 81 | 14.8 | 8.8×10−8 | 1.8×10−5 | |
| gp33 | 35 | 12.9 | 2.5×10−1 | 6.2×10−1 | |
| AsiA | 88 | 10.6 | 2.6×10−7 | 2.0×10−11 |
Analysis of the immunoisolated material from the tagged strain (lane 2) revealed the presence of all T4-encoded proteins that are known to associate directly with RNAP: MotA, an activator of T4 middle promoters20, gp55, a T4-encoded σ factor that controls the expression of T4 late genes21,22; RpbA, a protein with unknown function; gp33, an activator of T4 late genes that co-activates T4 late genes with σgp55; 22 and AsiA, a potent inhibitor of host-dependent transcription23,24 host RNAP appropriator, and coactivator of T4 middle genes20.
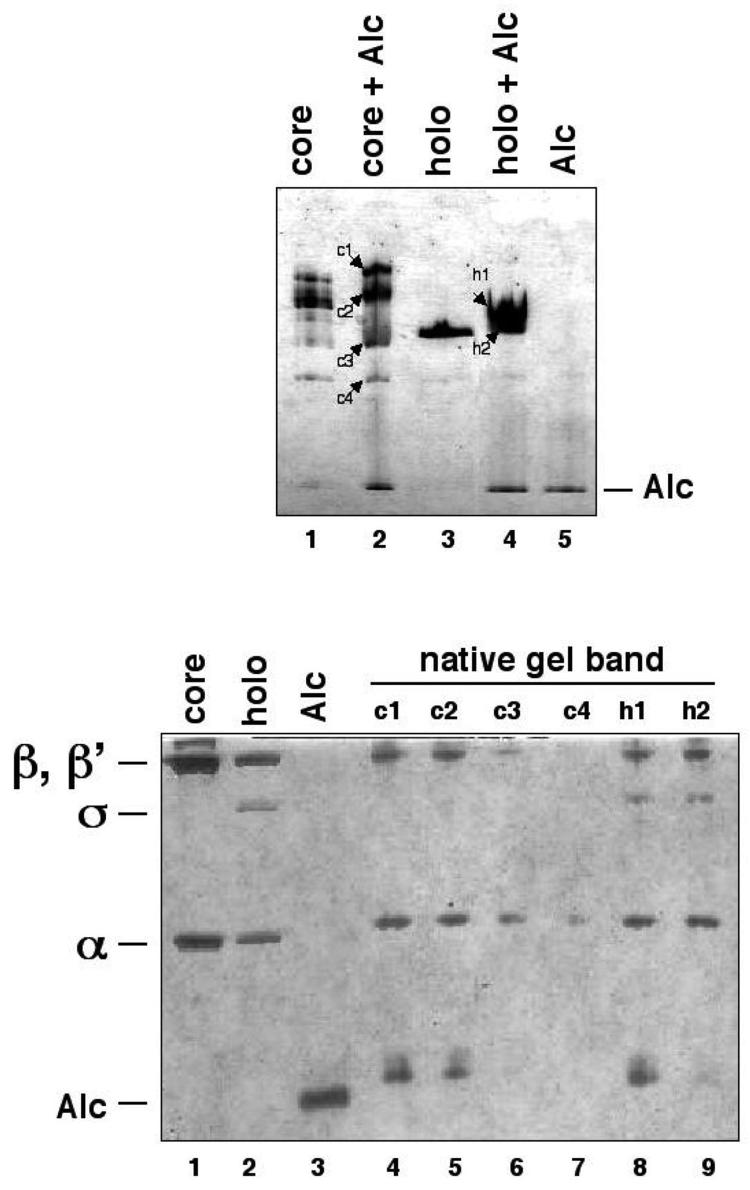
We also identified the T4-encoded transcription termination factor Alc. Alc increases the pool of host RNAP available for transcribing T4 DNA by selectively terminating RNAP transcription on Ec DNA without affecting transcription of T4 DNA3,4,5. Based on genetic studies, Alc is thought to bind directly to the β subunit dispensable region 1 (bDR1) of Ec RNAP, but no direct interaction between RNAP and Alc had been previously reported5,6. Since the immunoisolation experiment indicated an Alc/RNAP interaction, we tested this result using non-denaturing PAGE (Figure 2A). Under our conditions, purified core RNAP migrated as a diffuse set of bands (lane 1), the σ70-holoenzyme migrated as a single band (lane 3), and purified Alc migrated with the dye front (lane 5). From the gel it is clear that the bands labeled c1 and c2 appear to have a slightly lower electrophoretic mobility than the corresponding bands from the RNAP core sample, suggesting an interaction between core and Alc. With the mixture containing Alc and σ70-holoenzyme (lane 4), two poorly resolved bands are observed, one (labeled h2) has the same mobility as the σ70-holoenzyme alone, and another (h1) has a slightly decreased mobility compared to the σ70-holoenzyme alone.
Figure 2.
Non-denaturing PAGE and denaturing SDS-PAGE were used to probe the associations of Alc with the core enzyme and the σ70-holoenzyme. (A) Different combinations of Alc, core, and σ70-holoenzyme were incubated together and resolved under non-denaturing conditions.
(B) Bands due to the proteins separated by non-denaturing PAGE, labeled in (A), were excised and their composition revealed by denaturing SDS-PAGE.
The labeled bands were excised from the non-denaturing gel and their protein content established by denaturing SDS-PAGE (Figure 2B). It is apparent that the c1, c2, and h1 bands contained, in addition to the expected RNAP subunits, a band due to Alc (lanes 4, 5, and 8, respectively), thus establishing that Alc binds both core and σ70-holoenzyme. The material in bands c3, c4, and h2 contained little or no Alc (lanes 6, 7, and 9, respectively). The h2 band contained free σ70-holoenzyme; band c3 contained the RNAP assembly intermediate α2β, and band c4 contained the free α subunit. Thus, it appears that Alc fails to bind to α2β despite the fact that the genetically identified determinants for Alc binding are located in the β subunit3. This result suggests that Alc binding to RNAP may require determinants in both the β and β′ subunits.
Covalent modifications of Ec RNAP: T4-dependent ADP-ribosylation of the Ec RNAP α subunit
ADP-ribosyltransferases catalyze the transfer of the ADP-ribosyl moiety from substrate ADP-ribosyl-nicotinamide onto Arg or His residues of a target protein25. T4 encodes two ADP-ribosyltransferases, Alt and ModA, that ADP-ribosylate Arg 265 of the RNAP α subunit26-30. While ModA specifically modifies α-Arg 26530 of both α subunits, Alt modifies α-Arg 265 on only one of the two α subunits, multiple sites within Ec RNAP, and other host proteins31-33. The sites of Alt-dependent ADP-ribosylation within the β, β′, and σ70 subunits remain to be elucidated. Residue α-Arg 265 is involved in contacting both DNA and trans-acting factors34-36, thus, ADP-ribosylation of this key residue alters α/DNA and α/protein interactions necessary for transcription activation.
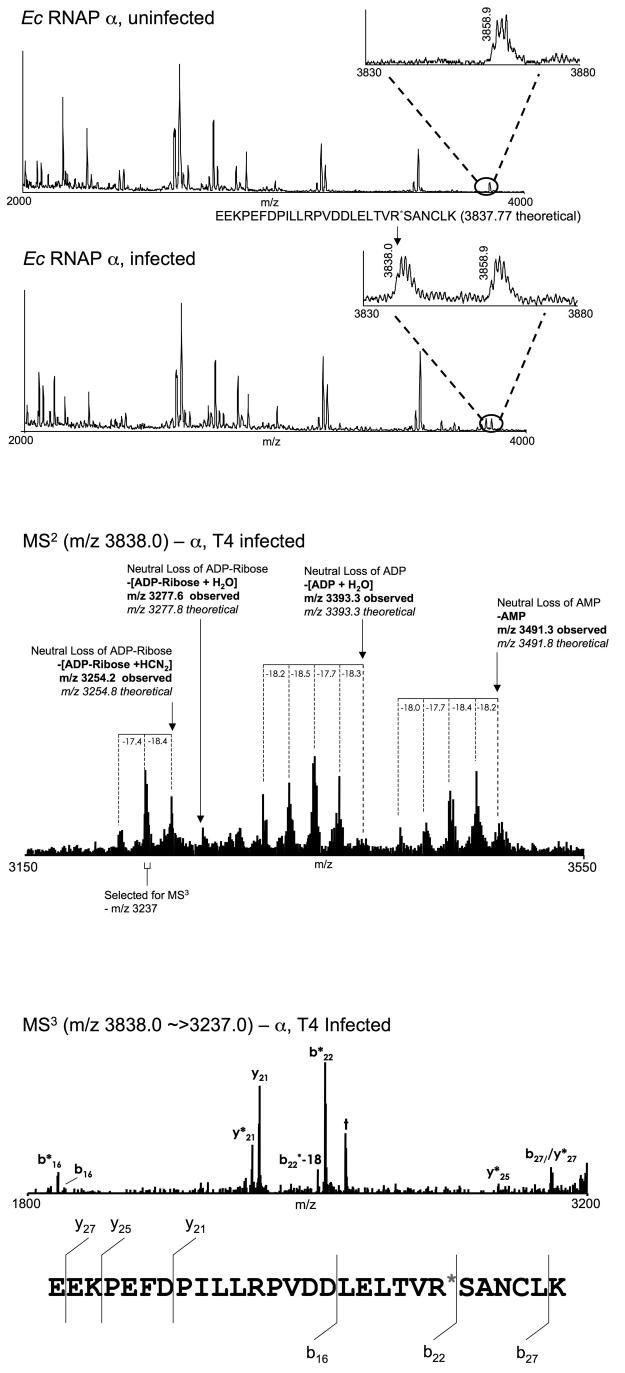
To determine the facility of mass spectrometric analysis for elucidating posttranslational modifications of RNAP, we sought to identify the ModA-dependent ADP-ribosylation of the Ec α subunit. To do this, tryptic peptides of the α subunit immunoisolated from both uninfected and T4-infected Ec cells were prepared and analyzed by MALDI MS. Using a subtractive approach, the resulting mass spectra were scanned for peptides present in the T4-infected sample, but absent from the uninfected sample. A peak due to a singly charged species at m/z 3838.0 was observed in the T4-infected sample but not the uninfected sample (Figure 3A), and thus is likely due to T4-dependent modification of the α subunit. The theoretical m/z for a singly protonated ADP-ribosylated tryptic peptide containing residues 244-271 is 3837.8. The match of both the theoretical and observed masses suggests that the species at m/z 3838.0 is due to α residues 244 to 271, encompassing an ADP-ribosyl group. This interpretation is further corroborated by the missed cleavage at α-Arg 265, suggesting that ADP-ribsoylation of this Arg residue may block tryptic cleavage.
Figure 3.
Identification of T4-dependent ADP-ribosylation of RNAP α subunit Arg 265. Bands due to the RNAP α subunit isolated from uninfected and T4-infected cells were excised, digested with trypsin, and analysed by MALDI MS. The resulting mass spectra were aligned and scanned for peptides present in the T4 infected sample, but absent from the uninfected sample. (A) Mass spectrum of tryptic peptides due to the RNAP α subunit isolated from uninfected Ec rpoC::2PrA cells (top panel) and Ec rpoC::2PrA cells infected with T4 (bottom panel). The appearance of a peak at 3838.0 m/z, with a mass that corresponds to a singly protonated ADP-ribsoylated peptide derived from α residues 244 to 271 in the spectrum of the infected phage that is absent in the uninfected sample spectrum suggests that this peptide is ADP-ribosylated in vivo in a T4 dependent manner. (B) The mass spectrum of the MS2 analysis of the species with an m/z of 3838.0 described in (A) illustrates neutral loss of AMP, ADP and ADP-ribose, together with various water/ammonia neutral losses. This fragmentation is consistent with the interpretation that the peptide ion at 3838.0 m/z is indeed an ADP-ribsoylated species. (C) MS3 analysis of the ions in the MS2 spectrum with a nominal m/z of 3237.0 (corresponding to the neutral loss of ADP-Ribose +HCN2 +water/ammonia) yielded backbone fragmentation providing partial sequence of the peptide. The fragmentation pattern is consistent with the interpretation that the original peptide ion at 3838.0 m/z is indeed derived from α residues 244 to 271. Observed are fragments corresponding to the preferential cleavage of the singly-protonated peptide ion carboxy-terminal to acidic residues and amino-terminal to Pro residues. Together, the MS, MS2, and MS3 spectra in panels (A), (B) and (C) provide strong evidence that α is ADP-ribosylated on an internal Arg residue within the tryptic peptide encoding residues 244-271.
Interrogation of this peptide ion using MALDI-ion-trap MS2 mass spectrometry yielded a family of strong peaks corresponding to fragmentation of the ADP-ribosyl modification. Peaks consistent with the neutral losses of adenosine monophosphate, adenosine diphosphate, and ADP-ribose, combined with multiple water/ammonia losses were observed (Figure 3B). MS3 of a peptide ion at m/z 3237.0 (loss of ADP-ribose + water) yields a fragmentation pattern consistent with residues 244-271 (Figure 3C). The expected preferential fragmentations carboxy-terminal to acidic amino acid residues (Asp and Glu) and amino-terminal to Pro residues are observed37. Our data reveal that mass spectrometry can be used to probe for phage-dependent covalent modifications of host proteins, and should allow the elucidation of those sites within Ec RNAP that are ADP-ribosylated by T4 Alt.
Conclusion
We have immunoisolated Ec RNAP from T4-infected cells and analyzed the T4-encoded co-isolating proteins using MALDI MS. The T4 phage system has been studied extensively and T4-encoded proteins that interact with and regulate the activity of host RNAP have been identified over the years by painstaking genetic and biochemical approaches. Our results show that optimized RNAP immunoisolations, coupled with mass spectrometric identification, allow rapid identification of many T4 proteins known to interact with Ec RNAP, as well as the novel identification of Alc as a binding partner, whose low affinity for RNAP prevented earlier identification. In addition, this method is also suitable for the identification of phage-dependent covalent modifications of the host transcription machinery, as demonstrated by our comprehensive mass spectrometric analysis of an ADP-ribosylated peptide. The ADP-ribosylation of proteins is an important process in both prokaryotes and eukaryotes. Indeed, many bacterial exotoxins are ADP-ribosyltransferases38, and function to manipulate and override host cellular processes. In eukaryotes, ADP-ribosylation is involved in regulating, amongst other things, the activity of histones and membrane traffic39,40. Elucidation of the MS2 and MS3 fragmentation characteristics of ADP-ribosylated peptides should facilitate the definition of control mechanisms and signaling pathways that are dependent upon ADP-ribosylation. Finally, we suggest that this methodology, coupled with phage genomic sequencing, will become an important tool for probing novel host/phage systems.
Acknowledgements
We are grateful to Betty Kutter, Sohail Malik, Ian Molineux, and Richard Wolf for many stimulating discussions and to Malcolm Twist for expert art work. This work was funded by grants from the National Institutes of Health: RR00862 (BTC), U54 RR022220 (MPR and BTC), R01 GM062427 (MPR), and GM61898 (SAD).
References
- 1.Matsuzaki S, Rashel M, Uchiyama J, Sakurai S, Ujihara T, Kuroda M, Ikeuchi M, Tani T, Fujieda M, Wakiguchi H, Imai S. Bacteriophage therapy: a revitalized therapy against bacterial infectious diseases. J. Infect. Chemother. 2005;11:211–9. doi: 10.1007/s10156-005-0408-9. [DOI] [PubMed] [Google Scholar]
- 2.Nechaev S, Severinov K. Bacteriophage-induced modifications of host RNA polymerase. Annu. Rev. Microbiol. 2003;57:301–22. doi: 10.1146/annurev.micro.57.030502.090942. [DOI] [PubMed] [Google Scholar]
- 3.Severinov K, Kashlev M, Severinova E, Bass I, McWilliams K, Kutter E, Nikiforov V, Snyder L, Goldfarb A. A non-essential domain of Escherichia coli RNA polymerase required for the action of the termination factor Alc. J. Biol. Chem. 1994;269:14254–9. [PubMed] [Google Scholar]
- 4.Drivdahl RH, Kutter EM. Inhibition of transcription of cytosine-containing DNA in vitro by the alc gene product of bacteriophage T4. J. Bacteriol. 1990;172:2716–27. doi: 10.1128/jb.172.5.2716-2727.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kashlev M, Nudler E, Goldfarb A, White T, Kutter E. Bacteriophage T4 Alc protein: a transcription termination factor sensing local modification of DNA. Cell. 1993;75:147–54. [PubMed] [Google Scholar]
- 6.Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Seraphin B. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 1999;17:1030–2. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- 7.Mooney RA, Landick R. Tethering sigma 70 to RNA polymerase reveals high in vivo activity of sigma factors and sigma 70-dependent pausing at promoter-distal locations. Genes Dev. 2003;17:2839–51. doi: 10.1101/gad.1142203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tackett AJ, Dilworth DJ, Davey MJ, O'Donnell M, Aitchison JD, Rout MP, Chait BT. Proteomic and genomic characterization of chromatin complexes at a boundary. J. Cell Biol. 2005;169:35–47. doi: 10.1083/jcb.200502104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cristea IM, Williams R, Chait BT, Rout MP. Fluorescent proteins as proteomic probes. Mol. Cell Proteomics. 2005;4:1933–41. doi: 10.1074/mcp.M500227-MCP200. [DOI] [PubMed] [Google Scholar]
- 10.Krutchinsky AN, Zhang W, Chait BT. Rapidly switchable matrix-assisted laser desorption/ionization and electrospray quadrupole-time-of-flight mass spectrometry for protein identification. J. Am. Soc. Mass Spectrom. 2000;11:493–504. doi: 10.1016/S1044-0305(00)00114-8. [DOI] [PubMed] [Google Scholar]
- 11.Krutchinsky AN, Kalkum M, Chait BT. Automatic identification of proteins with a MALDI-quadrupole ion trap mass spectrometer. Anal. Chem. 2001;73:5066–77. doi: 10.1021/ac010682o. [DOI] [PubMed] [Google Scholar]
- 12.Zhang W, Chait BT. Profound: an expert system for protein identification using mass spectrometric peptide mapping information. Anal. Chem. 2000;72:2482–9. doi: 10.1021/ac991363o. [DOI] [PubMed] [Google Scholar]
- 13.Field HI, Fenyo D, Beavis RC. RADARS, a bioinformatics solution that automates proteome mass spectral analysis, optimises protein identification, and archives data in a relational database. Proteomics. 2002;2:36–47. [PubMed] [Google Scholar]
- 14.Borukhov S, Goldfarb A. Recombinant Escherichia coli RNA polymerase: purification of individually overexpressed subunits and in vitro assembly. Protein Expr. Purif. 1993;4:503–11. doi: 10.1006/prep.1993.1066. [DOI] [PubMed] [Google Scholar]
- 15.Minakhin L, Severinov K. On the role of the Escherichia coli RNA polymerase sigma 70 region 4.2 and alpha-subunit C-terminal domains in promoter complex formation on the extended −10 galP1 promoter. J. Biol. Chem. 2003;278:29710–8. doi: 10.1074/jbc.M304906200. [DOI] [PubMed] [Google Scholar]
- 16.Aitchison JD, Rout MP, Marelli M,, Blobel G, Wozniak RW. Two novel related yeast nucleoporins Nup170p and Nup157p: complementation with the vertebrate homologue Nup155p and functional interactions with the yeast nuclear pore-membrane protein Pom152p. J. Cell Biol. 1995;131:1133–48. doi: 10.1083/jcb.131.5.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aitchison JD, Blobel G, Rout MP. Kap104p: a karyopherin involved in the nuclear transport of messenger RNA binding proteins. Science. 1996;274:624–7. doi: 10.1126/science.274.5287.624. [DOI] [PubMed] [Google Scholar]
- 18.Rout MP, Blobel G, Aitchison JD. A distinct nuclear import pathway used by ribosomal proteins. Cell. 1997;89:715–25. doi: 10.1016/s0092-8674(00)80254-8. [DOI] [PubMed] [Google Scholar]
- 19.Rout MP, Aitchison JD, Suprapto A, Hjertaas K, Zhao Y, Chait BT. The yeast nuclear pore complex: composition, architecture, and transport mechanism. J. Cell Biol. 2000;148:635–51. doi: 10.1083/jcb.148.4.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pande S, Makela A, Dove SL, Nickels BE, Hochschild A, Hinton DM. The bacteriophage T4 transcription activator MotA interacts with the far-C-terminal region of the sigma 70 subunit of Escherichia coli RNA polymerase. J. Bacteriol. 2002;184:3957–64. doi: 10.1128/JB.184.14.3957-3964.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Malik S, Zalenskaya K, Goldfarb A. Competition between sigma factors for core RNA polymerase. Nucleic Acids Res. 1987;15:8521–30. doi: 10.1093/nar/15.20.8521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nechaev S, Kamali-Moghaddam M, Andre E, Leonetti JP, Geiduschek EP. The bacteriophage T4 late-transcription coactivator gp33 binds the flap domain of Escherichia coli RNA polymerase. Proc. Natl. Acad. Sci. U.S.A. 2004;101:17365–70. doi: 10.1073/pnas.0408028101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Colland F, Orsini G, Brody EN, Buc H, Kolb A. the bacteriophage T4 AsiA protein: a molecular switch for sigma 70-dependent promoters. Mol Microbiol. 1998;27:819–29. doi: 10.1046/j.1365-2958.1998.00729.x. [DOI] [PubMed] [Google Scholar]
- 24.Severinova E, Severinov K, Darst SA. Inhibition of Escherichia coli RNA polymerase by bacteriophage T4 AsiA. J. Mol. Biol. 1998;279:9–18. doi: 10.1006/jmbi.1998.1742. [DOI] [PubMed] [Google Scholar]
- 25.Ueda K, Hayaishi O. ADP-ribosylation. Annu. Rev. Biochem. 1985;54:73–100. doi: 10.1146/annurev.bi.54.070185.000445. [DOI] [PubMed] [Google Scholar]
- 26.Goff CG. Chemical structure of a modification of the Escherichia coli ribonucleic acid polymerase alpha polypeptides induced by bacteriophage T4 infection. J. Biol. Chem. 1974;249:6181–90. [PubMed] [Google Scholar]
- 27.Horvitz HR. Control by bacteriophage T4 of two sequential phosphorylations of the alpha subunit of Escherichia coli RNA polymerase. J. Mol. Biol. 1974;90:727–38. doi: 10.1016/0022-2836(74)90536-1. [DOI] [PubMed] [Google Scholar]
- 28.Goff CG, Setzer J. ADP ribosylation of Escherichia coli RNA polymerase is nonessential for bacteriophage T4 development. J. Virol. 1980;33:547–9. doi: 10.1128/jvi.33.1.547-549.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koch T, Raudonikiene A, Wilkens K, Ruger W. Overexpression, purification, and chacterization of the ADP-ribosyltransferase (gpAlt) of bacteriophage T4: ADPribosylation of E. coli RNA polymerase modulates T4 “early” transcription. Gene Expr. 1995;4:253–64. [PMC free article] [PubMed] [Google Scholar]
- 30.Skorko R, Zillig W, Rohrer H, Fujiki H, Mailhammer R. Purification and properties of the NAD+:protein ADP-ribosyltransferase responsible for the T4-phage-induced modification of the alpha subunit of DNA-dependent RNA polymerase of Escherichia coli. Eur. J. Biochem. 1977;79:55–66. doi: 10.1111/j.1432-1033.1977.tb11783.x. [DOI] [PubMed] [Google Scholar]
- 31.Rohrer H, Zillig W, Mailhammer R. ADP-ribosylation of DNA-dependent RNA polymerase of Escherichia coli by an NAD+: protein ADP-ribosyltransferase from bacteriophage T4. Eur. J. Biochem. 1975;60:227–38. doi: 10.1111/j.1432-1033.1975.tb20995.x. [DOI] [PubMed] [Google Scholar]
- 32.Sommer N, Salniene V, Gineikiene E, Nivinskas R, Ruger W. T4 early promoter strength probe in vivo with unribosylated and ADP-ribosylated Escherichia coli RNA polymerase: a mutation analysis. Microbiology. 2000;146:2643–53. doi: 10.1099/00221287-146-10-2643. [DOI] [PubMed] [Google Scholar]
- 33.Depping R, Lohaus C, Meyer HE, Ruger W. The mono-ADPribosyltransferases Alt and ModB of bacteriophage T4: target proteins identified. Biochem. Biophys. Res. Commun. 2005;335:1217–23. doi: 10.1016/j.bbrc.2005.08.023. [DOI] [PubMed] [Google Scholar]
- 34.Gaal T, Ross W, Blatter EE, Tang H, Jia X, Krishnan VV, Assa-Munt N, Ebright RH, Gourse RL. DNA-binding determinants of the alpha subunit of RNA polymerase: novel DNA-binding domain architecture. Genes Dev. 1996;10:16–26. doi: 10.1101/gad.10.1.16. [DOI] [PubMed] [Google Scholar]
- 35.Benoff B, Yang H, Lawson CL, Parkinson G, Liu J, Blatter E, Ebright YW, Berman HM, Ebright RH. Structural basis of transcription activation: the CAP-alpha CTD-DNA complex. Science. 2002;297:1562–6. doi: 10.1126/science.1076376. [DOI] [PubMed] [Google Scholar]
- 36.Shah IM, Wolf RE., Jr. Novel protein-protein interaction between Escherichia coli SoxS and the DNA binding determinant of the RNA Polymerase alpha subunit: SoxS functions as a co-sigma factor and redeploys RNA polymerase from UP-element-containing promoters to SoxS-dependent promoters during oxidative stress. J. Mol. Biol. 2004;343:513–32. doi: 10.1016/j.jmb.2004.08.057. [DOI] [PubMed] [Google Scholar]
- 37.Qin J, Chait BT. Collision-induced dissociation of singly charged peptide ions in a MALDI-ion trap mass spectromter. Int. J. Mass Spectrom. Ion Processes. 1999;191:313–20. [Google Scholar]
- 38.Pallen MJ, Lam AC, Loman NJ, McBride A. An abundance of bacterial ADP-ribosyltransferases - implications for the origin of exotoxins and their human homologues. Trends Microbiol. 2001;9:302–7. doi: 10.1016/s0966-842x(01)02074-1. [DOI] [PubMed] [Google Scholar]
- 39.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–80. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 40.D’Souza-Schorey C, Chavrier P. ARF proteins: roles in membrane traffic and beyond. Nat. Rev. Mol. Cell Biol. 2006;7:347–58. doi: 10.1038/nrm1910. [DOI] [PubMed] [Google Scholar]